Key Points
Endogenous CyPG PGJ2 targets LSCs through PPARγ activation.
Selenium supplementation could serve as an adjunct therapy for CML.
Abstract
Supplementation with nontoxic doses of micronutrient selenium has been shown to alleviate chronic myelogenous leukemia (CML) via the elimination of leukemia stem cells (LSCs) in mice. This treatment provides a new and novel method for eliminating the LSCs that are otherwise not targeted by existing therapies. The antileukemic effect of selenium was dependent on the production of endogenous cyclopentenone prostaglandins (CyPGs), Δ-12 prostaglandin J2 (Δ12-PGJ2), and 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2). Here, we show that these endogenous CyPGs, produced by mice maintained on selenium-supplemented diets, alleviate the symptoms of CML through their ability to activate the nuclear hormone receptor, peroxisome proliferator activated receptor γ (PPARγ). GW9662, a potent PPARγ antagonist, blocked the antileukemic effect of selenium supplementation by significantly reducing CyPGs. This effect was mediated by an increase in 15-prostaglandin dehydrogenase (15-Pgdh) activity, which oxidizes and inactivates Δ12-PGJ2 and 15d-PGJ2. In contrast, treatment with the PPARγ agonist pioglitazone mimicked selenium supplementation. This treatment led to decreased 15-Pgdh activity and increased CyPG levels, which inhibited CML progression. Selenium-dependent activation of PPARγ mediated by endogenous CyPGs decreased Stat5 expression leading to the downregulation of Cited2, a master regulator of LSC quiescence. These studies suggest a potential role for selenium supplementation as an adjuvant therapy in CML.
Introduction
Chronic myeloid leukemia (CML) is caused by a reciprocal chromosomal translocation (t(9;22)(q34;q11)) to form the Philadelphia (Ph+) chromosome.1-4 As a result of this translocation, the fusion oncoprotein BCR-ABL is formed, which exhibits dysregulated ABL tyrosine kinase activity. BCR-ABL drives the development of a 2-stage disease. In the chronic phase, the disease is characterized by the overproduction of mature myeloid cells. This disease can then progress to a blast crisis stage that is an aggressive acute leukemia.2 Tyrosine kinase inhibitors (TKIs) that inhibit the BCR-ABL kinase activity are the standard of care for chronic-phase disease.3,5 As long as patients are maintained on these drugs the disease is held in check.6 Unfortunately, most patients relapse if taken off of TKI therapy because BCR-ABL kinase activity is not required to maintain CML leukemia stem cells (LSCs).7 Because TKIs do not target LSCs, there is a significant gap in our ability to treat CML.7-10
Previously, we demonstrated that cyclooxygenase 1 and 2 (Cox-1 and Cox-2)-derived cyclopentenone prostaglandins (CyPGs), including Δ-12 prostaglandin J2 (Δ12-PGJ2) and 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), are effective treatments for CML in a murine model of CML chronic-phase disease. These compounds are able to target CML-LSCs and result in complete remission of disease. Our work demonstrated that selenium supplementation shifts the arachidonic acid metabolism to more favor the production of CyPGs, instead of other prostaglandins, by a process termed “eicosanoid class switching.”11,12 The redox control of eicosanoid class switching, by selenoproteins, resulted in increased expression of hematopoietic-prostaglandin D synthase (H-Pgds),11 leading to the enhanced production of Δ12-PGJ2 and 15d-PGJ2. The observation that selenium promoted the production of CyPGs underlies the ability of selenium supplementation to prevent the progression of CML disease. Here, we show that increased production of CyPGs by selenium supplementation activates peroxisome proliferator activated receptor-γ (PPARγ)-dependent signaling.
The nuclear receptor PPARγ, a central regulator of metabolism, is activated by thiazolidinediones (TZDs), rosiglitazone and pioglitazone, as well as endogenous metabolites in the form of CyPGs including Δ12-PGJ2 and its dehydration product, 15d-PGJ2.11,13 These data extend the recent finding that pioglitazone, a PPARγ agonist, significantly reduced LSC burden and increased susceptibility of LSCs to treatment with imatinib and other TKIs. Here, we report that inhibition of CML progression by selenium supplementation relies on the activation of PPARγ by CyPGs, which in turn leads to decreased activation of Stat5a and a downregulation of Cited2 and Hif2α, 2 genes associated with stem cell quiescence. In addition to decreased Stat5 activity, we identify a novel mechanism regulating CyPG levels by the PPARγ-dependent regulation of 15-prostaglandin dehydrogenase (15-Pgdh) activity, which oxidizes and inactivates Δ12-PGJ2 and 15d-PGJ2. Taken together, these data present a new pathway where selenium-dependent production of CyPGs leads to LSC quiescence, through feed-forward activation of PPARγ and inhibition of Stat5a. In addition, selenium supplementation decreases 15-Pgdh, thus preventing the turnover of endogenous PPARγ-activating CyPGs. Thus, selenium supplementation results in an amplification of PPARγ signaling, inhibiting CML progression.
Methods
Selenium diet and mice
Three-week-old C57BL/6 mice (Taconic Biosciences, Hudson, NY) were randomly placed on selenium-adequate (Se-A; 0.08 ppm Se as selenite) or selenium-supplemented (Se-S; 0.4 ppm Se as selenite) semipurified diets (Harlan Teklad, Madison, WI) for at least 8 weeks. Diets were provided ad libitum and mice were maintained on Milli-Q water. Mice were housed 3 to 4 per cage in a temperature- and humidity-controlled room with a 12-hour light-dark cycle. All mice were 11 to 16 weeks of age at sacrifice. Mice over 40 g at time of bone marrow transplant were excluded. The Institutional Animal Use and Care Committee at The Pennsylvania State University preapproved all procedures.
Extraction and LC-MS/MS analysis of PGs from serum
Prostaglandins (PGs) were extracted from serum samples using a C-18 Sep-pak cartridge (Waters, Milford, MA). Briefly, serum samples were acidified with 2 N HCl and loaded on to the Sep-Pak cartridge; bound PGs were eluted with methanol, evaporated, and stored in ethyl acetate at −80°C until further analysis. After extraction, PGs were suspended in 70% methanol and analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) as described earlier.14 Quantitative analysis of Δ12-PGJ2, 15d-PGJ2, and 13,14-dihydro-15-keto-PGJ2 was performed using a calibration curve generated by respective standards in multiple reaction monitoring mode. Three transitions were used to detect each PG: Δ12-PGJ2 (332.8/188.9; 332.8/314.9; 332.8/271.3 m/z), 15d-PGJ2 (315.1/271.1; 315.1/203.0; 315.1/158.0 m/z), 13,14-dihydro-15-keto-PGJ2 (333.2/175.0; 333.2/207.2; 333.2/315.0 m/z). Data acquisition and analysis was performed using Analyst software (version 1.5; AB Sciex).
