Abstract
Dipeptidase 1 (DPEP1) is a zinc-dependent metalloproteinase that is fundamental in glutathione and leukotriene metabolism. DPEP1 was initially considered as a tumor suppressor gene in Wilms' tumor and breast cancer. However, it has been reported that DPEP1 is upregulated in colorectal cancers (CRCs) and high DPEP1 expression levels are associated with poorer patient survival. The role of DPEP1 genes in CRC, as well as their expression, requires investigation. Therefore, the present study investigated DPEP1 expression using reverse transcription-quantitative polymerase chain reaction or immunohistochemistry on surgically resected samples from CRC cases, and further examined the biological significance of DPEP1 by comparing the expression of the epithelial to mesenchymal transition (EMT) markers, including epithelial cadherin and Vimentin to clarify the function of DPEP1 in CRC, particularly in metastasis. The level of DPEP1 expression was identified to be significantly increased in tumorous tissue samples compared with that in non-tumorous tissue samples. In addition, increased DPEP1 mRNA expression levels were associated with positive lymph node metastasis in the included cohort. However, no positive correlations were observed between DPEP1 and EMT markers in the cohort. The results indiciates that further investigations into the upregulation of DPEP1 in colorectal carcinogenesis and the role of therapeutic or prognostic biomarkers are required.
Keywords: dipeptidase 1, colorectal cancer, lymph node metastasis, poorly differentiated histological types, epithelial to mesenchymal transition
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed types of cancer and is closely linked to aspects of the Western lifestyle (1,2). Early detection and surgical resection is the most effective measure to improve CRC-associated mortality (3). Despite recent advances in CRC treatment, such as chemotherapy and molecular targeted therapy, the prognosis remains poor in advanced CRC cases (1,2). Recurrence and metastasis are the main obstacles for improving the prognosis of postoperative CRC patients (2,4). In Japan, ~15% of patients with stage II and 30% of patients with stage III will develop recurrence within 5 years of surgical resection of CRC (5). The treatment of advanced CRC remains essentially palliative currently; therefore, it is necessary to understand the processes that contribute to tumor progression, particularly those that facilitate invasion and metastasis, to prevent CRC recurrence.
Cancer metastasis is a complex process in which malignant cancer cells disseminate from the primary tumor site to a secondary tumor at a distant site. During this multistep process, transition between epithelial and mesenchymal states occurs in cancer cells (6,7). Initially, metastasis is triggered by the epithelial to mesenchymal transition (EMT), which enhances cancer cell motility and intravasation into blood vessels (8). Notably, the reversible process of EMT, mesenchymal to epithelial transition (MET), is also observed at the metastatic site, and is involved in the metastatic process (6). EMT and MET are controlled by multiple molecular mechanisms, such as transcription factors, epigenetic modifications, alternative splicing, and miRNA networks, resulting in the modification of epithelial or mesenchymal gene expression levels (6,7). Among the molecular mechanisms, the downregulation and re-expression of epithelial (E-) cadherin are reportedly associated with EMT and MET, respectively (9). Accumulating evidence has revealed the important roles of EMT and MET in cancer metastasis; however, this modulation is complicated and further research is required.
Dipeptidase 1 (DPEP1), located on chromosome 16q24.3, is a zinc-dependent metalloproteinase, which is fundamental in glutathione and leukotriene metabolism (10). Leukotrienes are pro-inflammatory mediators that are associated with cancer development (11,12) and, as such, the dysregulation of DPEP1 potentially leads to the development of malignant tumors. Initially, loss of DPEP1 expression is associated with Wilms' tumor (13). Consistent with this, DPEP1 is considered to be a tumor suppressor gene in breast cancer and pancreatic ductal adenocarcinoma (14,15). However, DPEP1 is upregulated in CRC and its high expression level is associated with poorer patient survival (16). Furthermore, a recent report demonstrated that DPEP1 is highly expressed in CRC and promotes metastasis via regulation of E-cadherin expression levels (17). Therefore, further investigations are required to evaluate the expression of DPEP1 in an additional independent CRC cohort and the role of DPEP1 in CRC metastasis.
