Key Points
Aid loss leads to altered differentiation, transcription, and methylation in specific genetic loci in hematopoietic stem/progenitor cells.
Aid loss does not contribute to enhanced HSC self-renewal or cooperate with Flt3-ITD in myeloid leukemogenesis.
Abstract
Recent studies have reported that activation-induced cytidine deaminase (AID) and ten-eleven-translocation (TET) family members regulate active DNA demethylation. Genetic alterations of TET2 occur in myeloid malignancies, and hematopoietic-specific loss of Tet2 induces aberrant hematopoietic stem cell (HSC) self-renewal/differentiation, implicating TET2 as a master regulator of normal and malignant hematopoiesis. Despite the functional link between AID and TET in epigenetic gene regulation, the role of AID loss in hematopoiesis and myeloid transformation remains to be investigated. Here, we show that Aid loss in mice leads to expansion of myeloid cells and reduced erythroid progenitors resulting in anemia, with dysregulated expression of Cebpa and Gata1, myeloid/erythroid lineage-specific transcription factors. Consistent with data in the murine context, silencing of AID in human bone marrow cells skews differentiation toward myelomonocytic lineage. However, in contrast to Tet2 loss, Aid loss does not contribute to enhanced HSC self-renewal or cooperate with Flt3-ITD to induce myeloid transformation. Genome-wide transcription and differential methylation analysis uncover the critical role of Aid as a key epigenetic regulator. These results indicate that AID and TET2 share common effects on myeloid and erythroid lineage differentiation, however, their role is nonredundant in regulating HSC self-renewal and in myeloid transformation.
Introduction
Activation-induced cytidine deaminase (AID) has been a focus of intense study for over a decade, since it was shown to have a critical role in generating antibody diversity in B lymphocytes.1 AID initiates the somatic hypermutation process through deamination of cytidine to uridine in the recombined variable region, followed by removal of the uracil base by uridine DNA glycosylase and DNA repair by several error-prone base-excision repair (BER) and mismatch-repair enzymes.2 AID further induces the second step of antibody diversification, class-switch recombination, through deamination of bases in the switch region, causing double-strand breaks (DSBs) and recombination.3
Recent studies have uncovered a critical role for AID in active DNA demethylation in zebrafish, mouse embryos, and mammalian somatic cells.4-6 At the same time, a number of studies have identified critical enzymes involved in active DNA demethylation.5-8 The first step in demethylation is hydroxylation of 5-methylcytosine (5mC) to form 5-hydroxymethylcytosine (5hmC) and further oxidation to form 5-formylcytosine and 5-carboxylcytosine by the ten-eleven-translocation (TET) family members. The second step is deamination of 5mC or 5hmC to form 5-methyluracil or 5-hydroxymethyluracil by AID/apolipoprotein B messenger RNA (mRNA) editing enzyme, catalytic polypeptide-like (APOBEC) family members. The final step is replacement of 5-methyluracil, 5-hydroxymethyluracil, or 5-carboxylcytosine to cytosine by the uracil DNA glycosylase family of BER glycosylases.9 Taken together, these studies suggest AID acts downstream of TET family members in mediating active DNA demethylation.
Recent studies using high-throughput genome-wide sequencing have identified somatic deletions and loss-of-function mutations in the TET2 gene in 10% to 20% of patients with myelodysplastic syndromes/myeloproliferative neoplasms,10,11 in 10% to 20% of patients with acute myeloid leukemia (AML), and in 40% to 50% of patients with chronic myelomonocytic leukemia (CMML).12,13 In mouse models, conditional loss of Tet2 in the hematopoietic compartment leads to expansion of hematopoietic stem/progenitor cells (HSPCs; Lin−Sca-1+c-Kit+ [LSK]), enhanced hematopoietic stem cell (HSC) self-renewal capacity, and development of myeloproliferative disease.14-16 These data clearly indicate the critical role of TET2 in both HSC regulation and myeloid transformation.
AID was reported to induce DSBs at immunoglobulin heavy chain (IgH) translocating partners, including at c-myc/IgH in B-cell lymphoma.17 Whole-genome sequencing of a variety of human tumors uncovered mutation clusters of <10 kb in size termed “kataegis,”18 which are predominantly C to T transitions. Of note, kataegic mutations found in human lymphomas derived from germinal center B cells occurred frequently in WRCY hypermutation hotspots, indicating that AID is responsible for kataegic mutations in these cells.18 In addition, AID was reported to stabilize the pluripotent state in induced pluripotent stem cells (iPSCs) by removing epigenetic memory of pluripotency genes, suggesting the essential role of AID in iPSC reprogramming.19
Given that AID has multifaceted functions including a role in active DNA demethylation, in lymphomagenesis, and in iPSC regulation, we hypothesized that AID may regulate stem/progenitor self-renewal and differentiation and suppress myeloid transformation. Here, we show that Aid loss in mice leads to alterations in differentiation including increased myeloid expansion and attenuation in erythroid output, but does not induce alterations in self-renewal or in susceptibility to transformation. Consistent with these data, Aid loss in HSPCs causes transcriptional alteration of known regulators of erythropoiesis and enhanced DNA methylation in loci with a known role in transcriptional regulation. These data suggest that AID is a critical regulator in myeloid and erythroid lineage differentiation and a key epigenetic regulator that maintains transcriptional output.
Methods
Mice
Mice with the Aid knockout allele were previously described.2 The conditional Vav-Cre+ Tet2f/f mice and Flt3ITD mice were previously described.14,20 All animal procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the institutional animal care and use committees at Memorial Sloan Kettering Cancer Center.
Human samples
Human bone marrow (BM) samples were obtained from deidentified patients at Hospital for Special Surgery. All patients provided informed consent. Approval was obtained from the institutional review board at Memorial Sloan Kettering Cancer Center and Hospital for Special Surgery.
Statistical analysis
All statistical analyses were performed using the unpaired Student t test. P values <.05 were considered statistically significant.
Additional experimental procedures are described in the supplemental Methods (available on the Blood Web site).