Generation of experimental myeloid leukemia
The CML model was generated as previously described.15 Retroviral stocks were generated by transfecting HEK293T cells with pMIG-BCR-ABL (a gift from Dr. Warren Pear, University of Pennsylvania, Philadelphia, PA) and pEco for packaging using TransIT 293 reagent (Mirus Bio, Madison, WI). Transduction of hematopoietic stem cells (HSCs) was modified as previously described.16,17 Bone marrow cells were transduced with retrovirus, allowed to rest in conditioned media, and transplanted by retro-orbital injection (1 × 106 to 2 × 106 trypan blue–counted cells) into lethally irradiated mice. Transduced cells were passaged through mice at least 2 times prior to transplantation into experimental mice. Lineage-negative (Lin−) BCR-ABL cells (5 × 105; which represent undifferentiated HSCs expressing BCR-ABL), isolated using the EasySep mouse hematopoietic progenitor cell enrichment kit (Stem Cell Technologies, Vancouver, BC, Canada) were transplanted by retro-orbital injection into sublethally irradiated (475 rad) recipient mice. Disease progression was monitored by complete blood count (CBC) analysis, evaluating peripheral blood for leukocytosis on a Hemavet 950FS equipped with a veterinary software program. Green fluorescent protein (GFP) expression in the blood, by flow cytometry, was used as a secondary method to measure circulating CML cells. Mice were euthanized between day 14 through day 21 posttransplant, based on disease progression and humane end points.
Modulation of PPARγ activity in vivo
PPARγ agonist pioglitazone (5 mg/kg body weight) and antagonist GW9662 (1 mg/kg body weight) were purchased from Cayman Chemical (Ann Arbor, MI) and dissolved in a solution of hydroxypropyl-β-cyclodextrin (HPBCD; 30 mM in sterile water; Sigma-Aldrich, St Louis, MO) and administered by intraperitoneal (i.p.) injection daily, starting 1 day prior to BCR-ABL transplant, until mice were euthanized (∼21 days). HPBCD in sterile water was used as the vehicle control.
Flow cytometry of CML-LSCs
Flow cytometry was performed as previously described.15 Whole spleen and bone marrow were collected and red blood cells were lysed with ACK lysis buffer (155 mM NH4Cl, 12 mM KHCO3, 0.1mM EDTA-2Na). Isolated Lin− cells were stained for Ly-6A/E (Sca-1) and Cd117 (c-Kit) (BD Biosciences, San Jose, CA). Lin− white blood cells (WBCs) and the presence of LSCs were analyzed on Accuri C6 (BD Biosciences, San Jose, CA). Lin−GFP+c-Kit+Sca-1+ are hereafter designated as LSCs, unless otherwise indicated. When LSCs were used for protein and RNA analysis, Lin− cells were fluorescence-activated cell sorted (FACS) for the GFP+c-Kit+Sca-1+ population. In some assays where LSCs were limiting (as in Se-S CML mice), Lin−BCR-ABL cells from spleen and bone marrow that are enriched (>60%-70%) for the LSC population were used.
Western immunoblot analysis
Lin−BCR-ABL cell lysate was isolated with M-PER (ThermoFisher Scientific, Waltham, MA) and used in western immunoblotting analysis. Polyvinylidene difluoride membranes were incubated with the following antibodies: 15-Pgdh, thromboxane A synthase (Txas), and microsomal prostaglandin E synthase-1 (Ptges) (all from Cayman Chemical, Ann Arbor, MI), phospho-Stat5 and Stat5 (Cell Signaling Technologies, Danvers, MA), Cbp/P300-interacting transactivator with Glu/Asp-rich C-terminal domain 2 (Cited2; Abcam, Cambridge, United Kingdom), hypoxia-inducible factor 2α (Hif2α; R&D Systems, Minneapolis, MN), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh; Fitzgerald Industries, Acton MA). Appropriate secondary antibodies conjugated with horseradish peroxidase (Thermo Pierce, Rockport, IL) were used. Experiments were performed in at least biological triplicate and technical duplicate. Autoradiographs were subjected to densitometry analysis by Image J software (National Institutes of Health, Bethesda, MD).
Gene expression analysis
Quantitative reverse transcription polymerase chain reaction (qPCR) with TaqMan probes (Life Technologies, Carlsbad, CA) was performed to examine gene expression in Lin− BCR-ABL cells. Data were analyzed according to the method of Livak and Schmittgen with normalization to 18S ribosomal RNA (rRNA).18
LSC-CFUs
LSC–colony-forming units (LSC-CFUs) were plated and counted as previously described.15 Total splenocytes and/or bone marrow from leukemic mice were plated in M3231 Methocult (Stem Cell Technologies, Vancouver, BC, Canada). Media was supplemented with interleukin 3 (2.5 ng/mL; R&D Systems, Minneapolis, MN), stem cell factor (50 ng/mL; Gold Biotechnology, St. Louis, MO), growth differentiation factor 15 (Gdf15) (30 ng/mL; Biomatik, Wilmington, DE), and sonic hedgehog (Shh) (25 ng/mL; Gold Biotechnology). On days 7 to 10, colonies were counted. LSC-CFUs were defined based on their size (large, >100 cells at high magnification), shape (perfectly circular), density (dark brown/black color), and edge (clearly defined edge with limited differentiation). Any variation from LSC-CFU formation was also considered: in some cases, granulocyte-macrophage progenitor CFUs were observed.19 Each biological sample was plated in triplicate. The average number of LSC colonies was calculated for each biological replicate.