In the present study, DPEP1 expression levels were investigated using comprehensive gene expression analyses, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and immunohistochemical (IHC) staining in surgically resected CRC cases. In addition, the biological significance of DPEP1 was examined by comparing the expression levels of EMT markers to clarify the function of DPEP1 in CRC metastasis.
Materials and methods
Clinical samples of patients
A total of 78 surgical specimens were obtained from CRC patients who had undergone surgical resection at Fukushima Medical University Hospital (Fukushima, Japan) between January 2008 and December 2010. Specimens from all 78 cases were used for comprehensive gene expression analysis, specimens from five cases were used for protein expression analysis by western blotting, and specimens from 55 cases were used for IHC staining. Information regarding age, gender, TNM stage and pathological diagnosis, including lymphatic and venous invasion were retrospectively collected. The carcinomas at the time of primary tumor resection were staged according to the Union for International Cancer Control (UICC) TNM classification (the 7th classification) (18,19). Written informed consent was obtained from all patients and the current study was approved by the ethics committee of Fukushima Medical University.
Comprehensive gene expression analysis
DPEP1 expression data were obtained using custom microarray analysis, as previously described (20). Briefly, the surgical specimen was homogenized and mixed with ISOGEN® reagent (Nippon Gene Co., Ltd., Tokyo, Japan). Total RNA was subjected to purification of polyA(A)+ RNA using a MicroPoly(A)Purist kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Human reference RNA was prepared by mixing equal quantities of poly(A)+ RNA extracted from 22 human cancer cell lines (A431, A549, AKI, HBL-100, HeLa, HepG2, HL60, IMR-32, Jurkat, K562, KP4, MKN7, NK-92, Raji, RD, Saos-2, SK-N-MC, SW-13, T24, U251, U937 and Y79).
Synthetic polynucleotides (80-mers) representing 31,797 human transcripts (MicroDiagnostic, Inc., Tokyo, Japan) were arrayed on aminosilane-coated glass slides with a custom-made arrayer. RNA (2 µg) was subjected to reverse transcription using SuperScript II (Thermo Fisher Scientific, Inc.). Sample RNA was labeled using Cyanine 5-dUTP (PerkinElmer, Inc., Waltham, MA, USA) and the reference RNA was labeled using Cyanine 3-dUTP. Hybridization was performed with a Labeling and Hybridization kit (MicroDiagnostic, Inc.). Signals were measured with a GenePix 4000B scanner (Axon Instruments Inc., Union City, CA, USA) and processed into primary expression ratios. The primary expression ratios were then converted into log2 values and compiled into a matrix. An expression ratio of 1 (log ratio of 0) was assigned for spots that exhibited fluorescence intensities under the detection limits, and these were included in the signal calculation of the mean averages. Data were processed using MDI gene expression analysis software package (MicroDiagnostic, Inc.).
RT-qPCR
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from 5 µg of total RNA with a random hexamer using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Inc.). These cDNAs were used for the measurement of gene expression with a 7500 Real-time PCR system (Thermo Fisher Scientific, Inc.) using TaqMan probes. The assessors were blinded to the patient information and performed the experiments in triplicate. Taqman expression assays, DPEP1 (Hs01116752_m1) and β-actin (Hs99999903_m1) were purchased from Thermo Fisher Scientific, Inc. and β-actin served as an internal control. Relative DPEP1 gene expression was calculated using the 2−ΔΔCq method, according to the supplier's protocol (Thermo Fisher Scientific, Inc.) (21).
Western blotting
Surgical specimens were homogenized in a 100-mM Tris-HCl (pH 7.6) buffer containing 0.15 M NaCl, 5 mM EDTA, 1% Triton X-100, and 5% glycerol using a Polytron PT3100 homogenizer (Kinematica AG, Luzern, Switzerland). After centrifugation at 17,400 × g for 15 min at 4°C, the supernatants were collected. Next, 20 µg of each protein sample was run on SDS-polyacrylamide gels (5–15% gradient; Thermo Fisher Scientific, Inc.) and blotted onto Immun-Blot PVDF membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The blotted membranes were incubated with the following primary antibodies overnight at 4°C: Rabbit polyclonal anti-DPEP1 [(cat. no. HPA012783) Sigma-Aldrich; Merck KGaA, Darmstadt, Germany] at a dilution of 1:100, and mouse monoclonal anti-β-actin antibody (cat. no. sc-69879; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) served as a loading control at a dilution of 1:2,500. The blotted membranes were subsequently incubated with the appropriate horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG secondary antibody (cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) at a dilution of 1:5,000. Signals were detected using ImageQuant LAS4000 (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.).
