Abstract
Background
Expression and activity of heparanase, an endoglycosidase that cleaves heparan sulfate (HS) side chains of proteoglycans, is associated with progression and poor prognosis of many cancers which makes it an attractive drug target in cancer therapeutics.
Methods
In the present work, we report the in vitro screening of a library of 150 small molecules with the scaffold bearing quinolones, oxazines, benzoxazines, isoxazoli(di)nes, pyrimidinones, quinolines, benzoxazines, and 4-thiazolidinones, thiadiazolo[3,2-a]pyrimidin-5-one, 1,2,4-triazolo-1,3,4-thiadiazoles, and azaspiranes against the enzymatic activity of human heparanase. The identified lead compounds were evaluated for their heparanase-inhibiting activity using sulfate [35S] labeled extracellular matrix (ECM) deposited by cultured endothelial cells. Further, anti-invasive efficacy of lead compound was evaluated against hepatocellular carcinoma (HepG2) and Lewis lung carcinoma (LLC) cells.
Results
Among the 150 compounds screened, we identified 1,2,4-triazolo-1,3,4-thiadiazoles bearing compounds to possess human heparanase inhibitory activity. Further analysis revealed 2,4-Diiodo-6-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)phenol (DTP) as the most potent inhibitor of heparanase enzymatic activity among the tested compounds. The inhibitory efficacy was demonstrated by a colorimetric assay and further validated by measuring the release of radioactive heparan sulfate degradation fragments from [35S] labeled extracellular matrix. Additionally, lead compound significantly suppressed migration and invasion of LLC and HepG2 cells with IC50 value of ~5 μM. Furthermore, molecular docking analysis revealed a favourable interaction of triazolo-thiadiazole backbone with Asn-224 and Asp-62 of the enzyme.
Conclusions
Overall, we identified biologically active heparanase inhibitor which could serve as a lead structure in developing compounds that target heparanase in cancer.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-017-3214-8) contains supplementary material, which is available to authorized users.
Keywords: Heparanase inhibitors, triazolo-thiadiazoles, Metastasis, Anticancer activity
Background
The extracellular matrix (ECM) plays a prime role in maintaining the architecture and integrity of organs and tissues [1]. Collagen, fibronectin, laminin and several growth factors and cytokines interact with heparan sulfate proteolglycans (HSPGs) in the ECM and cell surface to maintain cellular framework and function [2, 3]. Heparanase is the predominant endoglycosidase that catalyzes the cleavage of heparan sulfate (HS) polysaccharide chains in HSPGs into smaller fragments and thereby modulates the functions of HS [4–10]. Heparanase degrades the linkage between glucuronic acid and N-sulfo glucosamine residues at restricted sites of HS yielding fragments of 4-7 kDa [6]. Heparanase activity contributes to disassembly and remodeling of basement membrane and ECM resulting in upregulated cell migration and invasion and release of HS-bound growth- and angiogenesis- promoting factors [7–9]. Notably, elevated levels of heparanase are positively correlated with triggered expression of MMP-9, hepatocyte growth factor (HGF) and vascular endothelial growth factor (VEGF) that are entangled with cancer progression [11–13]. Together, these and other results critically support the intimate involvement of heparanase in tumor progression and encourage the development of heparanase inhibitors as anti-cancer therapeutics [14–16].
Several heparin/HS mimetics were demonstrated as heparanase inhibitors and some have entered clinical trials [8, 15], among these are Muparfostat (PI-88), Roneparstat (SST0001), PG545, and necuparanib (M402) [8, 15]. Muparfostat is a mixture of sulfated di- to hexasaccharides which progressed to Phase III clinical trial in post-resection hepatocellular carcinoma. It displayed significant hematologic side effects when administered with docetaxel [17, 18]. PG545, a fully sulfated hexasaccharide conjugated with a lipophilic moiety, is a dual inhibitor of heparanase and angiogenesis, currently in phase-I clinical trials in patients with advanced solid tumors ([19], https://clinicaltrials.gov/ct2/show/NCT02042781). Roneparstat, N-acetylated glycol-split heparin, is in phase I clinical trial in myeloma patients (https://clinicaltrials.gov/ct2/show/study/NCT01764880, [20]. Similarly, necuparanib (glycol-split low molecular weight heparin) is in phase-I/II trial for pancreatic cancer in combination with nab-paclitaxel and gemcitabine (https://clinicaltrials.gov/ct2/show/NCT01621243, [21]). Given the diverse effects of heparin-like compounds, these studies indicate the significance of designing chemically novel, highly selective and biologically active heparanase inhibitors to potently target various types of cancers and possibly inflammatory diseases [8, 15]. Synthesis of heparanase-inhibiting small molecules has been reported [8, 16, 22], but none was advanced to preclinical and clinical studies [8]. We have previously reported the synthesis of various heterocylces with good anticancer activity [23–29]. The current saccharide-based compounds are not specific for heparanase leaving open the question as to how much of their anti-tumor effect is due specifically to blocking heparanase activity. Herein, we screened 150 small molecules with the scaffold bearing quinolones, oxazines, benzoxazines, isoxazoli(di)nes, pyrimidinones, quinolines, benzoxazines, and 4-thiazolidinones, thiadiazolo[3,2-a]pyrimidin-5-one, 1,2,4-triazolo-1,3,4-thiadiazoles, and azaspiranes for inhibition of human heparanase enzymatic activity. Selected molecules were tested for inhibition of cell migration and invasion. The most effective compound was examined for putative binding modes against the target enzyme using molecular docking analysis.
Methods
All solvents were of analytical grade and reagents were purchased from Sigma-Aldrich. 1H and 13C NMR spectra were recorded on a Varian and Bruker WH-200 (400 MHz) spectrometer in CDCl3 or DMSO-d6 as solvent, using TMS as an internal standard and chemical shifts are expressed as ppm. Mass spectra were determined on a Shimadzu LC-MS. High resolution mass spectra were determined on a Bruker Daltonics instrument. The elemental analyses were carried out using an Elemental Vario Cube CHNS rapid Analyzer. The progress of the reaction was monitored by TLC pre-coated silica gel G plates.
Heparanase
Active heparanase was produced in HEK 293 cells stably transfected with the human heparanase gene construct in the mammalian pSecTag vector. The enzyme was purified and kindly provided by Dr. Yi Zhang (Eli Lilly and Company, New York, NY) [30].
Cells
Mouse Lewis lung carcinoma (LLC; ATCC Cat. number: CRL-1642), human lung carcinoma (HCC827; ATCC Cat. number: CRL-2868), and human hepatocellular carcinoma (HepG2, Hep3B; ATCC Cat. number: HB-8065 and HB-8064, respectively) cell lines were obtained from the American Type Culture Collection and working stocks did not exceed four passages. The cell lines have recently been tested for mycoplasma contamination and authenticated using the Promega PowerPlex 16 HS kit. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with glutamine, pyruvate, antibiotics and 10% fetal calf serum in a humidified atmosphere containing 5% CO2 at 37 °C.
Real-time PCR
Total RNA was extracted with TRIzol (Sigma) and RNA (1 μg) was amplified using one step PCR amplification kit, according to the manufacturer’s (ABgene, Epsom, UK) instructions. The PCR primer sets utilized were: i) mouse heparanase - Forward: 5′ TTTGCAGCTGGCTTTATGTG 3′, Reverse: 5′ GTCTGGGCCTTTCACTCTTG 3′ (207 nucleotides); ii) mouse GAPDH - Forward: 5′ AGAACATCATCCCTGCATCC 3′, Reverse: 5′ AGCCGTATTCATTGTCATACC 3′ (348 nucleotides); iii) human heparanase - Forward: 5′ CCAGCCGAGCCACATCGCTC 3′, Reverse: 5′ ATGAGCCCCAGCCTTCTCCAT 3′ (550 nucleotides); iv) human GAPDH - Forward: 5′ ACAGTTCTAATGCTCAGTTGCTC 3′; Reverse: 5′ TTGCCTCATCACCACTTCTATT 3′ (360 nucleotides).