Results
Disrupted Aid expression and enzymatic activity in Aid knockout cells
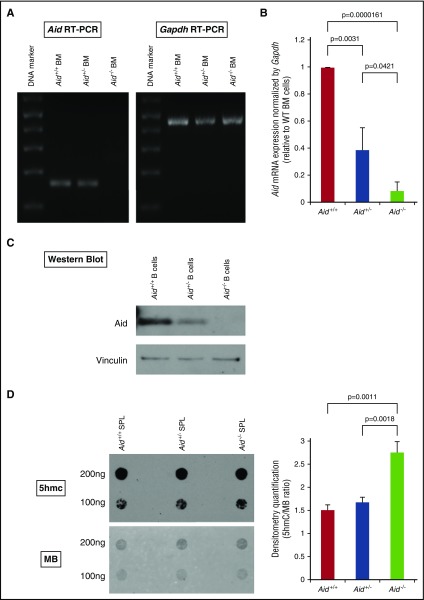
In order to explore the functional role of Aid in normal and malignant hematopoiesis, we used C57/Bl6 mice with a previously characterized Aid knockout allele.2 We first collected whole BM cells from either wild-type (WT), Aid+/−, or Aid−/− mice, performed semiquantitative reverse transcription polymerase chain reaction (RT-PCR), and confirmed decreased mRNA expression of Aid in Aid−/− BM cells compared with WT or Aid+/− cells (Figure 1A). Quantitative RT-PCR also showed a significant decrease in mRNA expression of Aid in both Aid+/− and Aid−/− cells compared with WT cells in an allele number–dependent manner (Figure 1B). Western blots against murine Aid further confirmed the absence of Aid expression in Aid−/− splenic B cells (Figure 1C). Because Aid is incorporated in active DNA demethylation by deamination of 5hmC to form 5-hydroxymethyluracil followed by cytosine replacement via BER,9 we performed dot-blot assays against 5hmC using DNA samples derived from either WT, Aid+/−, or Aid−/− splenic cells. Densitometry quantification confirmed a significant increase in 5hmC in Aid−/− samples compared with WT or Aid+/− hematopoietic cells (Figure 1D).
Figure 1.
Disrupted Aid expression and enzymatic activity in Aid knockout cells. (A) Whole BM cells were collected from WT, Aid+/−, and Aid−/− adult mice around 8 weeks old. Total RNA was extracted from these cells and mRNA expression of Aid and Gapdh was confirmed by semiquantitative RT-PCR. (B) mRNA expression levels of Aid were measured by RT-PCR using BM cells from WT, Aid+/−, and Aid−/− adult mice. Data were analyzed by the Δ cycle threshold (Ct) ratio technique using the murine Gapdh gene as a housekeeping gene. Results are presented as the ratio of the Aid+/− or Aid−/− value to the WT value. The data are mean ± standard deviation (SD) (n = 3 for each genotype). (C) CD43− naive splenic B cells were isolated from WT, Aid+/−, and Aid−/− adult mice and stimulated with lipopolysaccharide (LPS) and interleukin 4 (IL-4). Total protein was extracted from stimulated cells and western blot against Aid and Vinculin were performed. (D) Whole splenic cells were collected from WT, Aid+/−, and Aid−/− adult mice around 6 months old. Total DNA was extracted from these cells and dot-blot assay was performed to detect 5hmC. Membrane was stained with MB to detect DNA loading control. Blot density was quantified by Image J software using blots derived from 100 ng of DNA. Right bar graph shows the ratio of the 5hmC value to the MB value. The data are mean ± SD (n = 3 for each genotype). MB, methylene blue; SPL, spleen.
Aid loss causes myeloid expansion and anemia in vivo
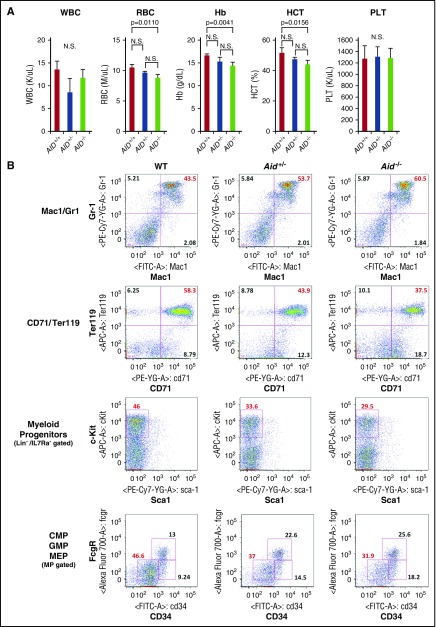
To investigate the impact of Aid on normal hematopoiesis, we assessed complete blood counts (CBCs) of WT, Aid+/−, and Aid−/− mice around 8 weeks old. Interestingly, Aid−/− mice showed significantly lower red blood cell (RBC) count, hemoglobin (Hb) level, and hematocrit (HCT) percentage compared with WT (Figure 2A; supplemental Table 2). Flow cytometric analysis of peripheral blood (PB) showed expansion of both Mac1+/Gr-1+ and Mac1+/Gr-1− myeloid cells in Aid−/− mice compared with WT (supplemental Figure 1A).
Figure 2.
Aid loss causes myeloid expansion and anemia in vivo. (A) PB samples were collected from WT, Aid+/−, and Aid−/− adult mice around 8 weeks old. White blood cell (WBC) counts, RBC counts, Hb levels, HCT percentages, and platelet counts were measured. The data are mean ± SD (WT and Aid+/−; n = 3, Aid−/−; n = 4). (B) Representative immunophenotype of BM granulocyte/monocyte lineage (Mac1/Gr1), erythroid lineage (CD71/Ter119), myeloid progenitors (Lineage−IL7Ra−Sca1−cKit+), and common myeloid progenitors (CMPs)/GMPs/MEPs derived from WT, Aid+/−, and Aid−/− adult mice around 8 weeks old. MP, myeloid progenitor; N.S., not significant; PLT, platelet.