IHC staining and evaluation
IHC staining was performed on paraffin-embedded histological sections (4-µm thick) using a polymer peroxidase method. Briefly, after deparaffinization and rehydration, the sections were treated with 0.3% hydrogen peroxide in methanol for 30 min to block endogenous peroxidase activity. Following rinsing in phosphate-buffered saline (PBS) (Thermo Fisher Scientific, Inc.), the sections were incubated with anti-DPEP1 antibody [cat. no. HPA012783 (dilution, 1:2,000); Sigma-Aldrich; Merck KGaA], Dako anti-E-cadherin antibody [cat. no. NCH-38 (dilution, 1:200); Agilent Technologies GmbH, Waldbronn, Germany], and anti-Vimentin antibody [cat. no. SP20 (dilution, 1:400); Nichirei Biosciences Inc., Tokyo, Japan] at 4°C overnight. Three further washes (5 min per wash) in PBS was followed by treatment with a peroxidase-labeled polymer, conjugated to goat anti-rabbit immunoglobulins [Dako EnVision+ System-HRP Labelled Polymer; ready-to-use (cat. no. K4003) Dako; Agilent Technologies] as the secondary antibody for 30 min at room temperature. The staining was visualized with diaminobenzidine, followed by counterstaining with hematoxylin. Expression of these proteins was evaluated as positive when the nucleus of the cancerous tissue and the total field of view were observed at a magnification of ×400. Blinded to the origination of the features and clinical outcomes, the staining of each specimen was evaluated. Stained cancer cells were counted per 1,000 cancer cells in the maximum field of cancer tissue by two investigators. The rate of positively stained cells was classified as follows: 0%, 0; 1–10%, 1; and 11–100%, 2, and the staining intensity was scored as 0 (negative), 1 (weak) and 2 (strong). The evaluation was expressed as a product of the score of positive rate and staining intensity. Positive staining was defined by a score of 2, while negative staining was scored at 0 or 1.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Fisher's extract test or χ2 test were performed to analze the contingency tables. In addition, the Mann-Whitney U test was conducted for comparison of the means of the two groups. The Kruskal-Wallis and one-way analysis of variance tests were used for comparisons between more than two groups. P<0.05 was considered to indicate a statistically significant difference and data are presented as the mean ± standard deviation.
Results
DPEP1 expression levels in CRC
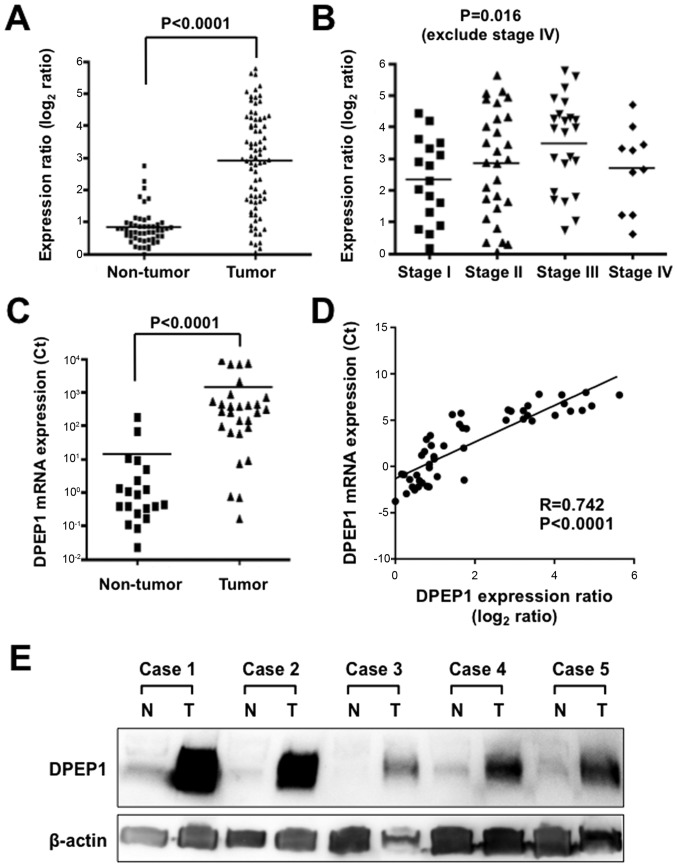