Preparation of Sulphated Ceria
Hydrous cerium oxide was prepared by the hydrolysis of cerium (III) nitrate hexahydrate with 1:1 ammonia. Cerium (III) nitrate was dissolved in double distilled water. To this clear solution, dilute (1:1) aqueous ammonia was added drop-wise from a burette with vigorous stirring until the pH of the solution reached 8.
The solution was boiled for 15 min and allowed to stand overnight. The mother liquor was then decanted and the precipitate was washed several times with distilled water till it is completely free of nitrate ions which was confirmed by brown ring test. The precipitate was filtered and dried overnight at 383 K for 16 h. The hydroxide obtained was sieved to get particles of 75-100 μm mesh size and immersed in (1:1) H2SO4 solution (2 mL/g) and subjected to stirring for 4 h. Excess water was evaporated and the resulting sample was oven dried at 383 K for 16 h, calcined at 823 K for 5 h and stored in vacuum desiccator.
General procedure for Microwave synthesis of 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol (2)
A mixture of methylbenzoate (1 mmol) and hydrazine hydrate (1 mmol) in 20 mL ethanol was irradiated in microwave at 700 W in a specially designed Teflon vessel containing lead acetate, until all the starting material was consumed (1-2 min, as monitored by TLC). To the above mixture (0.006 mmol) of KOH, CS2 (1 mmol) was added and further irradiated at 700 W for 1 min. Finally, hydrazine hydrate (2 mmol) was added drop wise to the above mixture and continued the irradiation at 700 W until a white solid appeared at the bottom (2-3 min). The lead acetate worked as a trap for H2S that was evolved during reaction. The solid obtained was dissolved in water (15-20 mL) and acidified with conc. HCl. The separated solid was filtered, dried and recrystallized to obtain pure 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol. Yield 78%, m.p. 232-234 °C; IR (KBr) γ/cm−1: 3310.07 (NH2 stretch), 3071.36 (aromatic CH stretch), 1472.38 (tautomeric C = S). 1H NMR: (400 MHz, DMSO-d6). δ:7.6-7.5 (m, 2H, ArH), 7.34-7.2 (m, 3H, ArH), 5.14 (s, 2H, NH2).
General procedure for the synthesis of 6-substituted-3-phenyl-(1,2,4)-triazolo(3,4-b)(1,3,4-thiadiazole (4a-4 h) by using SCe
To a mixture of 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol (1 mmol) and (3a-h) (1 mmol) in DMF (10 mL), SCe (20 mol%) and POCl3 (0.1 mmol) were added. The reaction mixture was refluxed for 10 h. Completion of the reaction was monitored by TLC and the catalyst was filtered and washed with water. Solvent was removed under reduced pressure and crushed ice was added to the concentrated mass. The pH of reaction mixture was adjusted to 8.0 using K2CO3 and KOH. The solid obtained was separated by filtration, washed with excess water, dried and recrystallized using appropriate solvent.
General procedure for the synthesis of 2-hydroxy-3,5-diiodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5a) and 2-hydroxy-5-iodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5b)
To 3a (1 eq) in DMF, EDC (1.1 eq) and HOBt (1.1 eq) was added and stirred at room temperature for 30 min. It was followed by the addition of amine (2) and stirred for 2 h. After completion of the reaction, it was diluted with water and the obtained solid was filtered and re-crystallized in appropriate solvent.
2,4-Diiodo-6-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)phenol (4a, DTP)
Yellow colored solid; 1H NMR (400 MHz, DMSO-d6) 8.37-8.35 (d, 2H), 8.26 (s, 1H), 7.85 (s, 1H), 7.69-7.63 (m, 2H), 7.54-7.52 (d, 1H), 4.92 (s, 1H); 13C NMR (DMSO-d6); 165.53, 154.53, 149.29, 148.83, 140.98, 137.51, 133.83, 132.45, 129.11, 128.64, 123.10, 122.44, 120.72, 96.18, 85.11; HRMS Calcd 568.840; Found: 568.840 (M + Na)+; Anal. Calcd for C15H8I2N4OS: C, 32.99; H, 1.48; N, 10.26; Found: C, 33.00; H, 1.49; N, 10.28.
6-(4-(1H-Imidazol-1-yl)phenyl)-3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazole (4b)
Pale yellow colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.46-8.44 (d, 2H), 7.81-7.77 (m, 2H), 7.53-7.49 (m, 3H), 7.39-7.34 (m, 3H), 7.27-7.24 (m, 2H); 13C NMR (DMSO-d6); 161.55, 149.29, 148.53, 140.98, 137.18, 137.11, 133.83, 132.48, 131.97, 129.11, 128.64, 128.18, 123.10, 122.43, 120.27; LCMS (MM:ES + APCI) 345.2 (M + H)+. Anal. Calcd for C18H12N6S: C, 62.77; H, 3.51; N, 24.40; Found: C, 62.79; H, 3.53; N, 24.43.
4-Iodo-2-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6-yl)phenol (4c, ITP)
Yellow colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.44-8.42 (d, 2H), 8.08-8.06 (d, 2H), 8.02-8.00 (m, 1H), 7.95-7.91 (m, 1H), 7.71 (s, 1H), 7.16-7.14 (d, 1H), 4.92 (s, 1H); 13C NMR (DMSO-d6) δ: 164.19, 159.73, 152.02, 147.46, 138.26, 133.27, 131.64, 129.40, 127.70, 124.93, 120.48, 119.82, 118.66, 88.23; HRMS Calcd 442.943; Found: 442.943 (M + Na)+; Anal. Calcd for C15H9IN4OS: C, 42.87; H, 2.16; N, 13.33; Found: C, 42.89; H, 2.17; N, 13.35.
6-(((R)-Tetrahydro-2H-pyran-2-yl)(phenyl)methyl)-3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazole (4d)
White colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.25-8.16 (d, 2H), 8.06 (m, 1H), 7.78-7.76 (m, 1H), 7.62-7.60 (m, 1H), 7.27-7.15 (m, 4H), 4.58-4.53 (m, 2H), 3.88-3.84 (m, 2H), 1.78-1.73 (m, 4H), 1.50-1.45 (m, 2H); 13C NMR (DMSO-d6) δ: 164.56, 149.30, 143.93, 141.04, 137.49, 132.82, 132.41, 130.23, 129.10, 128.10, 120.70, 80.11, 71.09, 43.59, 30.41, 30.33, 21.48; LCMS (MM:ES + APCI) 377.2 (M + H)+; Anal. Calcd for C21H20N4OS: C, 67.00; H, 5.35; N, 14.88; Found: C, 67.02; H, 5.37; N, 14.90.
2-(3-Phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)-1-p-tolylethanone (4e)
White colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.43-8.41 (m, 2H), 8.03-7.99 (m, 3H), 7.92 (m, 1H), 7.69 (m, 1H), 7.40-7.38 (m, 2H), 4.1 (s, 2H), 2.42 (m, 3H); 13C NMR (DMSO-d6) δ:192.83, 164.18, 159.42, 151.99, 146.87, 137.47, 132.28, 130.26, 125.66, 123.38, 121.01, 120.89, 48.13, 21.13; HRMS Calcd 357.078; Found: 357.078 (M + Na)+. Anal. Calcd for C18H14N4OS: C, 64.65; H, 4.22; N, 16.75; Found: C, 64.67; H, 4.25; N, 16.77.