In order to determine whether Aid loss induces a myeloproliferative phenotype as seen in Tet2−/− mice, we euthanized WT, Aid+/−, and Aid−/− adult mice and performed full analysis at 8 to 16 weeks of age. Although we observed a modest increase in liver size in Aid−/− mice, Aid loss did not cause splenomegaly or thymic enlargement (supplemental Figure 1B,D). We did not observe any statistically significant differences in total BM or spleen cell number between WT, Aid+/−, and Aid−/− mice (supplemental Figure 1C). Aid−/− mice showed a significantly higher percentage of granulomonocytic (GM) fraction (Mac1+/Gr-1+) and a lower percentage of erythroid progenitor fraction (CD71+/Ter119+) in BM compared with WT mice, which is consistent with the myeloid expansion and anemic phenotype (Figure 2B; supplemental Figure 2A). We observed a decreased percentage and total cell number of myeloid progenitors (Lineage−Sca-1−c-Kit+) in Aid+/− and Aid−/− mice compared with WT but no difference in LSK or long-term HSC fractions (CD150+/CD48− LSK, signaling lymphocyte activation molecule [SLAM] LSK) (Figure 2B; supplemental Figures 2B and 3A). In addition, both Aid+/− and Aid−/− mice had a significant decrease in the proportion and the number of megakaryocyte erythroid progenitors (MEPs; Lineage−Sca-1−c-Kit+CD34lowFcgRlow) in BM compared with WT, further supporting the erythroid differentiation defect induced by Aid loss (Figure 2B; supplemental Figures 2C and 3B). Consistent with the anemic phenotype in Aid−/− mice, immunohistochemistry staining for mouse Ter119 using femurs of WT and Aid−/− mice around 1 year old demonstrated a decreased number of mTer119+ erythroid cells in the epiphysis of the bone, but not in splenic red pulp (supplemental Figure 4A-B). In contrast to BM, splenic cells derived from Aid−/− mice demonstrated a significantly higher percentage of erythroid progenitors (CD71+/Ter119+) compared with WT, consistent with compensatory splenic erythropoiesis in the Aid−/− mice which was not sufficient to rescue the anemia phenotype (supplemental Figure 5). Taken together, these data indicate that Aid loss causes myeloid expansion and anemia due to altered myeloid/erythroid differentiation but does not cause myeloproliferative disease in vivo.
Aid loss does not affect HSC self-renewal but alters expression of myeloid/erythroid lineage-specific transcription factors
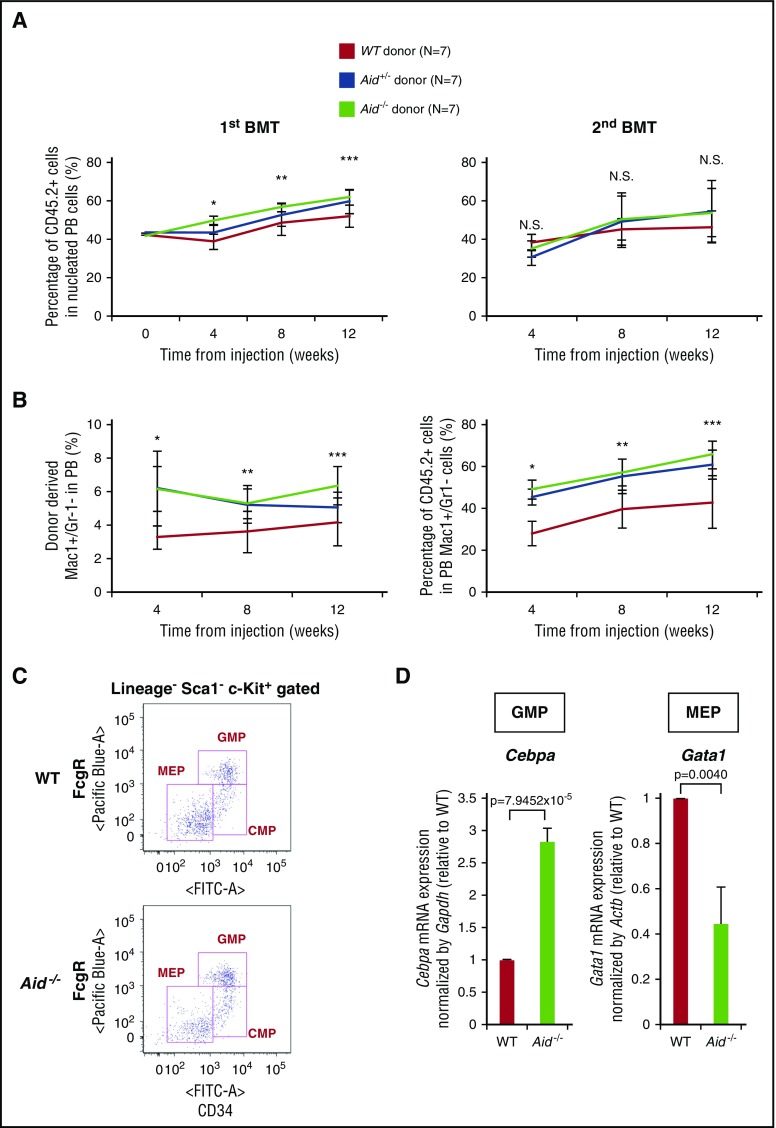
In order to explore the role of Aid on HSC self-renewal, we performed competitive repopulating assays. Whole BM cells from either WT, Aid+/−, or Aid−/− mice (CD45.2+ cells) were mixed with identical numbers of CD45.1+ WT BM cells and injected into lethally irradiated recipient mice (CD45.1). After injection, the percentage of CD45.2+ donor-derived PB cells as well as the myeloid compartment was analyzed by flow cytometry every 4 weeks. In primary transplant, Aid+/− and Aid−/− BM recipients showed a significant increase in chimerism compared with WT recipients in all indicated time points (Figure 3A). In contrast to the first recipients, Aid+/− and Aid−/− BM cells lost their competitive advantage in secondary transplantation studies (Figure 3A). Notably, the percentages of both the donor-derived myelomonocytic compartment (CD45.2+Mac1+Gr-1−) and CD45.2+ cells within Mac1+/Gr-1− cells were significantly higher in Aid+/− and Aid−/− recipients than in WT recipients, which is consistent with the myeloid expansion phenotype seen in the PB and BM of Aid−/− mice (Figure 3B). These data demonstrate that Aid loss does not alter HSC self-renewal capacity but alters differentiation toward myelomonocytic lineage in vivo.
Figure 3.