The expression level of DPEP1 mRNA in the current cohort was determined using comprehensive gene expression analysis data. The expression ratios of DPEP1 were compared between 78 tumorous tissue samples and 50 non-tumorous tissue samples. A significantly higher DPEP1 expression level was identified in tumorous tissue samples when compared with non-tumorous tissue samples (Fig. 1A; P<0.0001). DPEP1 expression levels and clinicopathological factors were then analyzed in the CRC specimens (Table I). Cases with poorly differentiated histological types (P=0.0001) and positive lymph node metastasis (P=0.024) showed significantly higher DPEP1 expression levels. However, the DPEP1 expression level was not associated with gender, age, tumor size, distant metastasis or stage. When the DPEP1 expression levels of the stage 1, 2 and 3 cases were compared, DPEP1 expression was observed to be significantly elevated in more advanced stage tumors, which is consistent with a role for DPEP1 in cancer progression (Fig. 1B; P=0.016).
Figure 1.
Expression levels of DPEP1 in CRC specimens. (A) Expression differences of DPEP1 between 78 tumorous and 50 non-tumorous tissue samples from the CRC cohort. The dot plot represents DPEP1 expression levels from microarray analysis, and a log2 scale of expression levels is presented. Horizontal bars indicate the mean expression values. P<0.0001, Mann-Whitney U-test. (B) Expression differences of DPEP1 between stage 1 (n=17), stage 2 (n=28) and stage 3 (n=23) (according to the 7th TNM classification) tumorous tissue samples from the CRC cohort. The dot plot represents DPEP1 expression levels from microarray analysis, and a log2 scale of the expression levels is presented. Horizontal bars indicate mean expression values. P=0.016, one-way analysis of variance test. (C) Expression differences of DPEP1 between 28 tumorous and 20 non-tumorous tissue samples from the CRC cohort. The dot plot represents DPEP1 expression levels from RT-qPCR analysis and the horizontal bars indicate the mean expression values. P<0.0001, Mann-Whitney U-test. (D) Correlation of DPEP1 mRNA expression between microarray (x-axis) and RT-qPCR (y-axis) analyses in the CRC patients. (E) Western blot analysis of DPEP1 in five representative paired samples of non-tumorous and tumorous tissue from CRC cases. β-actin served as a loading control. N, non-tumorous tissue; T, tumorous tissue; DPEP1, dipeptidase 1; CRC, colorectal cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table I.
Clinicopathological factors and DPEP1 mRNA expression levels (n=78).
| Characteristic | n | DPEP1 mRNA expression ratio | P-value |
|---|---|---|---|
| Age, years | 0.188 | ||
| <65 | 31 | 3.2±1.4 | |
| ≥65 | 47 | 2.7±1.6 | |
| Gender | 0.346 | ||
| Male | 53 | 3.0±1.6 | |
| Female | 25 | 2.7±1.5 | |
| TNM classificationa | 0.117 | ||
| I | 17 | 2.3±1.3 | |
| II | 28 | 2.8±1.7 | |
| III | 23 | 3.5±1.5 | |
| IV | 10 | 2.7±1.3 | |
| Histology | <0.0001 (tub1 vs. tub2) | ||
| Tub1b | 33 | 2.3±1.3 | |
| Tub2c | 37 | 3.7±1.4 | |
| Otherd | 8 | 1.6±1.3 | |
| Lymph node metastasis | 0.024 | ||
| Absent | 49 | 2.6±1.5 | |
| Present | 29 | 3.4±1.4 | |
| Distant metastasis | 0.435 | ||
| Absent | 74 | 2.9±1.5 | |
| Present | 4 | 2.3±1.6 |
Values are expressed as the mean ± standard deviation.