6-(3-4-Dimethoxybenzyl)-3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazole (4f)
Yellow colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.2 (d, 2H), 7.6-7.4 (m, 3H), 7.0 (s, 1H), 6.9 (d, 2H), 4.4 (s, 2H), 3.8 (s, 6H); LCMS (MM:ES + APCI) 353.2 (M + H)+; Anal.Calcd for C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90; Found: C, 61.39; H 4.59; N, 15.93.
3-(3-Phenyl--[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6-yl-)phenol (4 g)
White colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.32-8.31 (m, 2H), 8.13 (s, 1H), 7.94-7.87 (m, 3H), 7.65-7.59 (m, 2H), 7.46 (m, 1H), 4.91 (s, 1H); LCMS (MM:ES + APCI) 295.2 (M + H)+; Anal. Calcd for C15H10N4OS: C, 61.21; H, 3.42; N, 19.04; Found: C, 61.23; H, 3.44; N, 19.07.
3-Phenyl-6-styryl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazole (4 h)
White colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 8.25-8.22(d, 2H), 7.92-7.87 (m, 2H), 7.73-7.55 (m, 4H), 7.32-7.26 (m, 2H), 6.45-6.42 (m, 2H); 13C NMR (DMSO-d6) δ: 164.84, 159.58, 153.39, 145.42, 139.89, 131.08, 131.04, 130.53, 130.42, 130.31, 129.09, 125.84, 125.47, 118.08, 116.18, 115.90; HRMS Calcd 327.067; Found: 327.067 (M + Na)+; Anal. Calcd for C17H12N4S: C, 67.O8; H, 3.97; N, 18.41; Found: C, 67.09; H, 3.99; N, 18.44.
2-Hydroxy-3,5-diiodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5a, HTP)
Pale yellow colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 14.64 (s, NH), 12.30 (s, NH), 8.48 (s, 1H), 8.40 (s, 1H), 8.24-8.15 (m, 3H), 7.81-7.78 (m, 2H), 4.73 (s, 1H); 13C NMR (DMSO-d6) δ:181.47, 173.23, 153.47, 147.94, 145.12, 136.38, 134.36, 131.13, 129.09, 128.78, 128.21, 126.02, 125.58, 90.79, 72.33; HRMS Calcd 586.851; Found: 586.851 (M + Na)+; Anal.Calcd for C15H10I2N4O2S: C, 31.94; H, 1.79; N, 9.93; Found: C, 31.96; H, 1.81; N, 9.93.
2-Hydroxy-5-iodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5b)
Pale yellow colored solid; 1H NMR (400 MHz, DMSO-d6) δ: 12.5 (s, NH), 8.5 (s, 1H), 8.4 (m, 1H), 8.1 (m,1H), 7.8 (m, 3H), 7.6 (m,1H), 4.6 (s, 1H); LCMS (MM:ES + APCI) 438.4 (M + H)+; Anal. Calcd for C15H11IN4O2S: C, 41.11; H, 2.53; N, 12.78; Found: C, 41.12; H, 2.56; N, 12.80.
Spectral data of the compounds are presented in Additional file 1: Figure S1.
Colorimetric heparanase assay
The assay, carried out in 96 well microplates, measures the appearance of the disaccharide product of heparanase-catalyzed fondaparinux cleavage, colorimetrically using the tetrazolium salt WST-1 [31]. Briefly, assay solutions (100 μL) are composed of 40 mM sodium acetate buffer (pH 5.0) and 100 mM fondaparinux (Arixtra) with or without increasing concentrations of inhibitor. Recombinant heparanase was added to a final concentration of 140 pM, to start the assay. The plates are incubated at 37 °C for 18 h and the reaction is stopped by the addition of 100 μL solution containing 1.69 mM 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate (WST-1) in 0.1 M NaOH. The plates are developed at 60 °C for 60 min, and the absorbance is measured at 584 nm. In each plate, a standard curve constructed with D-galactose as the reducing sugar standard is prepared in the same buffer and volume over the range of 2–100 μM [31].
ECM degradation heparanase assay
The semi-quantitative heparanase assay was performed as described previously [32, 33]. Briefly, metabolically sulfate [35S] labeled ECM deposited by cultured endothelial cells and coating the surface of 35 mm tissue culture dishes [33], is incubated (3 h, 37 °C, pH 6.0, 1 mL final volume) with recombinant human heparanase (200 ng/mL) in the absence and presence of candidate small molecules. The ECM was also incubated (24 h, 37 °C, pH 6.0) with cell lysates (200 μg protein/dish) prepared by 3 cycles of freeze and thaw in reaction buffer, as described [32]. To evaluate the occurrence of proteoglycan degradation, the incubation medium is collected and applied for gel filtration on Sepharose 6B columns (0.9 × 30 cm). Fractions (0.2 mL) are eluted with PBS and counted for radioactivity. The excluded volume (Vo) is marked by blue dextran and the total included volume (Vt) by phenol red. Degradation fragments of HS side chains are eluted from Sepharose 6B at 0.5 < Kav < 0.8 (fractions 12-25) [32].
In vitro cytotoxicity assay
The antiproliferative effect of the compounds against LLC (Lewis lung carcinoma) and HepG2 (hepatocellular carcinoma) cells was determined by the MTT dye uptake method as described previously [34–36]. Briefly, cells (2.5 × 104/mL) were incubated in triplicate in a 96-well plate, in the presence of varying concentrations of test compounds at a volume of 0.2 mL, for different time intervals at 37 °C. Thereafter, 20 μL MTT solution (5 mg/mL in PBS) was added to each well. After 2 h incubation at 37 °C, 0.1 mL lysis buffer (20% SDS, 50% dimethylformamide) was added and incubated for 1 h at 37 °C, and the optical density (OD) at 570 nm was measured using a plate reader.
In vitro trans-well invasion/migration assay
Invasion of cells (LLC, HepG2) across a Matrigel™ coated membrane or migration through control uncoated inserts was assessed using 24-well plates (BD Biosciences, 8 μm pore size, insert size: 6.4 mm) according to the manufacturer’s protocol and as described earlier [37–39]. Briefly, single cell suspensions (1 × 106 cells/mL) were prepared by detaching and resuspending the cells in DMEM containing 0.1% BSA. Before adding the cells, the chambers were rehydrated for 2 h in an incubator at 37 °C. The lower chambers were filled with 600 μL DMEM containing chemo-attractant (10% FBS). After seeding the cells (2 × 105 in 200 μL of serum-free medium) into the upper chamber of triplicate wells with or without increasing concentrations of compounds, the chambers were incubated for 24 h (LLC) and 48 h (HepG2) at 37 °C. The non-invaded cells were removed from the upper surface of the membrane by scrubbing and cells that migrated through the filter were fixed, stained with Diff Quick solution, counted by examination of at least five microscopic fields and photographed.
Results
Chemical synthesis and characterization
In recent years, solid acid catalysts have gained considerable attention due to their high efficiency, eco-friendly, longer catalyst life, negligible equipment corrosion and their reusability. In present work we report the synthesis of novel 1,2,4-triazolo-1,3,4-thiadiazoles bearing compounds via sulfated ceria mediated cycalization reaction [40–42]. Initially we synthesized the sulphated ceria (SCe) catalyst as reported previously [43]. The powdered X-ray diffraction (PXRD), Burner- Ememett-Teller (BET) and Scanning Electron microscope patterns of SCe matched with the standard material.