Ablation of Aid does not affect long-term HSC self-renewal but causes altered expression of Cebpa and Gata1 in myeloid progenitors. (A) Percentage of donor-derived cells (CD45.2+ cells) in PB of primary recipients (left) and secondary recipients (right). The data are mean ± SD (n = 7 for each genotype). The asterisks indicate the P value: *(P = .0444 [WT vs Aid+/−], P = .0072 [Aid+/− vs Aid−/−], P = 3.3075 × 10−5 [WT vs Aid−/−]); **(P = .0071 [WT vs Aid−/−]); ***(P = .0467 [WT vs Aid+/−], P = .0035 [WT vs Aid−/−]). (B) Left graph shows the percentage of donor-derived Mac1+/Gr1− cells in PB of primary recipients. Asterisks indicate the P value: *(P = .0059 [WT vs Aid+/−], P = .0003 [WT vs Aid−/−]); **(P = .0246 [WT vs Aid+/−], P = .0134 [WT vs Aid−/−]); ***(P = .0368 [Aid+/− vs Aid−/−], P = .0088 [WT vs Aid−/−]). Right graph shows the percentage of donor-derived cells (CD45.2+ cells) in Mac1+/Gr1− cells in the PB of primary recipients. The data are mean ± SD (n = 7 for each genotype). Asterisks indicate the P value: *(P = 2.3610 × 10−5 [WT vs Aid+/−], P = 7.2218 × 10−6 [WT vs Aid−/−]); **(P = .0053 [WT vs Aid+/−], P = .0014 (WT vs Aid−/−]); ***(P = .0062 [WT vs Aid+/−], P = .0012 [WT vs Aid−/−]). (C) Gate plot for sorting myeloid progenitors from WT or Aid−/− adult mice around 12 months old. (D) Left bar graph shows the mRNA expression level of Cebpa in WT or Aid−/− GMP cells. Right bar graph shows the mRNA expression level of Gata1 in WT or Aid−/− MEP cells. Data were analyzed by the Δ Ct ratio technique using murine Gapdh or Actb gene as a housekeeping gene. Results are presented as the ratio of the Aid−/− value to the WT value. The data are mean ± SD (n = 3 for each genotype). BMT, bone marrow transplantation.
To delineate how Aid loss causes myeloid expansion and altered erythroid differentiation, we sorted myeloid progenitors from WT or Aid−/− mice around 12 months old and assessed mRNA expression of Cebpa, Pu.1, and Gata1, key myeloid/erythroid lineage-specific transcription factors (TFs) (Figure 3C). Aid−/− granulocyte monocyte progenitors (GMPs; Lineage−Sca-1−c-Kit+CD34hiFcgRhi) showed increased expression of Cebpa, and Aid−/− MEPs demonstrated decreased expression of Gata1 (Figure 3D; supplemental Figure 6). These data suggest that Aid loss in GMPs increases transcriptional output of Cebpa and ablation of Aid in MEPs decreases expression of Gata1, thereby causing skewed differentiation toward the myelomonocytic lineage and impaired erythroid differentiation.
Aid loss does not cooperate with Tet2 loss or Flt3-ITD in hematopoiesis and myeloid transformation
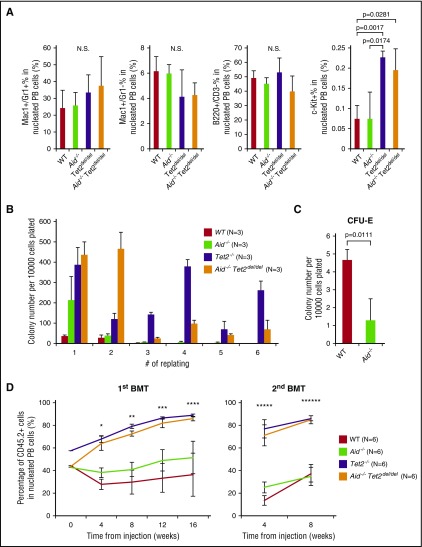
It was previously shown that AID cooperates with TET family members in active DNA demethylation.5-8 Given these observations and the impact of Aid loss on myeloid/erythroid fate specification, we hypothesized that Aid may act in concert with Tet2 to alter HSC regulation and myeloid transformation. To address this issue, we generated mice with both Aid and Tet2 loss in hematopoietic cells (Vav-Cre+Aid−/−Tet2f/f, Aid−/−Tet2del/del).2,14 CBC data revealed anemia in Aid−/−, Tet2del/del, and Aid−/−Tet2del/del mice compared with WT, however, there was no overall difference in blood counts between Tet2del/del and Aid−/−Tet2del/del mice (supplemental Figure 7). Flow cytometric analysis of PB revealed no significant difference in myeloid and lymphoid lineage proportions (Figure 4A). Aid−/−Tet2del/del as well as Tet2del/del mice demonstrated a higher c-Kit+ fraction in PB compared with WT or Aid−/− mice (Figure 4A). To investigate the effect of Aid/Tet2 dual loss on HSC self-renewal, we collected BM cells from WT, Aid−/−, Tet2del/del, and Aid−/−Tet2del/del mice and performed colony-forming unit (CFU) assays. Whereas WT and Aid−/− cells lost their replating capacity at third plating, both Tet2del/del and Aid−/−Tet2del/del cells were able to replate till sixth plating, suggesting that Aid loss does not affect the enhanced replating capacity of Tet2del/del cells (Figure 4B). Of note, Aid−/− cells generated fewer CFU-erythroid (CFU-E) colonies compared with WT cells in first plating, consistent with the anemia phenotype in Aid−/− mice (Figure 4C). Next, we examined the competitive repopulating capacity of WT, Aid−/−, Tet2del/del, and Aid−/−Tet2del/del BM cells in vivo. Consistent with CFU assays, PB chimerism of Aid−/−Tet2del/del recipients was similar to that of Tet2del/del recipients, which was significantly higher than that of WT or Aid−/− recipients (Figure 4D).
Figure 4.