ChIP
To examine binding of PPARγ to the PPAR response element (PPRE) in the Cd36 promoter, chromatin immunoprecipitation assay (ChIP) assays were performed as described.20 Briefly, LSCs (1 × 107) were subjected to various treatments (for 8 hours), crosslinked using 1% formaldehyde (10 minutes), and sonicated. Lysates were precleared using 20 μL of protein A/G plus-agarose beads and subjected to immunoprecipitation with PPARγ antibody (4 μg; Santa Cruz Biotechnology, Santa Cruz, CA) and 20 μL of protein A/G plus–agarose overnight at 4°C. A no-antibody control was included with each experiment. Immune complexes were washed 3 times each with low-salt wash buffer (20 mM Tris-HCl, pH 8.00 containing 0.1% sodium dodecyl sulfate, 1% Triton X-100, 2 mM EDTA, and 150 mM NaCl), high-salt wash buffer (above buffer with 500 mM NaCl), and 2 times with 10 mM Tris-(1mM) EDTA. Immune complexes were eluted and cross-links were reversed. DNA was extracted and used in qPCR with primers: Cd36 PPRE (forward primer), 5′-CAGGCTTTGTTGGGACAGAC-3′; (reverse primer), 5′-GCTAATTTGTGGTTGGTTGCCA-3′.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 6 (GraphPad Software, San Diego, CA). Unless noted, nonparametric tests, including 1-way analysis of variance and the unpaired 2-tailed Student t test, were used where appropriate. For BCR-ABL transplantation studies, at least 2 independent experiments were conducted. Data were compiled from these experiments for a total biological n value described in figure legends. For technical analysis (flow cytometry, qPCR, and western blot), samples were run in at least duplicate. The mean of the technical replicates was considered a biological n value for statistical analyses. PPARγ activation was considered as baseline for experiments: Se-A pioglitazone or Se-S vehicle were used as controls, unless otherwise noted, for t tests. For analysis of variance, all groups were compared, unless noted. Variation from these analyses is described in figure legends. Bars represent biological mean ± standard error of the mean (SEM): *P < .05, **P < .01, ***P < .001, #P < .0001.
Results
The protective effect of selenium supplementation is mediated by activation of PPARγ
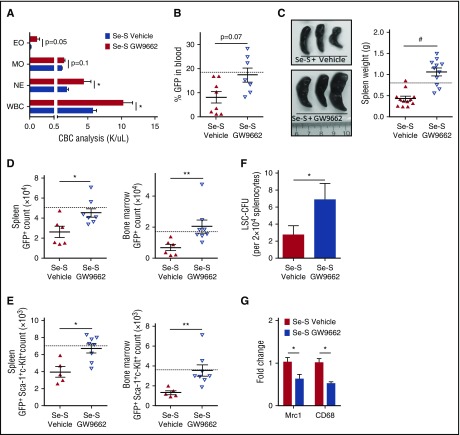
Previously, we demonstrated that selenium supplementation inhibits CML progression and blocks the maintenance of CML-LSCs as measured in secondary transplant assays.14 As with in vivo selenite administration, addition of selenium (as sodium selenite; 250-500 nM) to in vitro cultures increased the production of CyPGs through eicosanoid class switching. Because CyPGs are agonists of PPARγ, we sought to determine whether the effects of selenium supplementation are mediated by PPARγ signaling. To test this possibility, we used GW9662, a synthetic PPARγ antagonist. Overall, GW9662-treated Se-S BCR-ABL mice exhibited greater CML disease burden, when compared with vehicle-treated Se-S BCR-ABL mice (Figure 1A-C). CBC analysis conducted prior to euthanizing BCR-ABL mice indicated an increase in myeloid-derived leukocytes in the peripheral blood, upon treatment with GW9662. Neutrophils were significantly increased and there was a trend of increased monocytes and eosinophils (Figure 1A). Total WBCs were twice as high in Se-S GW9662 BCR-ABL mice as compared with Se-S vehicle BCR-ABL mice (Figure 1A). As GFP is coexpressed with BCR-ABL, GFP was used as a surrogate marker for total disease infiltration in blood and various tissues. Though the difference did not reach significance, there was over a twofold increase in peripheral blood GFP+ cells when comparing the mean GFP expression in Se-S vehicle vs Se-S GW9662 BCR-ABL mice (Figure 1B; supplemental Figure 1A, available on the Blood Web site). Splenomegaly was induced in Se-S GW9662 BCR-ABL mice, with over a twofold increase in splenic weight compared with the vehicle control BCR-ABL mice on the Se-S diet (Figure 1C).
Figure 1.
GW9662 inhibits Se-S protection in CML mice. Se-S mice were given daily i.p. injection of GW9662 (1 mg/kg) starting 1 day prior to BCR-ABL transplant, until euthanized. (A) CBC (K/μL blood), WBC, eosinophil (EO), monocyte (MO), neutrophil (NE) profile of Se-S BCR-ABL mice treated with or without GW9662 (n = 4). (B) Flow cytometry of peripheral blood GFP expressed as percentage of gated input in Se-S BCR-ABL transplanted mice (5 × 105 events collected, gated on forward scatter [FSC]) (n = 8). (C) Representative image and spleen weight of Se-S BCR-ABL mice treated with or without GW9662 (n = 11-12). (D) Flow cytometric analysis of GFP in the spleen (left) and bone marrow (right) of Se-S BCR-ABL mice treated with or without GW9662 (n = 6-8). Counts are shown. (E) LSC analysis by flow cytometry in the spleen (left) and bone marrow (right) of Se-S BCR-ABL mice treated with or without GW9662. Lin− cells were gated on the GFP+ population (shown in panel D). Total counts (GFP+Sca-1+c-Kit+) are shown. Gates are based on GFP control and fluorescence minus one controls (n = 5-8). (F) Lin− Se-S BCR-ABL splenocytes (2 × 104) were plated in technical triplicate in Methocult and counted on day 10. LSC-CFUs were counted and plotted as total counts (n = 8). (G) PPARγ inhibition with GW9662 in Lin− Se-S BCR-ABL splenocytes by qPCR (n = 4). In panels B through E, the dashed line represents untreated, Se-A BCR-ABL control. Bars represent average of each biological mean ± SEM. The Student 2-way t tests were performed to compare groups. Error bars represent biological mean ± SEM. *P < .05; **P < .01; #P < .0001.
Total disease burden was measured in the spleen and bone marrow by flow cytometric analysis of GFP. The number of GFP+ cells was significantly higher in Se-S GW9662 BCR-ABL mice (Figure 1D; supplemental Figure 1B-C). To further analyze the effect on the CML-LSC population upon GW9662 treatment, Lin−GFP+ cells from the spleen and bone marrow were analyzed for c-Kit and Sca-1 expression. CML-LSCs were significantly higher in both the spleen and bone marrow of Se-S GW9662 BCR-ABL mice when compared with Se-S BCR-ABL mice (Figure 1E; supplemental Figure 1B-C). Furthermore, functional CML-LSCs in the spleen were analyzed by their ability to form colonies in methylcellulose media. Lin− cells from GW9662-treated Se-S BCR-ABL mice formed significantly more LSC-CFUs as compared with vehicle-treated Se-S BCR-ABL mice (Figure 1F). GW9662 treatment of BCR-ABL Se-S mice significantly decreased 2 PPARγ downstream genes, Mrc1 and Cd68, in Lin− splenocytes confirming the inhibition of PPARγ signaling in the treated mice (Figure 1G). Taken together, these studies confirmed that PPARγ activation was essential for the protective effect in Se-S BCR-ABL mice.