The experimental strategy for the synthesis of starting material 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol (2) was achieved by Microwave method as reported recently (Scheme 1 , i) [36]. In order to synthesize the novel 1,2,4-triazolo-1,3,4-thiadiazoles, we focused on the efficiency of SCe in cyclisation reaction. To optimise the reaction conditions, we attempted reaction in the combination of 2 and 3-oxo-3-(p-tolyl)propanoic acid (3e) as a model reaction in different concentrations of SCe and the results are summarised in Additional file 1: Table S1. The ideal system for the cyclization was found to be 20 mol% of SCe in DMF (Additional file 1: Table S1, entry 8). We also observed incomplete conversions, when SCe was lower than 20 mol%, despite of longer reaction time. From the above reaction, we examined the generality of method by synthesizing series of 1,2,4-triazolo-1,3,4-thiadiazoles molecules (Scheme 1, ii).
Scheme 1.
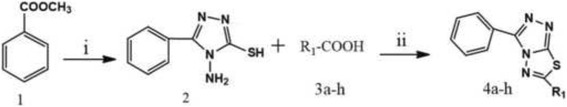
Schematic representation of new heparanase inhibitors used in this study. i) hydrazine hydrate, ethanol, MWI; CS2 and KOH, 5-6 min at 700 watt; ii) SCe (20 mol%), DMF, 10 h
Influence of SCe on cyclization
The modification of SCe with anions such as sulphate ions forms a super acidic catalyst which effectively catalyses the cyclization. Majority of reactions completed within 10 h and undissolved SCe was separated by simple filtration and finally furnished the product in good yield (Additional file 1: Table S1).
Plausible mechanism
The First step involves the protonation of acid followed by dehydration and simultaneous attack of nitrogen lone pair to the electron deficient acylium ion to form an intermediate. In the second step, the intermediate undergoes neighboring group participation with nucleophilic sulphur, which leads to the formation of C-S bond by the elimination of water molecule (Scheme 2). Finally, deprotonation results in the formation of the title products (4a-h).
Scheme 2.
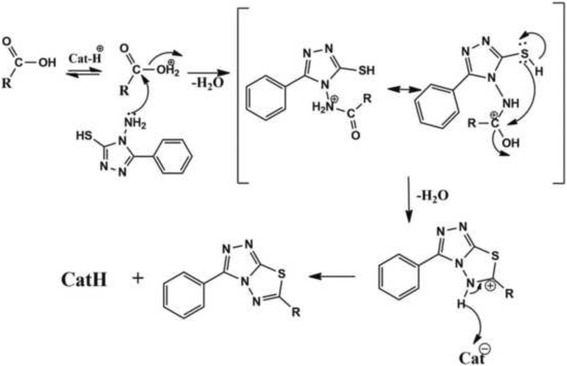
Plausible mechanism of cyclization and synthesis of title compounds
Re-usability of acid catalyst system
Experiment was performed to study the recyclability of the SCe system employing 2 with 3e to yield compound 4e (Scheme 1). After each run, catalyst was removed by filtration from the reaction mixture, washed thoroughly with acetone, dried and activated at 823 K and taken for next cycle. We observed significant reduction in the yield of the product after second run (Additional file 1: Table S2). It is important to note that this system is recyclable twice with the isolated yields above 70%.
Further, we synthesized the amide derivatives of 2 with corresponding mono and di iodo salicylic acid (3a and 3c) via HOBt/EDC amide formation reactions (Scheme 3) which resulted in the products 2-hydroxy-3,5-diiodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5a) and 2-hydroxy-5-iodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (5b). The compounds obtained were characterized by 1H NMR, 13C NMR, and mass spectral analysis (Additional file 1: Figure S1 – Spectral data). Detailed chemical characterization of the newly synthesized compounds is provided in the ‘methods’ section.
Scheme 3.
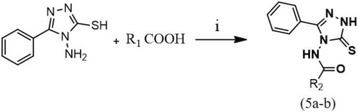
Synthetic scheme for the preparation of N-amino-triazole-amides. i) HOBt/EDC, DMF, RT, 2 h. R1 = 3a, 3c
In vitro screening of the small molecule library for inhibition of the catalytic activity of human heparanase
Initially we screened the entire library of small molecules with diverse structures for their in vitro inhibitory activity against recombinant human heparanase at different concentrations up to 20 μg/mL. We used a 96-well based colorimetric assay that measures the ability of recombinant heparanase to degrade fondaparinux (heparin derived pentasaccharide) in solution [31]. The assay measures the appearance of a disaccharide product of fondaparinux cleavage, using the tetrazolium salt WST-1 [31]. Compounds bearing triazolo-thiadiazole backbone displayed significant inhibitory activity, 2,4-Diiodo-6-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)phenol (DTP) being the lead and consistently active structure followed by 2-hydroxy-3,5-diiodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide (HTP) and 4-iodo-2-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6-yl)phenol (ITP) (Fig. 1a).
Fig. 1.
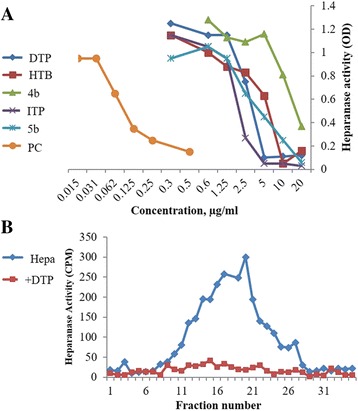
a Screening of compounds for inhibition of heparanase enzymatic activity applying the Fondaparinux heparanase assay. PC, positive control = N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide [22]. b Lead molecules which exhibited inhibitory activity against human heparanase were validated using a semi-quantitative assay that measures release of radioactive heparan sulfate fragments from an insoluble extracellular matrix as described in ‘Methods’ section. Briefly, sulfate [35S] labeled ECM was incubated (6 h, 37 °C, pH 6.0) with recombinant human heparanase (200 ng/mL) in the absence and presence of 10 μg/mL of the test compounds. Sulfate labeled material released into the incubation medium was subjected to gel filtration on Sepharose 6B. Compound DTP effectively inhibited the cleavage and release of heparan sulfate degradation fragments
In order to better resemble the in vivo situation, we applied as substrate metabolically sulfate [Na2 35SO4] labeled extracellular matrix (ECM) deposited by cultured endothelial cells [32]. This naturally produced substrate closely resembles the subendothelial basement membrane in its composition, biological function and barrier properties. Years of experience revealed that compounds that effectively inhibit the enzyme in this assay are also effective in preclinical animal models [20, 44, 45]. This semi-quantitative assay measures release of radioactive heparan sulfate (HS) degradation fragments from an insoluble extracellular matrix (ECM) that is firmly bound to a culture dish [32, 33]. Briefly, the ECM substrate is incubated with recombinant human heparanase in the absence and presence of candidate small molecules. The incubation medium is collected and subjected to gel filtration on Sepharose 6B. Degradation fragments of heparan sulfate side chains are eluted at 0.5 < Kav < 0.8, whereas nearly intact HSPG is eluted next to the void volume [32]. As demonstrated in Fig. 1b, compound DTP (10 μg/mL) completely inhibited the release of heparan sulfate degradation fragments. The other structural analogs were less effective (not shown). Thus, we have identified heparanase-inhibiting lead compound from a random screen of bioactive compounds.
Heparanase activity in various hepatocellular and lung carcinoma cell lines
Heparanase expression (RT-PCR) (Fig. 2a) and enzymatic activity (Fig. 2b) were examined in various hepatocellular carcinoma (human HepG2, Hep3B) and lung carcinoma (human HCC827, mouse LLC) cell lines. A relatively low expression level and enzymatic activity were noted in HepG2 cells as compared to the other cell lines which exhibited moderate-high heparanase enzymatic activity (Fig. 2b). HepG2 human hepatocellular carcinoma and LLC mouse Lewis lung carcinoma cells lines were selected for further experimentation, representing human and mouse cells expressing low (HepG2) and moderate-high (LLC) enzymatic activity, respectively.