UICC TNM 7th classification (18,19).
Well and
moderately differentiated tubular adenocarcinoma.
Solid-type poorly differentiated, mucinous and papillary adenocarcinoma or adenosquamous carcinoma. P-values were calculated using Mann-Whitney test or a Kruskal Wallis test, where appropriate. DPEP1, dipeptidase 1.
To further confirm the DPEP1 expression levels in CRC, RT-qPCR was performed for 28 randomly-selected samples of tumorous tissue and 20 samples of non-tumorous tissue from the cases used in Fig. 1A. DPEP1 mRNA expression was confirmed to be upregulated in tumorous tissue samples compared with non-tumorous tissue samples (Fig. 1C; P<0.0001). In addition, this DPEP1 mRNA expression data was positively correlated with the data from comprehensive gene expression analysis (R=0.742, P<0.0001), confirming the reliability of the microarray data (Fig. 1D).
In addition, DPEP1 protein expression was analyzed by western blotting in five representative non-tumorous/tumorous CRC tissue samples that showed high levels of DPEP1 mRNA expression (Fig. 1E). Consistent with the RT-qPCR results, DPEP1 protein was highly expressed in the tumorous tissue samples when compared with non-tumorous tissue samples. These results demonstrate the upregulation of DPEP1 mRNA, which resulted in the upregulation of DPEP1 protein expression.
IHC staining for DPEP1 and EMT markers
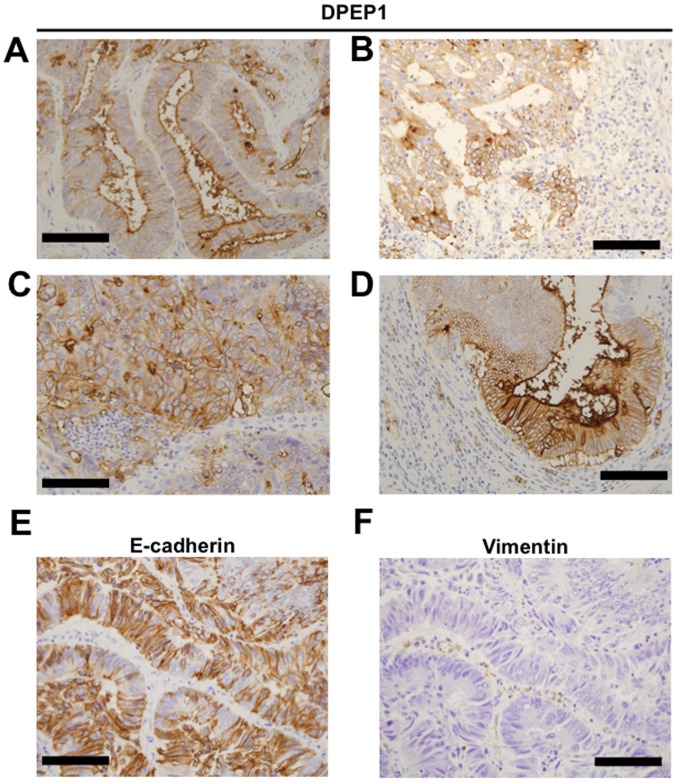
To investigate DPEP1 protein expression, IHC staining for DPEP1 was performed in 55 CRC specimens. Positive staining for DPEP1 at the apical cell surface (Fig. 2A), cytoplasm (Fig. 2B) or circumference of malignant cells (Fig. 2C) was observed in each of the CRC specimens. In addition, mixed patterns of the above localizations were observed in the specimens (Fig. 2D). As a result, the DPEP1 expression was observed to be positive in 45 cases (82%) and negative in 10 cases (18%). Based on the DPEP1 IHC staining intensity, the association between DPEP1 expression levels and clinicopathological factors was analyzed in the CRC patients (Table II). Consistent with the DPEP1 mRNA analysis, the positive expression rate of DPEP1 was significantly correlated with a poorer histology (P=0.021). However, DPEP1 expression was not associated with gender, age, lymph node metastasis, distant metastasis, or TNM stage classification.