Selenium-dependent inhibition of Stat5a activation requires PPARγ signaling
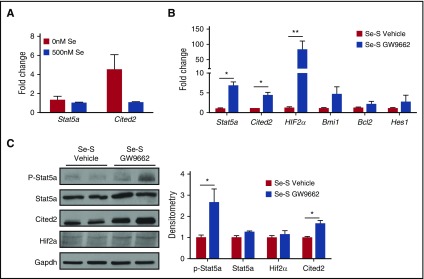
Stat5a and its downstream targets Cited2 and Hif2α play a key role in maintaining LSCs in CML. Recently, pioglitazone was shown to decrease Stat5a activation in CML cells.21 On the lines of our previous studies in macrophages where selenium treatment increased the activation of PPARγ,11 CML-LSCs treated with selenium (500 nM) increased the expression of prototypical PPARγ target genes, Cd36, Cd68, Mrc1 (Cd206), and Arg1 (supplemental Figure 2). More importantly, ChIP assay indicated increased nuclear translocation and binding of PPARγ in CML-LSCs to consensus PPRE (present in the Cd36 promoter) upon treatment of LSCs with selenium or Δ12-PGJ2 (as positive control), further corroborating the increased expression of Cd36 by endogenous activation of PPARγ (supplemental Figure 3). Similarly, when CML-LSCs were treated ex vivo with selenium (as sodium selenite), the expression of Stat5a and Cited2 was reduced, which is consistent with the decrease in LSCs we observed upon selenium treatment (Figure 2A). We next analyzed Se-S CML mice and Se-S CML mice treated with the PPARγ antagonist GW9662 for the expression of Stat5 and its downstream targets Cited2 and Hif2α. qPCR analysis of splenic Lin−BCR-ABL cells revealed a significant increase in the expression of Stat5a, and 2 of its downstream gene targets, Cited2 and Hif2α, in the GW9662-treated Se-S mice (Figure 2B). Stat5a was increased nearly sevenfold and Hif2α messenger RNA showed a 75-fold increase in Se-S GW9662 BCR-ABL samples. The mean expression of Cited2 was fourfold higher in GW9662 samples. In addition, Bcl2, another Stat5a target and regulator of apoptosis,22 was slightly increased in GW9662 samples. Bmi1 and Hes1 are 2 downstream targets of Cited2 that regulate HSC quiescence and have been shown to play a role in leukemia progression. GW9662 treatment also showed a trend of increased expression of Hes1 and Bmi1 when compared with Lin− splenocytes from Se-S BCR-ABL mice treated with vehicle (Figure 2B). At the protein level, the activation of Stat5a, in the form of phospho-(Y694)-Stat5a was increased in GW9662 samples (Figure 2C). Cited2 protein was significantly increased in Se-S GW9662 BCR-ABL samples. Hif2α protein was also increased upon GW9662 treatment in Se-S Lin− BCR-ABL cells. These studies demonstrate that treatment with selenium inhibits the activation of Stat5a and its downstream targets, through the activation of PPARγ.
Figure 2.
PPARγ inhibition by GW9662 increases Stat5a and downstream targets. Lin− splenocytes from Se-S BCR-ABL mice treated with or without GW9662 were analyzed for RNA and protein expression of Stat5a and downstream targets. (A) Lin−GFP+c-Kit+Sca-1+ FACS-sorted LSCs were treated with or without sodium selenite (500 nM) for 24 hours and the expression of Stat5a and Cited2 was examined by real-time PCR. (B) qPCR expression as fold change compared with Se-S vehicle for each gene and normalized to 18S rRNA expression (n = 4). (C) Western immunoblot analysis showing representative blot and densitometry (normalized to Se-S for each protein and relative to Gapdh) (n = 3-4). Bars represent biological mean ± SEM. All analysis was done in technical triplicate. *P < .05; **P < .01.
PPARγ signaling prevents the turnover of CyPGs in Se-S BCR-ABL mice
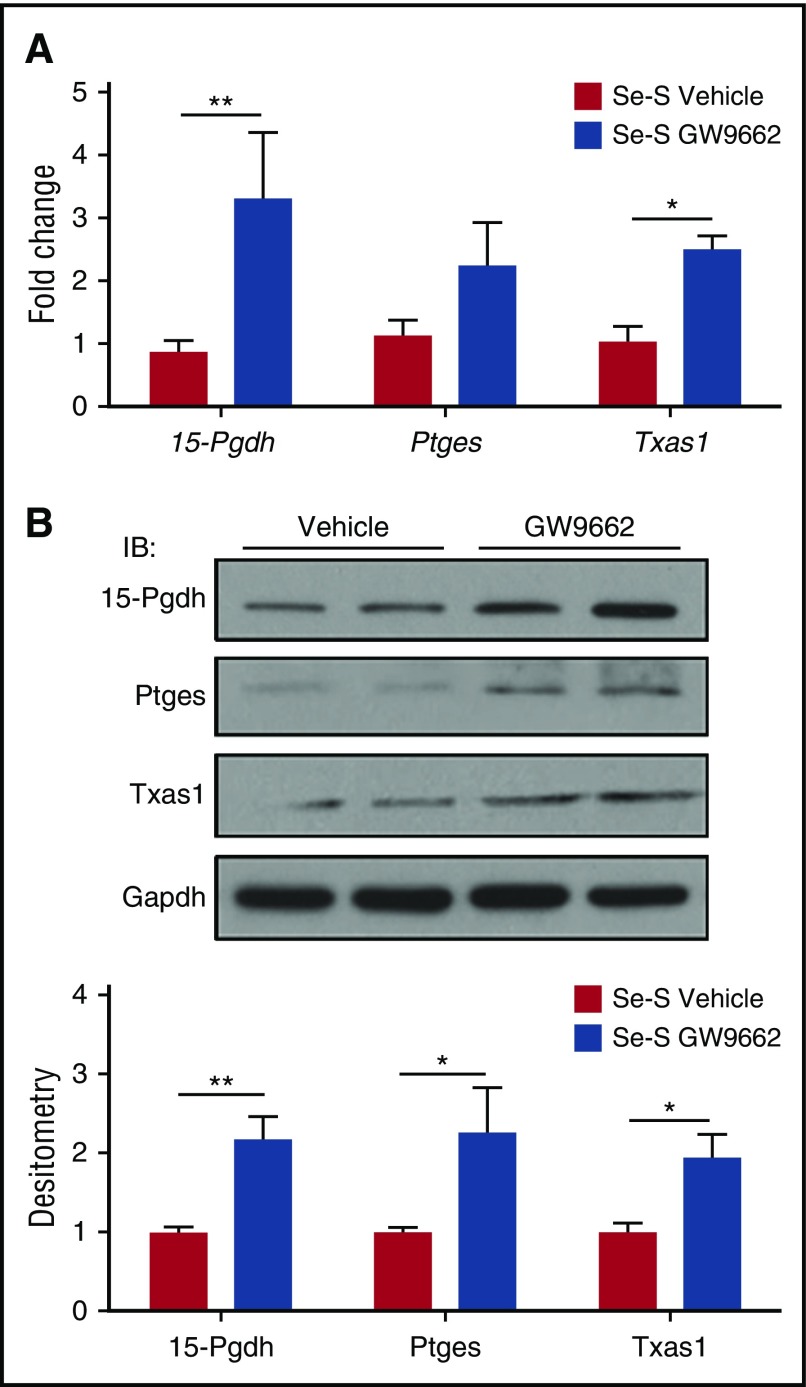