Fig. 2.
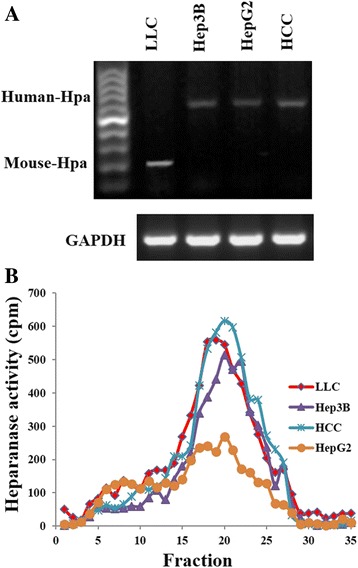
Heparanase expression and activity in various hepatocellular and lung carcinoma cell lines. Mouse Lewis lung carcinoma (LLC), human lung carcinoma (HCC827 = HCC), and human hepatocellular carcinoma (HepG2, Hep3B) cells maintained in culture were subjected to RT-PCR (a) and heparanase activity (b) assays, as described in ‘Methods’
DTP suppresses the proliferation of LLC and HepG2 cells
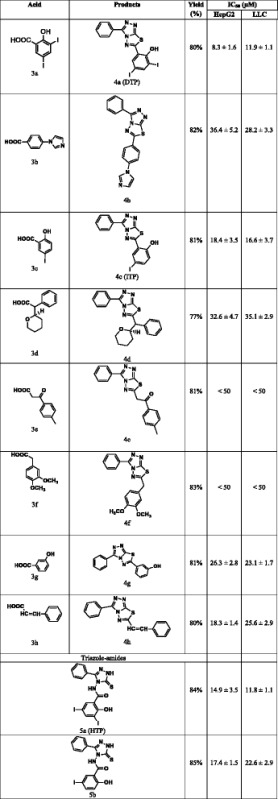
Given the overexpression of heparanase in hepatocellular and lung carcinoma cancer cell lines, we next analyzed the effect of triazolo-thiadiazoles on LLC (Lewis Lung carcinoma) and HepG2 (hepatocellular carcinoma) cell proliferation using the MTT assay [46–48]. Paclitaxel and DMSO were used as reference drug and vehicle control, respectively. Among the tested triazolo-thiadiazoles, DTP was found to exert an antiproliferative effect with IC50 value of 11.9 and 8.3 μM against LLC and HepG2, respectively (Table 1). Thus, structure activity relationship of the lead anticancer agent revealed that phenolic and iodine substituents on the core triazolo-thiadiazole nucleus were found to increase the inhibitory activity towards the proliferation of cancer cells. Notably, the exo-conjugation to the triazolo-thiadiazole core structure also enhances the cytotoxicity. The hydrophobic substituents on the core structure were found to be ineffective against proliferation of cancer cells.
Table 1.
Characterization and anti-proliferative activity of the newly synthesized small molecules that are used for the in vitro heparanase enzyme inhibition studies
DTP inhibits migration and invasion of LLC cells
The involvement of heparanase in cancer metastasis is clearly demonstrated in various types of cancer [9, 14, 32]. We investigated the effect of DTP on LLC and HepG2 cell migration and invasion applying trans-well filters (8 μM pore size) that were either uncoated or coated with Matrigel, respectively. LLC (Fig. 3) and HepG2 (Fig. 4) cells migrated through uncoated filters and invaded through Matrigel in response to stimulation with FBS. DTP significantly suppressed cell migration (Figs. 3a and 4a) and invasion (Figs. 3b and 4b) in a dose dependent manner, yielding nearly 50% inhibition at 5 μM. This effect is likely attributed to inhibition of heparanase enzymatic activity by DTP. Heparin was used as positive control.
Fig. 3.
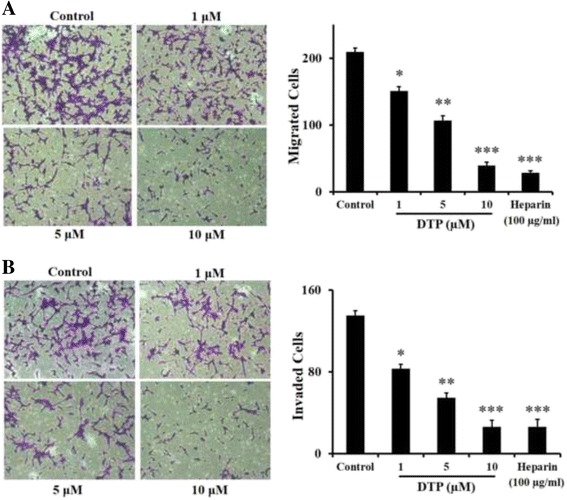
Effect of DTP on LLC cell migration and Invasion. LLC cells were plated on BD BioCoat™ chambers (BD Biosciences) and cell migration (without Matrigel coat) (a) and invasion (with Matrigel coat) (b) were measured as described in ‘Methods’. The effect of lead compound DTP (1–10 μM) or heparin (100 μg/mL) on cell migration and invasion is demonstrated by representative photomicrographs (magnification: ×10) and the respective bar graphs. Data are represented as mean ± S.E. *P < 0.1; **P < 0.05. ***P < 0.01
Fig. 4.
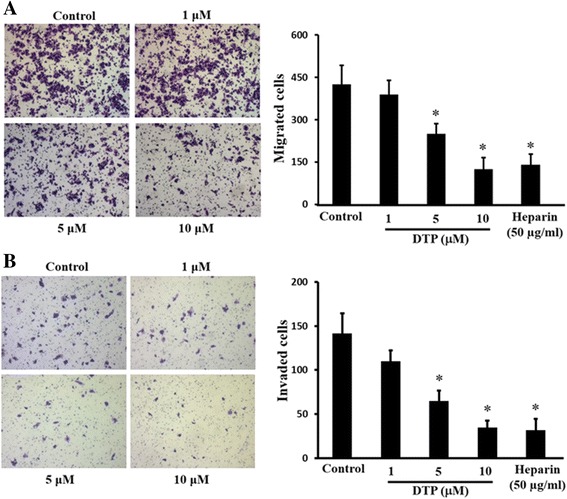
Effect of DTP on HepG2 cell migration and Invasion. HepG2 cells were plated on BD BioCoat™ chambers (BD Biosciences) and cell migration (without Matrigel coat) (a) and invasion (with Matrigel coat) (b) were measured as described in ‘Methods’. The effect of lead compound DTP (1–10 μM) or heparin (100 μg/mL) on cell migration and invasion is demonstrated by representative photomicrographs (magnification: ×5) and the respective bar graphs. Data are represented as mean ± S.E. *P < 0.05
Rationalizing SAR trends via protein-ligand interactions
In order to perform virtual screening, a recently published X-ray crystal structure for human heparanase was obtained from the Protein Data Bank (PDB:5E97; Glycoside Hydrolase ligand structures 1, 1.63 Å resolution) [49]. The structure was loaded into MOE [50] and corrected using the Structure Preparation tool before running Protonate 3D. The Site Finder tool identified the active site containing Glu-343 and Glu-225 that were identified as the catalytic nucleophile and acid-base of Heparanase [45, 49]. Compound structures were loaded into MOE and energy minimised before carrying out rigid receptor docking (triangle matcher, London dG Forcefield refinement, GBVI/WSA dG rescoring).
The 52 docked poses that included the three active compounds DTP, HTP, and ITP did not appear to explain the experimentally observed trend in SAR. However, docking results revealed a similar interaction pattern between active compounds ITP and DTP, with poses that interact favourably with both Asn-224 and Asp-62 due to the triazolo-thiadiazole backbone (Fig. 5). For compound HTP, this interaction profile was found to be slightly less favourable, interacting instead with Asn-224 and the active site acid-base Glu-343.