Aid loss does not cooperate with Tet2 loss in hematopoietic regulation. (A) The percentage of PB granulocyte/monocyte lineage (Mac1/Gr1), lymphoid lineage (B220/CD3), and hematopoietic progenitor fraction (cKit+) derived from WT, Aid−/−, Tet2del/del, and Aid−/−Tet2del/del adult mice around 10 months old. The data are mean ± SD (n = 3 for each arm). (B) Methylcellulose serial CFU assay using BM cells from WT, Aid−/−, Tet2del/del, and Aid−/−Tet2del/del mice. Absolute number of colonies is shown at different platings. The data are mean ± SD (n = 3 for each arm). (C) The number of CFU-E colony derived from WT or Aid−/− BM cells in first plating. The data are mean ± SD (n = 3 for each arm). (D) Donor BM cells were collected from either WT, Aid−/−, Tet2del/del or Aid−/−Tet2del/del adult mouse (1.0 × 106 lysed BM cells per genotype, CD45.2+, 8 weeks old), mixed with support BM cells (1.0 × 106 lysed BM cells, CD45.1+, 8 weeks old) and injected to lethally irradiated recipient mice (CD45.1, n = 6 for each genotype). Percentage of donor-derived cells (CD45.2+ cells) in PB of primary recipients (left) and secondary recipients (right). The data are mean ± SD. Asterisks indicate the P value: *(P = 7.0736 × 10−9 [WT vs Tet2del/del], P = 8.6984 × 10−8 [Aid−/− vs Tet2del/del], P = 4.0864 × 10−7 [WT vs Aid−/−Tet2del/del], P = 7.4401 × 10−6 [Aid−/− vs Aid−/−Tet2del/del]); **(P = 1.3386 × 10−6 [WT vs Tet2del/del], P = 5.9805 × 10−8 [Aid−/− vs Tet2del/del], P = 4.5634 × 10−6 [WT vs Aid−/−Tet2del/del], P = 3.9577 × 10−7 [Aid−/− vs Aid−/−Tet2del/del]); ***(P = 3.0646 × 10−6 [WT vs Tet2del/del], P = 2.2681 × 10−6 [Aid−/− vs Tet2del/del], P = 1.0519 × 10−5 [WT vs Aid−/−Tet2del/del], P = 1.5423 × 10−5 [Aid−/− vs Aid−/−Tet2del/del]); ****(P = .0023 [WT vs Tet2del/del], P = .0020 [Aid−/− vs Tet2del/del], P = .0030 [WT vs Aid−/−Tet2del/del], P = .0030 [Aid−/− vs Aid−/−Tet2del/del]); *****(P = 2.8775 × 10−7 [WT vs Tet2del/del], P = 1.8491 × 10−6 [Aid−/− vs Tet2del/del], P = 1.4464 × 10−6 [WT vs Aid−/−Tet2del/del], P = 1.0054 × 10−5 [Aid−/− vs Aid−/−Tet2del/del]); ******(P = 8.9093 × 10−7 [WT vs Tet2del/del], P = 8.1382 × 10−7 [Aid−/− vs Tet2del/del], P = 1.6057 × 10−6 [WT vs Aid−/−Tet2del/del], P = 1.4062 × 10−6 [Aid−/− vs Aid−/−Tet2del/del]).
Given we have shown that Tet2 loss cooperates with Flt3-ITD to induce AML in vivo,20 we hypothesized that Aid loss may contribute to myeloid transformation in concert with Tet2-cooperating disease alleles. To address this issue, we crossed Aid−/− mice with Flt3ITD/ITD mice to generate Aid−/− Flt3ITD/ITD mice. CBC measurement of PB samples derived from mice at 4 to 6 months of age showed no significant difference in CBC parameters between Flt3ITD/ITD and Aid−/−Flt3ITD/ITD mice (supplemental Figure 8). Flt3+/ITD and Aid+/−Flt3+/ITD mice at 8 months of age also showed no difference in CBC parameters (supplemental Figure 9A). In addition, flow cytometric analysis of PB revealed no significant difference in myeloid and progenitor cell proportions between Flt3+/ITD and Aid+/−Flt3+/ITD mice at 1 year old (supplemental Figure 9B). These observations are in contrast to the previous report showing that Tet2 haploinsufficiency combined with Flt3-ITD results in lethal myeloid leukemia around 1 year old.20 Taken together, these results suggest that Aid loss does not cooperate with Tet2 loss or Flt3-ITD in HSC regulation and myeloid transformation.
AID knockdown in human BM cells leads to skewed differentiation toward the myelomonocytic lineage
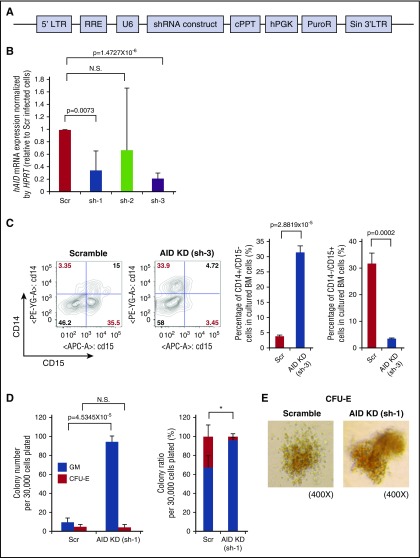
Given Aid loss-induced expansion of the myelomonocytic compartment in both primary Aid−/− mice and recipients of Aid−/− cells, we explored whether knockdown of human AID has similar effects on human BM cells. For this purpose, we used lentiviral short-hairpin constructs against human AID (Figure 5A). We isolated human BM CD34+ cells, transduced them with the lentiviral vector containing short-hairpin construct against human AID or control vectors, and confirmed significant knockdown efficiency by RT-PCR (Figure 5B). Transduced human BM CD34+ cells were cultured in serum-free medium for 5 days followed by flow cytometric analysis of the GM marker (CD14/CD15). The percentage of the CD14+/CD15− monocytic compartment was significantly higher in AID-silenced (sh-3 transduced) cells than in scramble cells, whereas the percentage of the CD14−/CD15+ granulocytic compartment was lower in AID-silenced (sh-3 transduced) cells (Figure 5C). In methylcellulose (H4434) assays, AID-silenced (sh-1 transduced) cells generated a higher number of GM colonies than scramble cells (Figure 5D). Although the number of CFU-E colonies was similar in these 2 arms, the percentage of the CFU-E colony was significantly lower in AID-silenced (sh-1 transduced) cells compared with scramble cells (Figure 5D). We also performed the same experiment using another hairpin which effectively silenced AID (sh-3) and confirmed a significantly higher number of GM colonies and a lower percentage of CFU-E colonies in AID-silenced (sh-3 transduced) cells (supplemental Figure 10). Of note, CFU-E colonies derived from both scramble cells and AID-silenced cells showed reddish discoloration, suggesting that these colonies are hemoglobinized (Figure 5E; supplemental Figure 10). These findings are consistent with the expansion of the myelomonocytic compartment seen in Aid−/− mice and demonstrate that silencing of human AID has a similar effect on human hematopoietic cell differentiation with a key role in regulating myeloid fate commitment.
Figure 5.