The protective effect of selenium supplementation in BCR-ABL mice is mediated by production of endogenous CyPGs, which results from the increased expression of H-PGDS and the consequent production of Δ12-PGJ2 and 15d-PGJ2.14 Here, we examined whether the effect of increased disease burden in Se-S GW9662 mice was due to a shift in the eicosanoid cascade away from the production of CyPGs. There was no difference in H-PGDS expression at both the RNA and protein level (data not shown). However, qPCR analysis in splenic Lin−BCR-ABL cells revealed a significant increase in 15-Pgdh, an enzyme responsible for the degradation of many lipid mediators, including Δ12-PGJ2 and 15d-PGJ2 (Figure 3A). There was also a significant increase in Txas1 and a trend of increased expression of Ptges (Figure 3A). However, at the protein level, the differences were more obvious and significant. 15-Pgdh, Ptges, and Txas1 expression were all significantly higher in Se-S GW9662 Lin−BCR-ABL cells, with a twofold increase in expression compared with the Lin−BCR-ABL cells from vehicle-treated mice (Figure 3B). These data suggest that inhibition of PPARγ with GW9662 in Se-S BCR-ABL mice interferes with the production of CyPGs that act as endogenous PPARγ ligands.
Figure 3.
Inhibition of PPARγ represses expression of Ptges-1 and Txas1 in Se-S CML progenitor cells. Lin− splenocytes from Se-S BCR-ABL mice treated with or without GW9662 were analyzed for RNA and protein expression. Se-S mice were given daily i.p. injection of GW9662 (1 mg/kg) starting 1 day prior to BCR-ABL transplant until euthanized (n = 4). (A) qPCR expression as fold change compared with Se-S vehicle for each gene and normalized to 18S rRNA expression. (B) Western blot analysis showing representative blot and densitometry (normalized to Se-S for each protein and relative to GAPDH). Bars represent biological mean ± SEM. All analyses were done in technical triplicate. *P < .05; **P < .01.
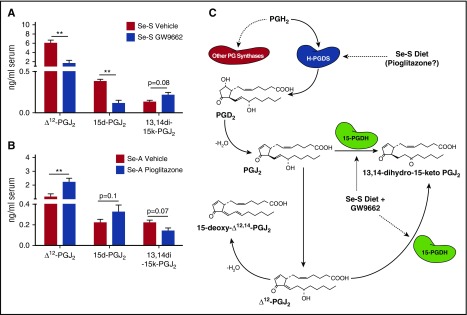
Inhibition of PPARγ signaling leads to decreased serum levels of Δ12-PGJ2 and 15d-PGJ2 in selenium-supplemented BCR-ABL mice
The observation of increased expression of 15-Pgdh was of particular interest due to its ability to degrade Δ12-PGJ2 and 15d-PGJ2. Serum was collected from Se-S BCR-ABL mice and analyzed for these CyPGs by LC-MS/MS. Consistent with increased expression of 15-Pgdh, we observed a significant decrease in both Δ12-PGJ2 and 15d-PGJ2 in the serum of Se-S GW9662-treated mice (Figure 4A). There was a 3.5-fold decrease in Δ12-PGJ2 and a threefold decrease in 15d-PGJ2 when mice were treated with GW9662 (Figure 4A). The upregulation of 15-Pgdh observed in Se-S GW9662 BCR-ABL mice (Figure 3) was accompanied by an increased in 13,14-dihydro-15-keto-PGJ2, which is the product of the 15-Pgdh catalyzed reaction of PGJ2 (Δ13-PGJ2) or Δ12-PGJ2 (Figure 4A). Consistent with the decrease in CyPGs observed when PPARγ signaling is inhibited, treatment of Se-A BCR-ABL mice with pioglitazone resulted in a significant increase in Δ12-PGJ2 and a trend of increased production of 15d-PGJ2 in Se-A (Figure 4B). The breakdown product, 13,14-dihydro-15-keto-PGJ2, was decreased in Se-A pioglitazone BCR-ABL mice as would be expected by the decrease in 15-Pgdh expression (Figure 4B-C).
Figure 4.
Endogenous Δ12-PGJ2and 15d-PGJ2concentrations are affected when PPARγ is targeted in CML mice. Lipid extracts from the serum of BCR-ABL mice were analyzed by LC-MS/MS for Δ12-PGJ2, 15d-PGJ2, and 13,14-dihydro-15-keto PGJ2 (13,14di-15k-PGJ2). (A) Se-S BCR-ABL mice treated with or without GW9662. Se-S mice were given daily i.p. injection of GW9662 (1 mg/kg) starting 1 day prior to BCR-ABL transplant until euthanized (n = 3). (B) Pioglitazone (5 mg/kg) was given by i.p. injection daily starting 1 day prior to BCR-ABL transplant (n = 5-6). Bars represent biological mean ± SEM (*P < .05). (C) Schematic of selenium supplementation and GW9662 involvement in endogenous PPARγ ligand synthesis in BCR-ABL mice. Se-S diet increases H-Pgds, leading to subsequent increase in Δ12-PGJ2 and 15d-PGJ2. Pioglitazone may also increase CyPGs. GW9662 in Se-S BCR-ABL mice increases 15-Pgdh, which leads to breakdown of Δ12-PGJ2 and 15d-PGJ2.