Fig. 5.
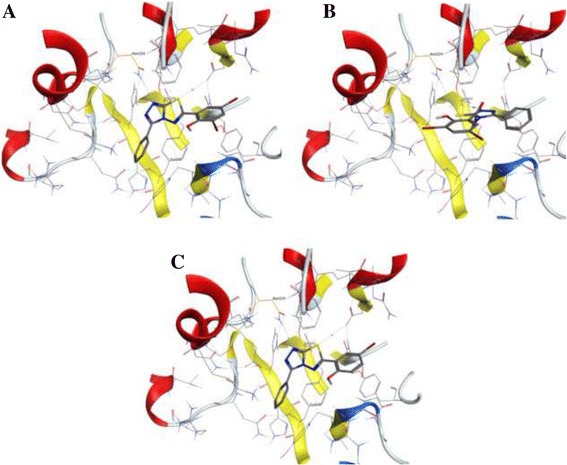
Selected docked poses for active compounds DTP, HTP and ITP (a, b and c, respectively), showing similar active site interaction modes. DTP and ITP are shown to interact with both Asn-224 and Asp-62 via the triazolo-thiadiazole backbone, and HTP is shown to interact with Asn-224 and the active site acid-base Glu-343
Although these compounds do not appear to be more favourable than the other docked compunds, the presence of iodine substituents found on all hit compounds may preferentially lower the phenols’ pKA sufficiently to allow for deprotonation of the ligands in protein environment.
Discussion
Human heparanase is an endoglucuronidase that cleaves heparan sulfate chains thereby regulating multiple biological activities that together enhance tumor growth, metastasis and angiogenesis [7–10, 14, 32]. Heparanase is expressed by most types of cancer and has emerged as a valid target for anti-cancer therapy [8, 15]. Heparanase represents a druggable target because: (i) there is only a single enzymatically active heparanase expressed in humans, (ii) the enzyme is present in low levels in normal tissues but dramatically elevated in tumors where it is associated with poor prognosis and reduced postoperative survival time, and (iii) heparanase deficient mice appear normal [51]. Thus, properly designed heparanase inhibitors will likely have few, if any, negative side effects. Development of heparanase inhibitors has focused predominantly on carbohydrate-based compounds with heparin-like properties [8, 15, 44]. These compounds bind to the heparin-binding domains that flank the enzyme active site of heparanase thereby inhibiting cleavage of heparan sulfate. Four different heparin mimics are currently in clinical trials in human cancer patients. However all of these mimics have the disadvantage that they are not specific for heparanase and likely interact with different heparin-binding proteins with unknown consequences and off target effects [8, 15]. Therefore, even if they prove efficacious in patients it will be impossible to attribute their effect solely to heparanase inhibition. In addition three of the four mimics are heterogeneous in their structure adding further to their uncertainty as viable drugs for use in humans [8]. A number of heparanase-inhibiting small molecules were reported [8, 16, 22], but none entered clinical testing.
Heparanase expressed in cancer cells and cells of the tumor microenvironment provides a most appropriate therapeutic molecular target and could serve a decisive role in cancer regime. In addition to remodeling of ECM, human heparanase regulates multiple signaling cascades involved in tumor cell survival, angiogenesis and metastasis [7, 8, 14, 15, 44, 52]. The positive correlation of heparanase with progression of malignancies makes it an attractive target in the treatment of cancer. It is hoped that our identification of a lead molecule and the recently resolved crystal structure of the heparanase protein [49] will accelerate rational design of heparanase-inhibiting small molecules endowed with considerably improved binding affinity, specificity, pharmacokinetics and efficacy in xenograft cancer models. Selected molecules exerting little or no side effects will then be examined for oral availability and anti cancer effect in combination with currently available treatments, applying patient derived xenograft models and, at a later stage, animal models of other diseases shown to be causally related to heparanase [53–58].
Conclusions
In a search for small molecule inhibitors that can interfere with the catalytic activity of human heparanase, we report the synthesis and biological evaluation of a library of synthetic small molecules and identification of triazolo-thiadiazole derivative as a potent inhibitor of human heparanase. The identified lead structure displayed antiproliferative activity and suppressed the migration and invasion of cancer cells in correlation with inhibition of heparanase enzymatic activity. Further development of this novel class of heparanase inhibitors and optimization to maximize their affinity, pharmacokinetics and oral availability will provide a unique opportunity for development of innovative anti-cancer therapeutics. Moreover, because heparanase helps drive the progression of other diseases (e.g., diabetes, diabetic nephropathy, arthritis, colitis, sepsis, atherosclerosis) [53–58], these drugs hold potential to impact public health.
Acknowledgements
CPB thanks S. Jayasree for her guidance in synthesizing the catalyst. IV thanks S. Feld for excellent technical assistance.
Funding
This research was supported by the Israel Science Foundation (ISF) and the University Grants Commission (UGC) India, awarded to IV and KSR within the ISF-UGC joint research program framework (grant No. 2277/15). The research was also supported by University Grants Commission (41-257-2012-SR), Vision Group Science and Technology, Department of Science and Technology (NO. SR/FT/LS-142/2012) to Basappa. KSR thanks DST Indo-Korea [INT/Indo-Korea/122/2011-12] and Institution of Excellence, University of Mysore for financial support. CDM thanks the University of Mysore for Department of Science and Technology-Promotion of University Research and Scientific Excellence (DST-PURSE) Research Associate fellowship. IV is a research Professor of the Israel Cancer Research Fund (ICRF). The funding agencies did not participate in the design of the study and collection, analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyses during this study are included in this article and its Additional file 1.
Authors’ contributions
CPB, CDM, SR, DJM, JEF, UB and B carried out the chemistry, biological and molecular studies. CDM, AB, IV, B and KSR interpreted the results and assisted in manuscript preparation. AB, IV, B and KSR provided the tools, reagents for research and prepared the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study does not involve animal studies and human data.
Consent for publication
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BSA
Bovine serum albumin
- DMEM
Dulbecco’s Modified Eagle Medium
- DTP
2,4-Diiodo-6-(3-phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)phenol
- ECM
Extracellular matrix
- HGF
Hepatocyte growth factor
- HS
Heparan Sulfate
- HSPG
heparan sulphate proteolglycan
- HTP
2-Hydroxy-3,5-diiodo-N-(3-phenyl-5-thioxo-1H-1,2,4-triazol-4(5H)-yl)benzamide
- ITP
4-Iodo-2-(3-phenyl-[1, 2, 4]triazolo[3,4b][1, 3, 4]thiadiazol-6-yl)phenol.
- LLC
Lewis lung carcinoma
- MOE
Molecular operating environment
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NMR
Nuclear magnetic resonance
- OD
Optical density
- SAR
Structural activity relationship
- SCe
sulphated ceria
- TLC
Thin layer chromatography
- VEGF
Vascular endothelial growth factor
Additional file
Optimisation of mol% of SCe catalyst, and selection of medium for cyclization reaction. To optimize the reaction conditions for the synthesis of novel 1,2,4-triazolo-1,3,4-thiadiazoles, the reaction was performed in combination of 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol and 3-oxo-3-(p-tolyl)propanoic acid as a model reaction in different concentrations of SCe and solvent. The optimal system for cyclization was 20 mol% of SCe in DMF. Table S2. Evaluation of the reuse of SCe for cyclization reaction. The recyclability of the SCe system was evaluated by employing 4-amino-5-phenyl-4 h-1,2,4-triazole-3-thiol with 3-oxo-3-(p-tolyl)propanoic acid to yield 2-(3-Phenyl-[1, 2, 4]triazolo[3,4-b][1, 3, 4]thiadiazol-6yl)-1-p-tolylethanone. The catalyst was removed by filtration after each run and thoroughly washed with acetone, dried and activated at 823 K and taken for the next cycle. There was a significant reduction in the yield of the product after the second run using SCe. Figure S1. Spectral data. Scanned copy of 1H NMR, 13C NMR, and mass spectra of the indicated compounds. (DOCX 4202 kb)
Contributor Information
C. P. Baburajeev, Email: cpbrajeevnambiar@yahoo.co.in
Chakrabhavi Dhananjaya Mohan, Email: cd.mohan@yahoo.com.