Figure 2.
Immunohistochemical staining of DPEP1 in CRC. Representative image of DPEP1 staining of (A-D) tumorous tissue samples, (E) E-cadherin and (F) Vimentin. (A) Apical, (B) cytoplasmic, (C) circumference and (D) mixed staining patterns for DPEP1 in CRC. (E) Positive staining pattern for E-cadherin in CRC. (F) Negative staining pattern for Vimentin in CRC. Scale bar=100 µm. DPEP1, dipeptidase 1; CRC, colorectal cancer; E-cadherin, epithelial cadherin.
Table II.
Clinicopathological factors and DPEP1 expression levels (n=55) observed by IHC staining.
| DPEP1 IHC staining | ||||
|---|---|---|---|---|
| Characteristic | n | Positive, n=45 (%) | Negative, n=10 (%) | P-value |
| Age, years | 0.056 | |||
| <65 | 23 | 22 (48.9) | 1 (10.0) | |
| ≥65 | 32 | 23 (51.1) | 9 (90.0) | |
| Gender | 0.780 | |||
| Male | 35 | 29 (64.4) | 6 (60.0) | |
| Female | 19 | 16 (35.6) | 4 (40.0) | |
| TNM classificationa | 0.086 | |||
| I | 14 | 10 (22.2) | 4 (40.0) | |
| II | 18 | 14 (31.1) | 4 (40.0) | |
| III | 16 | 15 (33.3) | 1 (10.0) | |
| IV | 7 | 6 (13.3) | 1 (10.0) | |
| Histology | 0.021 (tub1 vs. tub2) | |||
| Tub1b | 24 | 17 (37.8) | 7 (70.0) | |
| Tub2c | 26 | 25 (55.6) | 1 (10.0) | |
| Otherd | 5 | 3 (6.7) | 2 (20.0) | |
| Lymph node metastasis | 0.231 | |||
| Absent | 33 | 25 (55.6) | 8 (80.0) | |
| Present | 22 | 20 (44.4) | 2 (20.0) | |
| Distant metastasis | 1.000 | |||
| Absent | 48 | 39 (86.7) | 9 (90.0) | |
| Present | 7 | 6 (13.3) | 1 (10.0) | |
Values are expressed as the mean ± standard deviation.
UICC TNM 7th classification (18,19).
Well and
moderately differentiated tubular adenocarcinoma.
Solid-type poorly differentiated, mucinous and papillary adenocarcinoma or adenosquamous carcinoma or adenosquamous carcinoma. P-values were calculated using Fisher's exact test or a χ2 test, where appropriate. DPEP1, dipeptidase 1; IHC, immunohistochemical.
The expression levels of EMT markers, E-cadherin and Vimentin were observed in patients with CRC (Fig. 2E and F) and the impact of DPEP1 expression on EMT markers (Table III) was evaluated. However, EMT status was not identified to be associated with DPEP1 expression levels in the present study.
Table III.
Epithelial to mesenchymal transition status and DPEP1 mRNA expression levels in colorectal cancer patients (n=51).
| Immunohistochemical staining | |||
|---|---|---|---|
| E-cadherin | Vimentin | n | DPEP1 mRNA expression ratio (means ± standard deviation) |
| Positive | Positive | 0 | - |
| Positive | Negative | 45 | 2.9±2.0 |
| Negative | Positive | 0 | - |
| Negative | Negative | 6 | 3.1±1.4 |
DPEP1, dipeptidase 1; E-cadherin, epithelial cadherin.
Discussion
In the present study, the tumor expression of DPEP1 was identified to be upregulated at the mRNA and protein levels in CRC. Increased DPEP1 mRNA expression levels were identified to be associated with positive lymph node metastasis and poorer tumor histology in CRC patients. Furthermore, in the IHC staining analysis, upregulated DPEP1 expression was associated with poorer tumor histology. These findings were consistent with previous studies demonstrating that DPEP1 is highly expressed in CRC compared with matched normal mucosa, indicating a significant role of DPEP1 in CRC development (16,17). While the current results indicate the oncogenic role of DPEP1 in CRC, no significant associations between DPEP1 expression levels and patient prognosis, as well as EMT status, were observed in the current cohort.