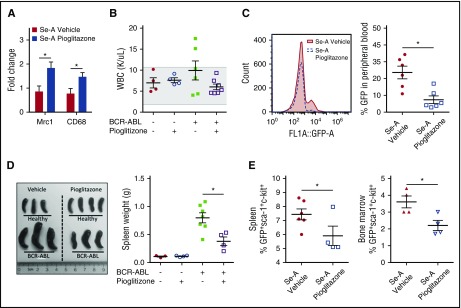
Pioglitazone decreases murine BCR-ABL CML disease burden in selenium adequate mice
Previous work from Prost et al showed that pioglitazone, a potent PPARγ agonist, acts in concert with TKIs to inhibit CML-LSC maintenance.21 Given that treatment of mice with pioglitazone increased the levels of CyPGs, we investigated whether PPARγ activation by pioglitazone without the addition of TKIs could rescue the leukemic mice. The efficacy of pioglitazone was confirmed by its ability to increase the expression of PPARγ target genes, Mrc1 and Cd68 (Figure 5A). Treatment of Se-A BCR-ABL mice with pioglitazone decreased the overall burden of the disease as shown in Figure 5B-E. Most importantly, pioglitazone treatment significantly decreased the CML-LSC burden in the spleen and bone marrow of treated mice. In addition, it had no effect in healthy (nonleukemic) mice in terms of WBC counts (Figure 5B). Furthermore, the difference in total WBC counts was not significant between Se-A BCR-ABL vehicle- and pioglitazone-treated mice. However, all pioglitazone-treated BCR-ABL mice on Se-A diet had WBC counts in the normal range, whereas Se-A vehicle BCR-ABL mice showed an increased trend in WBCs above the normal range (Figure 5B). GFP+ cells in peripheral blood indicated differences, as GFP is coexpressed with BCR-ABL.16 There was a significant decrease in total peripheral blood GFP in Se-A pioglitazone-treated BCR-ABL mice as compared with vehicle-treated (Figure 5C). In addition, splenomegaly, which is a commonly observed feature of murine BCR-ABL CML,23 was also decreased upon treatment of BCR-ABL Se-A mice with pioglitazone (Figure 5D). However, the spleen size was not affected by pioglitazone in healthy Se-A mice (Figure 5D). More importantly, the LSC-like population (GFP+c-kit+Sca-1+) was significantly decreased with pioglitazone treatment both in the spleen and bone marrow of Se-A pioglitazone-treated mice (Figure 5E). Overall, these findings supported the notion that PPARγ activation with pioglitazone treatment was protective in experimental BCR-ABL CML in otherwise susceptible Se-A mice.
Figure 5.
Pioglitazone activation of PPARγ reduces CML burden in Se-A mice. Pioglitazone (5 mg/kg) was given to Se-A mice by i.p. injection daily starting 1 day prior to BCR-ABL transplant, until euthanized. The same duration of treatment was given for healthy pioglitazone mice. Peripheral blood, spleen, and bone marrow were used to measure disease burden. LSC-like cells (GFP+Sca-1+c-Kit+) were determined from total bone marrow and spleen. (A) Confirmation of PPARγ activation with pioglitazone treatment. qPCR expression as fold change in Se-A BCR-ABL splenocytes. Gene expression was normalized to 18S rRNA expression and Se-A vehicle samples were used as control (n = 5). (B) Total WBC (K/μL blood) count in peripheral blood of healthy (n = 4) and BCR-ABL (n = 7) transplanted mice. The shaded box indicates the healthy range. (C) Peripheral blood GFP expressed as percentage of gated input in Se-A BCR-ABL transplanted mice (500 000 events collected, gated on FSC). Representative histogram is shown along with individual data points (n = 6). (D) Representative image and spleen weight in healthy (n = 3-4) and BCR-ABL transplanted (n = 4-7) mice. (E) LSC population evaluation by flow cytometry. LSC population in the spleen (n = 4-6) and bone marrow (n = 4) of BCR-ABL mice. Individual data points are shown from 2 independent experiments. Error bars represent mean ± SEM. *P < .05.
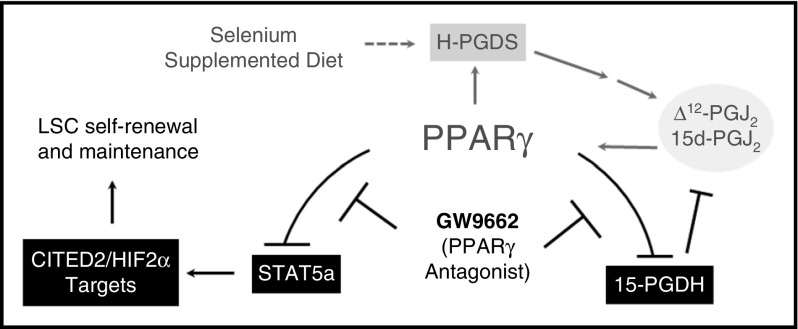
Taken together, our data implicate CyPGs and the activation of PPARγ as being essential in the positive feedback loop where the increased Δ12-PGJ2 and 15d-PGJ2 production (in Se-S mice) leads to increased PPARγ-dependent signaling and decreased CyPG metabolism by 15-Pgdh (Figure 6).
Figure 6.
PPARγ activation is central in the protective effect of Se-S in CML mice. Se-S diet increases expression of H-Pgds, leading to increased production of the PPARγ ligands Δ12-PGJ2 and 15d-PGJ2. PPARγ activation feeds this cycle forward. GW9662 inhibits PPARγ-dependent decrease in Stat5a activation and downstream transcription factors (including Cited2 and Hif2α) implicated in the maintenance of the LSC population. Antagonism of PPARγ by GW9662 interrupts the positive feedback loop activated by selenium supplementation. Solid lines indicate a direct relationship; dashed lines, indirect.