Shobith Rangappa, Email: shobithrangappa@gmail.com.
Daniel J. Mason, Email: djm234@cam.ac.uk
Julian E. Fuchs, Email: jf544@cam.ac.uk
Andreas Bender, Email: ab454@cam.ac.uk.
Uri Barash, Email: ubarash@yahoo.com.
Israel Vlodavsky, Email: vlodavsk@mail.huji.ac.il.
Basappa, Email: salundibasappa@gmail.com.
Kanchugarakoppal S. Rangappa, Email: rangappaks@yahoo.com
References
- 1.Lu P, Takai K, Weaver VM, Werb Z. Extracellular Matrix Degradation and Remodeling in Development and Disease. Cold Spring Harb Perspect Biol. 2011; 3(12):10.1101/cshperspect.a005058 a005058. [DOI] [PMC free article] [PubMed]
- 2.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Ann Review Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15(5):1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Brit J Pharmacol. 2007;151(1):1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JP, Vlodavsky I. Heparin, heparan sulfate and heparanase in inflammatory reactions. Thromb Haemost. 2009;102(5):823–828. doi: 10.1160/TH09-02-0091. [DOI] [PubMed] [Google Scholar]
- 6.Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32(5):223–227. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Arvatz G, Shafat I, Levy-Adam F, Ilan N, Vlodavsky I. The heparanase system and tumor metastasis: is heparanase the seed and soil? Cancer Metastasis Rev. 2011;30(2):253–268. doi: 10.1007/s10555-011-9288-x. [DOI] [PubMed] [Google Scholar]
- 8.Rivara S, Milazzo FM, Giannini G. Heparanase: a rainbow pharmacological target associated to multiple pathologies including rare diseases. Fut Med Chem. 2016;8(6):647–680. doi: 10.4155/fmc-2016-0012. [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I, Beckhove P, Lerner I, Pisano C, Meirovitz A, Ilan N, Elkin M. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 2012;5(2):115–132. doi: 10.1007/s12307-011-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vreys V, David G. Mammalian heparanase: what is the message? J Cellular Mol Med. 2007;11(3):427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66(3):1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 12.Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J Biol Chem. 2011;286(8):6490–6499. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283(47):32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem & Cell Biol. 2006;38(12):2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Pisano C, Vlodavsky I, Ilan N, Zunino F. The potential of heparanase as a therapeutic target in cancer. Biochem Pharmacol. 2014;89(1):12–19. doi: 10.1016/j.bcp.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simizu S, Ishida K, Osada H. Heparanase as a molecular target of cancer chemotherapy. Cancer Sci. 2004;95(7):553–558. doi: 10.1111/j.1349-7006.2004.tb02485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khasraw M, Pavlakis N, McCowatt S, Underhill C, Begbie S, de Souza P, Boyce A, Parnis F, Lim V, Harvie R, et al. Multicentre phase I/II study of PI-88, a heparanase inhibitor in combination with docetaxel in patients with metastatic castrate-resistant prostate cancer. Annals Oncol. 2010;21(6):1302–1307. doi: 10.1093/annonc/mdp524. [DOI] [PubMed] [Google Scholar]
- 18.https://clinicaltrials.gov/ct2/show/record/NCT01402908.
- 19.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, Brandt R, Ferro V, Bytheway I. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104(4):635–642. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, Penco S, Pisano C, Carminati P, Tortoreto M, et al. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res. 2011;17(6):1382–1393. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, Zhao G, Smith S, Galcheva-Gargova Z, Karlgren J, et al. M402, a Novel Heparan Sulfate Mimetic, Targets Multiple Pathways Implicated in Tumor Progression and Metastasis. PLoS One. 2011;6(6):e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y-J, Miao H-Q, Pan W, Navarro EC, Tonra JR, Mitelman S, Camara MM, Deevi DS, Kiselyov AS, Kussie P, et al. N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide derivatives as small molecule heparanase inhibitors. Bioorganic & Med Chem Lett. 2006;16(2):404–408. doi: 10.1016/j.bmcl.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 23.Keerthy HK, Mohan CD, Sivaraman Siveen K, Fuchs JE, Rangappa S, Sundaram MS, Li F, Girish KS, Sethi G. Basappa et al. Novel synthetic biscoumarins target tumor necrosis factor-alpha in hepatocellular carcinoma in vitro and in vivo. J Biol Chem. 2014;289(46):31879–31890. doi: 10.1074/jbc.M114.593855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharathkumar H, Mohan CD, Ananda H, Fuchs JE, Li F, Rangappa S, Surender M, Bulusu KC, Girish KS, Sethi G, et al. Microwave-assisted synthesis, characterization and cytotoxic studies of novel estrogen receptor alpha ligands towards human breast cancer cells. Bioorganic & Med Chem Lett. 2015;25(8):1804–1807. doi: 10.1016/j.bmcl.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 25.Rakesh KS, Jagadish S, Vinayaka AC, Hemshekhar M, Paul M, Thushara RM, Sundaram MS, Swaroop TR, Mohan CD, Basappa, et al. A new ibuprofen derivative inhibits platelet aggregation and ROS mediated platelet apoptosis. PloS one. 2014;9(9):107182. doi: 10.1371/journal.pone.0107182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roopashree R, Mohan CD, Swaroop TR, Jagadish S, Raghava B, Balaji KS, Jayarama S. Basappa, Rangappa KS. Novel synthetic bisbenzimidazole that targets angiogenesis in Ehrlich ascites carcinoma bearing mice. Bioorganic & Med Chem Lett. 2015;25(12):2589–2593. doi: 10.1016/j.bmcl.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Keerthy HK, Garg M, Mohan CD, Madan V, Kanojia D, Shobith R, Nanjundaswamy S, Mason DJ, Bender A, Basappa, et al. Synthesis and characterization of novel 2-amino-chromene-nitriles that target Bcl-2 in acute myeloid leukemia cell lines. PLoS One. 2014;9(9):e107118. doi: 10.1371/journal.pone.0107118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohan CD, Srinivasa V, Rangappa S, Mervin L, Mohan S, Paricharak S, Baday S, Li F, Shanmugam MK, Chinnathambi A, et al. Trisubstituted-Imidazoles Induce Apoptosis in Human Breast Cancer Cells by Targeting the Oncogenic PI3K/Akt/mTOR Signaling Pathway. PLoS One. 2016;11(4):e0153155. doi: 10.1371/journal.pone.0153155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baburajeev CP, Mohan CD, Patil GS, Rangappa S, Pandey V, Sebastian A, Fuchs JE, Bender A, Lobie PE. Basappa et al. Nano-cuprous oxide catalyzed one-pot synthesis of a carbazole-based STAT3 inhibitor: a facile approach via intramolecular C-N bond formation reactions. RSC Adv. 2016;6(43):36775–36785. doi: 10.1039/C6RA01906D. [DOI] [Google Scholar]