The associations between EMT status and DPEP1 levels were investigated in a CRC in the present study. The results are inconsistent with those of a recent study, indicating that DPEP1 promotes CRC metastasis through regulation of E-cadherin expression (17). E-cadherin is a transmembrane protein and acts as an anchor between neighboring cells to form adherens junctions (7,22). Therefore, it is reasonable that loss of E-cadherin promotes cancer metastasis. In addition to E-cadherin, Vimentin and neural (N-) cadherin serve as classical EMT markers for diagnosing whether tumor cells are an epithelial phenotype or mesenchymal phenotype. It is well known that downregulation of E-cadherin, and upregulation of Vimentin and N-cadherin are key markers for EMT, and that re-upregulation of E-cadherin is a key marker for MET and is also a necessary process of metastasis (6).
EMT and MET are regulated by multiple modulators, such as transcription factors, epigenetic modifications, alternative splicing, and miRNA networks (6,7). Transcription factors, such as snail family transcriptional repressor 1, snail family transcriptional repressor 2, twist family bHLH transcription factor 1, zinc finger E-box binding homeobox 1 and zinc finger E-box binding homeobox 2, have previously been investigated (7). Furthermore, there are continued efforts to identify a novel EMT regulator to further understand cancer progression and metastasis via an EMT process (23). However, as the EMT process may be a ‘druggable’ target by specific inhibitors, candidate therapeutic targets are being developed. Sorafenib is one example of a drug that inhibits EMT via histone modifications that occur during EMT in lung adenocarcinoma cell lines (24). Currently, sorafenib is used for cases with an activating mutation of A-Raf proto-oncogene, serine/threonine kinase (25); with further research, sorafenib may be administered to CRC patients. In addition, an activating mutation in fibroblast growth factor receptor 4, which enhances EMT in colon cancer cells, is also hypothesized to be a therapeutic target of specific inhibitors for CRC (26,27). Further experimental studies or mice studies investigating the functional role of DPEP1 are required to reveal whether DPEP1 may be a candidate therapeutic target for CRC.
In conclusion, the current study revealed that the level of DPEP1 expression was significantly increased in tumorous cells, and no positive correlation was observed between DPEP1 and EMT markers. The present results prompt further investigations into the efficacy of DPEP1 as a biomarker or therapeutic target in CRC.
Acknowledgements
The present study was partially supported by grants for translational research programs from the New Energy and Industrial Technology Development Organization (Tokyo, Japan).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Mayer RJ, Venook AP, Schilsky RL. Progress against GI cancer during the American Society of Clinical Oncology's first 50 years. J Clin Oncol. 2014;32:1521–1530. doi: 10.1200/JCO.2014.55.4121. [DOI] [PubMed] [Google Scholar]
- 4.Venook AP, Weiser MR, Tepper JE. Colorectal cancer: All hands on deck. Am Soc Clin Oncol Educ Book. 2014;34:83–89. doi: 10.14694/EdBook_AM.2014.34.83. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe T, Itabashi M, Shimada Y, Tanaka S, Ito Y, Ajioka Y, Hamaguchi T, Hyodo I, Igarashi M, Ishida H, et al. Japanese Society for Cancer of the Colon and Rectum: Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20:207–239. doi: 10.1007/s10147-015-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, Willbanks A, Sarkar S. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae KM, Parker NN, Dai Y, Vieweg J, Siemann DW. E-cadherin plasticity in prostate cancer stem cell invasion. Am J Cancer Res. 2011;1:71–84. [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa H, Inazawa J, Inoue K, Misawa S, Kashima K, Adachi H, Nakazato H, Abe T. Assignment of the human renal dipeptidase gene (DPEP1) to band q24 of chromosome 16. Cytogenet Cell Genet. 1992;59:258–260. doi: 10.1159/000133263. [DOI] [PubMed] [Google Scholar]