AID knockdown in human BM cells causes skewed differentiation toward myelomonocytic lineage. (A) Schema of short hairpin RNA (shRNA) construct against human AID cloned into pLKO.1 vector. (B) Human BM CD34+ cells were transduced with 3 lentiviral vectors expressing short hairpins (sh) against human AID (hAID), and mRNA expression was measured by RT-PCR. Data were analyzed by the Δ Ct ratio technique using human HPRT gene as a housekeeping gene. Results are presented as the ratio of the sh-AID value to the scramble value. The data are mean ± SD (n = 3 for each hairpin). (C) Scramble or sh-3 transduced human BM CD34+ cells were cultured in serum-free StemSpan medium supplemented with IL-3, FLT3L, stem cell factor (SCF), and granulocyte colony-stimulating factor (G-CSF) to monitor GM differentiation in vitro. The left figure shows the representative immunophenotype of CD14/CD15 in cultured cells. The right bar graph shows the percentage of CD14+/CD15− or CD14−/CD15+ cells in cultured cells. The data are mean ± SD (n = 3 for each arm). (D) Scramble or sh-1 transduced human BM CD34+ cells were cultured in methylcellulose medium (H4434) for 9 days and colony numbers were counted. Left bar graph shows the number of GM and CFU-E colonies in each arm. Right bar graph shows the percentage of GM and CFU-E colony in each arm. Thirty thousand cells were plated per well in triplicate. Data are representative of 2 independent experiments. The data are mean ± SD. *P = .0172 (percentage of CFU-E in Scr vs AID-KD). (E) Representative images of CFU-E colony derived from scramble or AID-silenced (sh-1 transduced) cells in indicated magnification. 5′LTR, 5′ long terminal repeat; cPPT, central polypurine tract; hPGK, human phosphoglycerate kinase; KD, knockdown; PuroR, puromycin resistance gene; RRE, Rev response element; Scr, scramble; Sin 3′LTR, self-inactivating 3′ long terminal repeat; U6, human U6 promoter.
Deletion of Aid alters transcription and methylation of hematopoietic regulators and tumor suppressors
Given the putative role of AID/APOBEC family members in mutation clusters called “kataegis,”18 we performed whole-exome sequencing (WES) using DNA from either WT or Aid−/− nucleated PB cells and compared the distribution of somatic missense mutations. The result did not show any clusters of C>T or C>G mutations in both arms, suggesting that there is no functional link between kataegic mutations in coding regions and the hematopoietic phenotype seen in Aid−/− mice (supplemental Figure 11).
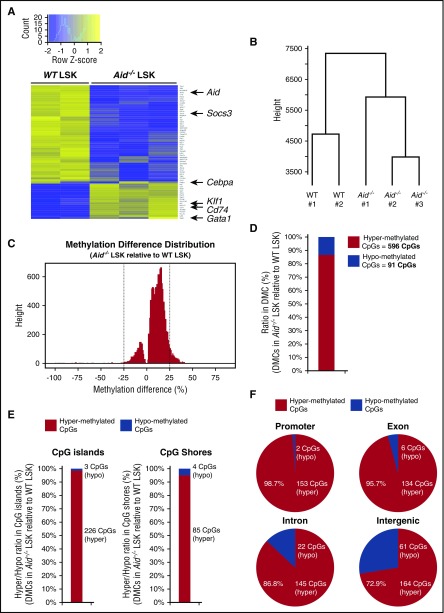
To determine whether Aid loss causes alteration of genome-wide transcription and methylation profile, we performed RNA sequencing (RNA-seq) and enhanced reduced representation bisulfite sequencing (eRRBS) on LSK cells from WT and Aid−/− mice. Differential gene expression analysis of RNA-seq data revealed statistically significantly differentially expressed genes (adjusted P value of <.05 and |log2 fold change| > 1 [Figure 6A; supplemental Figure 12]). These include decreased expression of Aid in Aid−/− LSKs and dysregulated expression of known cytokine signaling regulators and master regulator of erythropoiesis including Socs3, Cd74, and Klf1 (Figure 6A). In contrast to the expression pattern of myeloid/erythroid lineage-specific TFs in myeloid progenitors, Aid−/− LSK cells showed a trend toward higher expression of Gata1 and variable Cebpa expression among each sample (Figure 6A), which might be due to differential transcriptional regulation in LSK and myeloid progenitor cells in Aid−/− mice. Hierarchical clustering and 2-dimensional embedding by t-distributed stochastic neighbor embedding21 demonstrated clear separation of Aid−/− LSKs from WT LSKs (Figure 6B; supplemental Figure 13A).
Figure 6.
Deletion of Aid leads to transcriptional alteration of some key regulators of erythropoiesis and enhanced DNA methylation in gene regulatory elements. (A) Differential gene expression analysis from RNA-seq data of WT and Aid−/− BM LSK cells (WT, n = 2; Aid−/−, n = 3). (B) Dendrogram of the hierarchical clustering based on RNA-seq profiles from WT and Aid−/− LSK cells (WT, n = 2; Aid−/−, n = 3). (C) Methylation difference distribution of CpGs (q < 0.01 only) in Aid−/− LSK cells relative to WT LSK cells based on eRRBS methylation profiles. Pattern clearly shows shift toward more hypermethylated phenotype. DMCs were called as the CpGs with q < 0.01 and >25% change in either direction (as shown by the vertical lines). (D) The ratio and the exact number of hypermethylated and hypomethylated CpGs within total DMCs in Aid−/− LSK cells relative to WT LSK cells. (E) The ratio and the exact number of hypermethylated and hypomethylated CpGs within DMCs localized in CpG islands or CpG shores in Aid−/− LSK cells relative to WT LSK cells. (F) The ratio and the exact number of hypermethylated and hypomethylated CpGs within DMCs localized in coding regions in Aid−/− LSK cells relative to WT LSK cells.