- 30.Blich M, Golan A, Arvatz G, Sebbag A, Shafat I, Sabo E, Cohen-Kaplan V, Petcherski S, Avniel-Polak S, Eitan A, et al. Macrophage activation by heparanase is mediated by TLR-2 and TLR-4 and associates with plaque progression. Arteriosclerosis Thromb & Vascular Biol. 2013;33(2):e56–e65. doi: 10.1161/ATVBAHA.112.254961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond E, Li CP, Ferro V. Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal Biochem. 2010;396(1):112–116. doi: 10.1016/j.ab.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nature Med. 1999;5(7):793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 33.Vlodavsky I. Preparation of Extracellular Matrices Produced by Cultured Corneal Endothelial and PF-HR9 Endodermal Cells. Curr protocols Cell Biol. 1999; 10.14. 11-10.14. 14. [DOI] [PubMed]
- 34.Bharathkumar H, Mohan CD, Rangappa S, Kang T, Keerthy HK, Fuchs JE, Kwon NH, Bender A, Kim S. Basappa et al. Screening of quinoline, 1,3-benzoxazine, and 1,3-oxazine-based small molecules against isolated methionyl-tRNA synthetase and A549 and HCT116 cancer cells including an in silico binding mode analysis. Organic & Biomol Chem. 2015;13(36):9381–9387. doi: 10.1039/C5OB00791G. [DOI] [PubMed] [Google Scholar]
- 35.Ashwini N, Garg M, Mohan CD, Fuchs JE, Rangappa S, Anusha S, Swaroop TR, Rakesh KS, Kanojia D, Madan V, et al. Synthesis of 1,2-benzisoxazole tethered 1,2,3-triazoles that exhibit anticancer activity in acute myeloid leukemia cell lines by inhibiting histone deacetylases, and inducing p21 and tubulin acetylation. Bioorg & Medicinal Chem. 2015;23(18):6157–6165. doi: 10.1016/j.bmc.2015.07.069. [DOI] [PubMed] [Google Scholar]
- 36.Baburajeev CP, Dhananjaya Mohan C, Ananda H, Rangappa S, Fuchs JE, Jagadish S, Sivaraman Siveen K, Chinnathambi A, Ali Alharbi S, Zayed ME, et al. Development of Novel Triazolo-Thiadiazoles from Heterogeneous "Green" Catalysis as Protein Tyrosine Phosphatase 1B Inhibitors. Scientific Reports. 2015;5:14195. doi: 10.1038/srep14195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohan CD, Bharathkumar H, Bulusu KC, Pandey V, Rangappa S, Fuchs JE, Shanmugam MK, Dai X, Li F, Deivasigamani A, et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J Biol Chem. 2014;289(49):34296–34307. doi: 10.1074/jbc.M114.601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basappa M. S, Kavitha CV, Purushothaman A, Nevin KG, Sugahara K, Rangappa KS. A small oxazine compound as an anti-tumor agent: a novel pyranoside mimetic that binds to VEGF, HB-EGF, and TNF-alpha. Cancer Lett. 2010;297(2):231–243. doi: 10.1016/j.canlet.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Sulaiman NB, Mohan CD, Basappa S, Pandey V, Rangappa S, Bharathkumar H, Kumar AP, Lobie PE, Rangappa KS. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int J Oncol. 2016;49(3):1221–1229. doi: 10.3892/ijo.2016.3615. [DOI] [PubMed] [Google Scholar]
- 40.Hara M, Yoshida T, Takagaki A, Takata T, Kondo JN, Hayashi S, Domen K. A carbon material as a strong protonic acid. Angew Chem Int Ed. 2004;43(22):2955–2958. doi: 10.1002/anie.200453947. [DOI] [PubMed] [Google Scholar]
- 41.Toda M, Takagaki A, Okamura M, Kondo JN, Hayashi S, Domen K, Hara M. Green chemistry: biodiesel made with sugar catalyst. Nature. 2005;438(7065):178. doi: 10.1038/438178a. [DOI] [PubMed] [Google Scholar]
- 42.Mo X, López DE, Suwannakarn K, Liu Y, Lotero E, Goodwin JG, Lu C. Activation and deactivation characteristics of sulfonated carbon catalysts. J Catalysis. 2008;254(2):332–338. doi: 10.1016/j.jcat.2008.01.011. [DOI] [Google Scholar]
- 43.Devi KS, Jayashree S. Modified ceria as a substitute for sulfuric acid in the liquid phase nitration of toluene. React Kinet Mech Catal. 2013;108(1):183–192. doi: 10.1007/s11144-012-0513-6. [DOI] [Google Scholar]
- 44.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13(20):2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 45.Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, Parish CR. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry. 2000;39(51):15659–15667. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- 46.Neelgundmath M, Dinesh KR, Mohan CD, Li F, Dai X, Siveen KS, Paricharak S, Mason DJ, Fuchs JE, Sethi G, et al. Novel synthetic coumarins that targets NF-kappaB in Hepatocellular carcinoma. Bioorganic & Med Chem Lett. 2015;25(4):893–897. doi: 10.1016/j.bmcl.2014.12.065. [DOI] [PubMed] [Google Scholar]
- 47.Anusha S, Anandakumar BS, Mohan CD, Nagabhushana GP, Priya BS, Rangappa KS. Basappa, T CG. Preparation and use of combustion-derived Bi2O3 for the synthesis of heterocycles with anti-cancer properties by Suzuki-coupling reactions. RSC Adv. 2014;4(94):52181–52188. [Google Scholar]
- 48.Anusha S, Mohan CD, Ananda H, Baburajeev CP, Rangappa S, Mathai J, Fuchs JE, Li F, Shanmugam MK, Bender A, et al. Adamantyl-tethered-biphenylic compounds induce apoptosis in cancer cells by targeting Bcl homologs. Bioorg Med Chem Lett. 2016;26(3):1056–1060. doi: 10.1016/j.bmcl.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 49.Wu L, Viola CM, Brzozowski AM, Davies GJ. Structural characterization of human heparanase reveals insights into substrate recognition. Nature Struc Biol. 2015;22(12):1016–1022. doi: 10.1038/nsmb.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. C. G. Molecular Operating Environment version 2014.0901.
- 51.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS One. 2009;4(4):e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Bioch Sci. 2009;34(10):511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parish CR, Freeman C, Ziolkowski AF, He YQ, Sutcliffe EL, Zafar A, Rao S, Simeonovic CJ. Unexpected new roles for heparanase in Type 1 diabetes and immune gene regulation. Matrix Biol. 2013;32(5):228–233. doi: 10.1016/j.matbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 54.Gil N, Goldberg R, Neuman T, Garsen M, Zcharia E, Rubinstein AM, van Kuppevelt T, Meirovitz A, Pisano C, Li JP, et al. Heparanase is essential for the development of diabetic nephropathy in mice. Diabetes. 2012;61(1):208–216. doi: 10.2337/db11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li RW, Freeman C, Yu D, Hindmarsh EJ, Tymms KE, Parish CR, Smith PN. Dramatic regulation of heparanase activity and angiogenesis gene expression in synovium from patients with rheumatoid arthritis. Arthritis & Rheumatism. 2008;58(6):1590–1600. doi: 10.1002/art.23489. [DOI] [PubMed] [Google Scholar]
- 56.Lerner I, Hermano E, Zcharia E, Rodkin D, Bulvik R, Doviner V, Rubinstein AM, Ishai-Michaeli R, Atzmon R, Sherman Y, et al. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J Clin Invest. 2011;121(5):1709–1721. doi: 10.1172/JCI43792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt EP, Yang Y, Janssen WJ, Gandjeva A, Perez MJ, Barthel L, Zemans RL, Bowman JC, Koyanagi DE, Yunt ZX, et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nature Med. 2012;18(8):1217–1223. doi: 10.1038/nm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlodavsky I, Blich M, Li JP, Sanderson RD, Ilan N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013;32(5):241–251. doi: 10.1016/j.matbio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyses during this study are included in this article and its Additional file 1.