- 11.Jeong CH, Bode AM, Pugliese A, Cho YY, Kim HG, Shim JH, Jeon YJ, Li H, Jiang H, Dong Z. [6]-Gingerol suppresses colon cancer growth by targeting leukotriene A4 hydrolase. Cancer Res. 2009;69:5584–5591. doi: 10.1158/0008-5472.CAN-09-0491. [DOI] [PubMed] [Google Scholar]
- 12.Bellamkonda K, Chandrashekar NK, Osman J, Selvanesan BC, Savari S, Sjölander A. The eicosanoids leukotriene D4 and prostaglandin E2 promote the tumorigenicity of colon cancer-initiating cells in a xenograft mouse model. BMC Cancer. 2016;16:425. doi: 10.1186/s12885-016-2466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Austruy E, Cohen-Salmon M, Antignac C, Béroud C, Henry I, Nguyen VC, Brugières L, Junien C, Jeanpierre C. Isolation of kidney complementary DNAs down-expressed in Wilms' tumor by a subtractive hybridization approach. Cancer Res. 1993;53:2888–2894. [PubMed] [Google Scholar]
- 14.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green AR, Krivinskas S, Young P, Rakha EA, Paish EC, Powe DG, Ellis IO. Loss of expression of chromosome 16q genes DPEP1 and CTCF in lobular carcinoma in situ of the breast. Breast Cancer Res Treat. 2009;113:59–66. doi: 10.1007/s10549-008-9905-8. [DOI] [PubMed] [Google Scholar]
- 16.Eisenach PA, Soeth E, Röder C, Klöppel G, Tepel J, Kalthoff H, Sipos B. Dipeptidase 1 (DPEP1) is a marker for the transition from low-grade to high-grade intraepithelial neoplasia and an adverse prognostic factor in colorectal cancer. Br J Cancer. 2013;109:694–703. doi: 10.1038/bjc.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Lee SJ, Cho HJ, Kim TW, Kim JT, Kim JW, Lee CH, Kim BY, Yeom YI, Lim JS, et al. Dehydropeptidase 1 promotes metastasis through regulation of E-cadherin expression in colon cancer. Oncotarget. 2016;7:9501–9512. doi: 10.18632/oncotarget.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobin LH, Compton CC. TNM seventh edition: what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–5339. doi: 10.1002/cncr.25537. [DOI] [PubMed] [Google Scholar]
- 19.Sobin LH, Gospodarowicz MK, Wittekind C, editors. International Union Against Cancer (UICC), corp-author TNM Classification of Malignant Tumors. 7th edition. Wiley-Blackwell; Oxford, UK: 2009. [Google Scholar]
- 20.Okabe N, Ezaki J, Yamaura T, Muto S, Osugi J, Tamura H, Imai J, Ito E, Yanagisawa Y, Honma R, et al. FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. Int J Oncol. 2015;46:999–1006. doi: 10.3892/ijo.2015.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Chen YL, Ji G, Fang W, Gao Z, Liu Y, Wang J, Ding X, Gao F. Sorafenib inhibits epithelial-mesenchymal transition through an epigenetic-based mechanism in human lung epithelial cells. PLoS One. 2013;8:e64954. doi: 10.1371/journal.pone.0064954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, Schiller J, Villalona-Calero MA, Meyerson M, Carbone DP. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. J Clin Invest. 2014;124:1582–1586. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R, Li J, Xie K, Zhang T, Lei Y, Chen Y, Zhang L, Huang K, Wang K, Wu H, et al. FGFR4 promotes stroma-induced epithelial-to-mesenchymal transition in colorectal cancer. Cancer Res. 2013;73:5926–5935. doi: 10.1158/0008-5472.CAN-12-4718. [DOI] [PubMed] [Google Scholar]
- 27.Hagel M, Miduturu C, Sheets M, Rubin N, Weng W, Stransky N, Bifulco N, Kim JL, Hodous B, Brooijmans N, et al. First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway. Cancer Discov. 2015;5:424–437. doi: 10.1158/2159-8290.CD-14-1029. [DOI] [PubMed] [Google Scholar]