Differential methylation analysis of eRRBS data using methylKit22 for finding single-nucleotide differences (q value < 0.01 and methylation percentage difference of at least 25%) revealed overall hypermethylation phenotype in Aid−/− LSKs compared with WT LSKs with a total of 687 differentially methylated cytosine guanine dinucleotides (CpGs; DMCs) (Figure 6C-D), most of which are hypermethylated (596 CpGs of 687 CpGs) (Figure 6D). Moreover, over 90% of DMCs in CpG islands and CpG shores in Aid−/− LSKs relative to WT LSKs were hypermethylated (Figure 6E). Dominant hypermethylation was also observed in coding regions, especially in promoters and exons, in Aid−/− LSKs compared with WT LSKs (Figure 6F). The background distribution of all CpGs revealed the highest proportion in CpG islands (supplemental Figure 13B). Of note, the proportion of CpG islands, CpG shores, promoters, and exons within hypermethylated CpGs in Aid−/− LSKs relative to WT LSKs were substantially higher than those within hypomethylated CpGs (supplemental Figure 13C). In order to seek correlation between methylation status and gene expression, we focused on 270 genes with differentially methylated regions, which show average methylation difference over 10% in either coding sequence, intron, or promoter regions (supplemental Table 5). Although there was no significant overall correlation between gene expression and methylation, we identified 27 genes within this gene list which show overall hypermethylation and trend toward decreased expression in Aid−/− LSKs compared with WT LSKs (supplemental Figure 14). These include Igfbp6 which functions as a tumor suppressor by inhibiting IGF-II–induced cell proliferation and survival,23 and Dok3, a known tumor suppressor that inhibits tyrosine kinase signaling.24,25 Taken together, these data clearly indicate that Aid loss alters transcription of known regulators of erythropoiesis and promotes DNA methylation at loci with a role in transcriptional regulation.
Discussion
Recent studies reported TET family members and AID cooperate in active DNA demethylation processes.5-9 Deletion of Tet2 in the hematopoietic compartment leads to enhanced HSC self-renewal and skewed differentiation toward myelomonocytic lineage in mice.14-16 AID has also been reported to stabilize the pluripotent state in iPSC reprogramming,19 demonstrating that AID is a critical player in stem cell biology. Taken together, these studies led us to hypothesize that AID might also regulate the hematopoietic system, especially in terms of HSC self-renewal and differentiation. In this study, we investigated the effect of Aid loss on both normal and malignant hematopoiesis, which allowed us to make some important observations. First, Aid loss leads to myeloid expansion and anemia due to altered erythroid differentiation and increased myeloid fate commitment, but this did not result in a myeloproliferative disease in vivo. Second, silencing of AID skewed differentiation toward the myelomonocytic lineage in human BM cells. Third, Aid loss does not contribute to enhanced HSC self-renewal or cooperate with Flt3-ITD in myeloid leukemogenesis. Finally, disruption of Aid causes altered transcription and methylation of regulators of erythropoiesis and tumor suppressors in BM HSPCs.
We showed that homozygous deletion of Aid in vivo leads to expansion of the myelomonocytic lineage. Given that AID cooperates with TET family members in DNA demethylation, these findings are consistent with previous reports showing that mice with Tet2 deletion demonstrated skewed differentiation toward myelomonocytic lineage.14-16 In addition, Aid−/− mice developed anemia with disrupted erythroid differentiation, which is further supported by the observation of decreased mTer119+ erythroid cells in Aid−/− BM. This result is also consistent with the erythroid phenotype seen in Tet2−/− mice.15,16 Of note, Aid−/− myeloid progenitors showed increased expression of Cebpa and decreased expression of Gata1, which may provide a molecular basis of altered myeloid/erythroid differentiation in Aid−/− cells. Taken together, these common features of hematopoietic differentiation seen in both Tet2−/− and Aid−/− mice might be due to deregulated DNA demethylation processes which may affect expression of key myeloid/erythroid lineage-specific TFs, although with less severity and transformation potential than observed with Tet2 loss. It is noteworthy that human BM cells with silenced AID also showed skewed differentiation toward the myelomonocytic lineage in vitro. This is particularly relevant given a recent study in vitro that suggested differentiation was skewed toward monocytic CD14+ populations in TET2-silenced human cord blood cells.26
Our study demonstrated that Aid loss did not confer enhanced replating capacity or competitive advantage on HSC in in vitro CFU assays or in in vivo competitive repopulating assays, in contrast to Tet2 loss.14-16 In addition, Aid−/− mice did not develop lethal myeloproliferative disease in vivo within 2 years of observation (data not shown). Of note, Aid loss did not cooperate with Flt3-ITD to cause aggressive myeloid leukemia. Given that Tet2−/− mice develop myeloid neoplasms reminiscent of human CMML14 and that Tet2 loss cooperates with Flt3-ITD to induce AML in vivo,20 these results are in sharp contrast to Tet2 loss. Several possibilities may explain the differential effect of Aid loss and Tet2 loss on HSC regulation and myeloid transformation. Guo et al have recently reported that Apobec1 cooperates with Tet1 in neuronal activity–induced, region-specific, active DNA demethylation and subsequent gene expression in the dentate gyrus in mouse adult brain.8 Moreover, within the AID/APOBEC family, APOBEC3 has been reported to possess cytidine deaminase activity.27 These data suggest that Apobec1 and Apobec3 may function as redundant factors of Aid in active DNA demethylation processes. The exact biological significance of 5hmC remains to be delineated. Mendonca et al recently demonstrated that DNA modified with 5hmC generally exhibits an open chromatin configuration.28 In addition, 5hmC was reported to accumulate at DNA damage foci induced by irradiation and colocalize with major DNA damage response proteins 53BP1 and γH2AX, indicating that the TET family members and 5hmC play essential roles in ensuring genome integrity.29 Given that Tet2 loss causes decreased 5hmC whereas Aid loss induces accumulation of 5hmC (Figure 1D), it is possible that Tet2 loss and Aid loss demonstrate a different effect on hematopoiesis through the unique and specific biological role of 5hmC independent of DNA demethylation.