Discussion
The production of endogenous CyPGs, Δ12-PGJ2 and 15d-PGJ2, serves as a key factor in the antileukemic effect of selenium supplementation in experimental models of CML.14 Our previous work demonstrated that treatment with indomethacin to inhibit Cox-1 and Cox-2 or an inhibitor of H-Pgds, such as HQL-79, blocked the antileukemic properties of selenium supplementation.11 These data showed that the Cox-H-Pgds pathway of arachidonic acid metabolism was essential for the endogenous production of CyPGs that eliminate LSCs. Here, we extended those findings to identify a causal relationship between cellular selenium levels and the activation of PPARγ. These data show that supplementation of dietary selenium leads to increased production of CyPGs, which mediate the activation of PPARγ in CML-LSCs. The activation of this signaling pathway decreased the expression of Stat5 and its downstream targets that maintain CML-LSC quiescence. Though CyPGs can also bind to G-protein–coupled receptors (DP1 and DP2/CRTH2) at the cell surface,24 we focused here on intracellular binding to PPARγ. Additional studies will be needed to determine whether the eicosanoid-specific cell surface receptors are also involved in this signaling mechanism.
Blocking PPARγ signaling with the antagonist GW9662 abrogated the effects of selenium supplementation and led to an increase in Stat5a phosphorylation. It is known that protein phosphatase-2A (PP2A), Shp-2, and dual-specificity phosphatase-4 regulate Stat5a activity.25-27 Preliminary analysis suggests that PP2A could be a target of PPARγ (data not shown). In fact, PP2A-activating drugs eradicate TKI-resistant CML cells,28 meaning that PPARγ-dependent activation of protein phosphatase (such as PP2A) could dephosphorylate Stat5a to make it less active. Cited2, a target of Stat5a in CML-LSCs, is known to regulate metabolic activity in addition to maintaining the quiescent LSC population.22,29 Conditional deletion of Cited2 in murine HSCs led to increased reactive oxygen species (ROS) by decreasing glycolytic metabolism and increasing mitochondrial activity.30 These observations corroborate well with our previously reported findings where selenium supplementation increased ROS in LSCs by increasing the expression of NADPH oxidase 1 (Nox1) and other p53 downstream targets, cytochrome C oxidase (Sco2), Tp53-induced glycolytic and apoptotic regulator (Tigar), and ribonucleotide reductase (Rm2b; P53r2).31,32 In addition, it is plausible that CyPGs cause changes in intracellular iron to contribute to increased ROS despite the increased levels of selenoproteins and other antioxidant enzymes.14
Our studies also identify a new target for PPARγ signaling, the suppression of 15-Pgdh activity. This observation adds to our previous data showing that selenium supplementation (250-500 nM) induces a change in arachidonic acid metabolism to increase CyPG production. The ability of CyPGs to induce PPARγ signaling, which in turn decreases the levels of 15-Pgdh, demonstrates that increases in dietary selenium induce an amplification loop that leads to increased levels of CyPGs and increased PPARγ signaling. Sorted LSCs cultured with 250 nM selenite and GW9662 or HQL-79 blocked the ability of selenium to decrease LSC-derived colonies and impact the selenium-dependent decrease in the expression of Hif2α, suggesting that LSCs contribute endogenous CyPGs upon selenium treatment (supplemental Figures 4 and 5). This is in agreement with our previous data on the expression of the enzymatic machinery (comprising Cox-1, Cox-2, and H-Pgds) in sorted LSCs that produce Δ12-PGJ2 in a selenium-dependent manner.14
Surprisingly, GW9662 treatment led to an upregulation of 15-Pgdh expression resulting in the increased oxidation of Δ12-PGJ2 to 13,14-dihydro-15-keto-PGJ2 (Figures 3 and 4), suggesting that selenium-mediated downregulation of 15-Pgdh may be important to sustain high-enough levels of Δ12-PGJ2 and 15d-PGJ2. In fact, this is the first report of the detection of 13,14-dihydro-15-keto-PGJ2 in the serum. Our data clearly show that 13,14-dihydro-15-keto-PGJ2 was unable to activate Cd36 in primary murine bone marrow–derived macrophages, even at high nanomolar concentrations, suggesting that this metabolite may represent an inactivation pathway of proapoptotic CyPGs (supplemental Figure 6). Also referred to as the “eicosanoid reductase” due to its wide specificity, 15-Pgdh is known to inactivate many bioactive lipids that include anti-inflammatory and proresolving lipoxins (to their 15-oxo derivatives) in addition to other eicosanoids.33,34 Thus, our data support the notion that low levels of 15-Pgdh in selenium supplementation might even help in timely initiation and resolution to synergize with events that include clearance of apoptotic cells (such as LSCs) by phagocytosis, where bioactive lipid mediators such as lipoxin A4 are known to play a key role.35
Although pioglitazone clearly affected the progression of leukemia in the CML mouse model, clinically it is unclear whether the long-term use of pioglitazone would be feasible given its association with an increase in the risk of certain cancers, including bladder cancer.36 Pioglitazone is the only commonly available TZD as other members of this family of drugs have health risks such as myocardial infarction.37 The ability to activate PPARγ with endogenous CyPGs through the manipulation of diet may be a promising alternative to TZD treatment in CML patients. Furthermore, our studies suggest that 15-Pgdh inhibitors may also complement CyPGs in the treatment of CML.
In conclusion, we demonstrate that the antileukemic effect of selenium is mediated through activation of PPARγ by endogenous CyPGs. The reduction of the LSC population, and overall reduction in CML burden, was achieved without the addition of TKIs. These findings suggest new options for the development of novel treatments for CML. Dietary supplementation with selenium is sufficient for activation of PPARγ in CML-LSCs, thus eliminating the off-target effects seen with synthetic PPARγ agonists. This treatment coupled with 15-Pgdh inhibitors to further amplify the activation of PPARγ signaling could be efficacious in treating CML. In addition, these treatments could be combined with TKI therapies to effectively eliminate the CML-LSC population and alleviate CML disease burden.
Acknowledgments
The authors thank Ruth Nissly at the Huck Institutes' Flow Cytometry and Microscopy Core Facility, and current and former members of the Prabhu laboratory for valuable suggestions and help.
This work was supported, in part, by grants from the National Institutes of Health (NIH) National Cancer Institute (R01CA162665); NIH National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK077152); US Department of Agriculture–National Institute of Food and Agriculture Hatch project numbers 4605 (K.S.P.) and 4581 (R.F.P.); and the NIH National Center for Advancing Translational Sciences in the form of UL1 TR000127 and TL1 TR000125 (E.R.F.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: E.R.F. conducted all experiments and prepared figures; D.B.T. analyzed samples by LC-MS/MS, and provided data in supplemental Figures 2 through 6; L.L.G. assisted with sample collection; M.D.Q. prepared retrovirus; K.S.P., E.R.F., and R.F.P. planned the study and wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: K.S.P. and R.F.P. are inventors on patents (Δ12-Prostaglandin J3) assigned to the Penn State Research Foundation and are scientific founders of Nemean Pharma, Inc. K.S.P.'s and R.F.P.’s interests were reviewed and are managed by the Penn State Office for Research Protections in accordance with their conflict-of-interest policies. The remaining authors declare no competing financial interests.
Correspondence: K. Sandeep Prabhu, The Pennsylvania State University, 115 Henning Building, University Park, PA 16802; e-mail: ksprabhu@psu.edu; and Robert F. Paulson, The Pennsylvania State University, 115 Henning Building, University Park, PA 16802; e-mail: rfp5@psu.edu.
References
- 1.Goldman JM, Melo JV. Targeting the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1084-1086. [DOI] [PubMed] [Google Scholar]
- 2.Apperley JF. Chronic myeloid leukaemia. Lancet. 2015;385(9976):1447-1459. [DOI] [PubMed] [Google Scholar]
- 3.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353(2):172-187. [DOI] [PubMed] [Google Scholar]
- 4.Hughes T, Deininger M, Hochhaus A, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108(1):28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hehlmann R, Hochhaus A, Baccarani M; European LeukemiaNet. Chronic myeloid leukaemia. Lancet. 2007;370(9584):342-350. [DOI] [PubMed] [Google Scholar]
- 6.Chu S, McDonald T, Lin A, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011;118(20):5565-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson E, Holyoake T. The chronic myeloid leukemia stem cell. Clin Lymphoma Myeloma. 2009;9(suppl 4):S376-S381. [DOI] [PubMed] [Google Scholar]
- 8.Hamad A, Sahli Z, El Sabban M, Mouteirik M, Nasr R. Emerging therapeutic strategies for targeting chronic myeloid leukemia stem cells. Stem Cells Int. 2013;2013:724360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham SM, Jørgensen HG, Allan E, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319-325. [DOI] [PubMed] [Google Scholar]
- 10.Chomel J-C, Bonnet M-L, Sorel N, et al. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118(13):3657-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi UH, Kaushal N, Ravindra KC, et al. Selenoprotein-dependent up-regulation of hematopoietic prostaglandin D2 synthase in macrophages is mediated through the activation of peroxisome proliferator-activated receptor (PPAR) gamma. J Biol Chem. 2011;286(31):27471-27482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson SM, Lei X, Prabhu KS. Selenium levels affect the IL-4-induced expression of alternative activation markers in murine macrophages. J Nutr. 2011;141(9):1754-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83(5):803-812. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi UH, Kaushal N, Hegde S, et al. Selenium suppresses leukemia through the action of endogenous eicosanoids. Cancer Res. 2014;74(14):3890-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finch ER, Kudva AK, Quickel MD, et al. Chemopreventive effects of dietary eicosapentaenoic acid supplementation in experimental myeloid leukemia. Cancer Prev Res (Phila). 2015;8(10):989-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92(10):3780-3792. [PubMed] [Google Scholar]
- 17.Hegde S, Kaushal N, Ravindra KC, et al. Δ12-prostaglandin J3, an omega-3 fatty acid-derived metabolite, selectively ablates leukemia stem cells in mice. Blood. 2011;118(26):6909-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402-408. [DOI] [PubMed] [Google Scholar]
- 19.Pessina A, Albella B, Bueren J, et al. Prevalidation of a model for predicting acute neutropenia by colony forming unit granulocyte/macrophage (CFU-GM) assay. Toxicol In Vitro. 2001;15(6):729-740. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3(4):698-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prost S, Relouzat F, Spentchian M, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature. 2015;525(7569):380-383. [DOI] [PubMed] [Google Scholar]
- 22.Hoelbl A, Schuster C, Kovacic B, et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2(3):98-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koschmieder S, Schemionek M. Mouse models as tools to understand and study BCR-ABL1 diseases. Am J Blood Res. 2011;1(1):65-75. [PMC free article] [PubMed] [Google Scholar]
- 24.Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol Rev. 1999;79(4):1193-1226. [DOI] [PubMed] [Google Scholar]
- 25.Ross JA, Cheng H, Nagy ZS, Frost JA, Kirken RA. Protein phosphatase 2A regulates interleukin-2 receptor complex formation and JAK3/STAT5 activation. J Biol Chem. 2010;285(6):3582-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Wen R, Yang S, et al. Identification of Shp-2 as a Stat5A phosphatase. J Biol Chem. 2003;278(19):16520-16527. [DOI] [PubMed] [Google Scholar]
- 27.Huang CY, Lin YC, Hsiao WY, Liao FH, Huang PY, Tan TH. DUSP4 deficiency enhances CD25 expression and CD4+ T-cell proliferation without impeding T-cell development. Eur J Immunol. 2012;42(2):476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neviani P, Harb JG, Oaks JJ, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013;123(10):4144-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korthuis PM, Berger G, Bakker B, et al. CITED2-mediated human hematopoietic stem cell maintenance is critical for acute myeloid leukemia. Leukemia. 2015;29(3):625-635. [DOI] [PubMed] [Google Scholar]
- 30.Du J, Li Q, Tang F, et al. Cited2 is required for the maintenance of glycolytic metabolism in adult hematopoietic stem cells. Stem Cells Dev. 2014;23(2):83-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puzio-Kuter AM. The role of p53 in metabolic regulation. Genes Cancer. 2011;2(4):385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107-120. [DOI] [PubMed] [Google Scholar]
- 33.Hughes D, Otani T, Yang P, et al. NAD+-dependent 15-hydroxyprostaglandin dehydrogenase regulates levels of bioactive lipids in non-small cell lung cancer. Cancer Prev Res (Phila). 2008;1(4):241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vunta H, Davis F, Palempalli UD, et al. The anti-inflammatory effects of selenium are mediated through 15-deoxy-Delta12,14-prostaglandin J2 in macrophages. J Biol Chem. 2007;282(25):17964-17973. [DOI] [PubMed] [Google Scholar]
- 35.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510(7503):92-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis JD, Habel LA, Quesenberry CP, et al. Pioglitazone use and risk of bladder cancer and other common cancers in persons with diabetes. JAMA. 2015;314(3):265-277. [DOI] [PubMed] [Google Scholar]
- 37.Woodcock J, Sharfstein JM, Hamburg M. Regulatory action on rosiglitazone by the U.S. Food and Drug Administration. N Engl J Med. 2010;363(16):1489-1491. [DOI] [PubMed] [Google Scholar]