Alexandrov et al have reported that AID is responsible for kataegic mutations in various human cancers.18 However, WES using DNA from either WT or Aid−/− nucleated PB cells failed to show any clusters of C>T or C>G mutations in Aid−/− cells. These data suggest that, although our Aid−/− mice model is a useful tool to elucidate the effect of Aid loss in normal and malignant hematopoiesis, it does not recapitulate the “Kataegis” phenotype observed in human cancers. We demonstrated that Aid−/− LSKs show altered expression of regulators of erythropoiesis and tumor suppressors, with concomitant DNA hypermethylation at some of these loci. Of note, enhanced DNA methylation in Aid−/− LSKs was preferentially seen at loci with potential capacity for transcriptional regulation, including CpG islands, CpG shores, and promoters, suggesting that DNA hypermethylation status in these regions caused by Aid loss may lead to epigenetic alterations which modulate transcriptional output. Indeed, CpG islands and particularly CpG shores have been described to be critical regulatory regions for hematopoiesis.30 Consistent with this notion, we observed both hypermethylation and a trend toward decreased expression of known tumor suppressors in Aid−/− HSPCs. Although it is possible that this finding is due to the loss of active DNA demethylation machinery in Aid−/− cells,9 there may be other possible mechanisms such as the potential loss of the passive DNA demethylation machinery in Aid−/− cells. In addition, bisulfite sequencing cannot discriminate between 5mC and 5hmC, raising the possibility that hypermethylation seen in some genetic loci in Aid−/− cells may include hydroxymethylation. Further analysis and studies comparing the transcriptomes and methylomes of WT, Aid−/−, and Tet2del/del LSKs may provide important insights for mechanisms and targets of epigenetic regulation critical for HSC biology.
In summary, we demonstrate that Aid is a critical regulator of myeloid and erythroid lineage differentiation in both the murine and human context. We also showed that, in contrast to Tet2, Aid does not have an essential role in HSC self-renewal or in myeloid transformation. Moreover, RNA-seq and eRRBS revealed that Aid modulates transcription and methylation of a subset of key hematopoietic regulators and tumor suppressors. Our data suggest that myeloid and erythroid lineage differentiation is regulated at least in part by epigenetic gene regulation through active DNA demethylation, whereas HSC self-renewal and myeloid transformation might be regulated through a novel biological role of 5hmC, which may be in part independent of DNA demethylation or regulated by other enzymes which mediate demethylation in HSCs. Further studies are required to elucidate the exact molecular targets of DNA demethylation relevant for hematopoietic cell differentiation and to uncover the unique biological significance of 5hmC, AID, and other enzymes involved in DNA demethylation downstream of Tet2 in HSC regulation and myeloid transformation.
Acknowledgments
The authors thank Ritu Kumar, Bharat Vaidyanathan, and members of the Levine Laboratory for technical support. The authors also thank Caroline Sheridan, Tak Lee, and Yaseswini Neelamraju for technical support and the Weill Cornell Medicine Epigenomics core for sequencing services.
This work was supported by the Starr Cancer Consortium, Tri-Institutional Stem Cell Consortium, a Leukemia & Lymphoma Scholar Award, and National Institutes of Health (NIH) National Cancer Institute (NCI) R01CA173636 (R.L.L.); NIH National Institute of Allergy and Infectious Diseases 1R01AI072194 and 1R01AI124186 (J.C.); Leukemia & Lymphoma Society (LLS) SCOR 9328-16, LLS SCOR 7006-13, NIH NCI R01CA198089, and the Chemotherapy Foundation (A.M.); NIH NCI K08CA169055 and an American Society of Hematology (ASH) award (ASHAMFDP-20121) under the ASH–Harold Amos Medical Faculty Development Program (AMFDP) partnership with the Robert Wood Johnson Foundation (F.E.G.-B.); NIH National Institute of General Medical Sciences Grant T32 GM007739 and NIH NCI Fellowship F30CA18349 (A.S.M.); the ASH Senior Research Training Award for Fellows and a New York State Council on Graduate Medical Education Empire Clinical Research Investigator Program Fellowship (B.D.); the Translational Research Oncology Training Program from the Iris & Junming Le Foundation (E.P.); an LLS special fellowship and NIH NCI K08CA181507-01A1 (A.H.S.); and a Sumitomo Life Welfare and Culture Foundation Foreign Medical Research Grant, an Astellas Foundation for Research on Metabolic Disorders Foreign Medical Research Grant, and a Clinical Scholars Biomedical Research Training Program Fellowship from the Charles A. Dana Foundation (H.K.). The flow cytometric analysis in this study and use of other core services was funded in part through NIH NCI Cancer Center Support Grant P30CA008747.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.K., A.S.M., C.M., F.R., B.D., E.P., and R.L.L. designed experiments and interpreted results; H.K., A.S.M., C.M., F.R., F.E.G.-B., E.P., and R.L.L. wrote and edited the manuscript; H.K., K.S., A.N., C.M., F.R., B.D., F.E.G.-B., and A.S.M. performed experiments; and A.H.S., A.M., J.C., and R.L.L. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ross L. Levine, Human Oncology and Pathogenesis Program, Center for Epigenetics Research, Center for Hematologic Malignancies, Leukemia Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, Rockefeller Research Laboratories, RRL-401C, 430 East 67th St, New York, NY 10065; e-mail: leviner@mskcc.org.
References
- 1.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10(11):1147-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553-563. [DOI] [PubMed] [Google Scholar]
- 3.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12(7):517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146(6):866-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289-2301. [DOI] [PubMed] [Google Scholar]
- 11.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838-842. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quivoron C, Couronné L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25-38. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Cai X, Cai CL, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118(17):4509-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135(6):1028-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer [published correction appears in Nature. 2013;502(7470):258]. Nature. 2013;500(7463):415-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R, DiMenna L, Schrode N, et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013;500(7460):89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Maaten L, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9(11):2579-2605. [Google Scholar]
- 22.Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach LA, Fu P, Yang Z. Insulin-like growth factor-binding protein-6 and cancer. Clin Sci (Lond). 2013;124(4):215-229. [DOI] [PubMed] [Google Scholar]
- 24.Honma M, Higuchi O, Shirakata M, et al. Dok-3 sequesters Grb2 and inhibits the Ras-Erk pathway downstream of protein-tyrosine kinases. Genes Cells. 2006;11(2):143-151. [DOI] [PubMed] [Google Scholar]
- 25.Berger AH, Niki M, Morotti A, et al. Identification of DOK genes as lung tumor suppressors. Nat Genet. 2010;42(3):216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronier E, Almire C, Mokrani H, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118(9):2551-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9(6):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendonca A, Chang EH, Liu W, Yuan C. Hydroxymethylation of DNA influences nucleosomal conformation and stability in vitro. Biochim Biophys Acta. 2014;1839(11):1323-1329. [DOI] [PubMed] [Google Scholar]
- 29.Kafer GR, Li X, Horii T, et al. 5-hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Reports. 2016;14(6):1283-1292. [DOI] [PubMed] [Google Scholar]
- 30.Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338-342. [DOI] [PMC free article] [PubMed] [Google Scholar]