Abstract
Channelrhodopsins (ChRs) are light-activated ion channels widely employed for photostimulation of excitable cells. This study focuses on ReaChR, a chimeric ChR variant with optimal properties for optogenetic applications. We combined electrophysiological recordings with infrared and UV-visible spectroscopic measurements to investigate photocurrents and photochemical properties of ReaChR. Our data imply that ReaChR is green-light activated (λmax = 532 nm) with a non-rhodopsin-like action spectrum peaking at 610 nm for stationary photocurrents. This unusual spectral feature is associated with photoconversion of a previously unknown light-sensitive, blue-shifted photocycle intermediate L (λmax = 495 nm), which is accumulated under continuous illumination. To explain the complex photochemical reactions, we propose a symmetrical two-cycle-model based on the two C15=N isomers of the retinal cofactor with either syn- or anti-configuration, each comprising six consecutive states D, K, L, M, N, and O. Ion conduction involves two states per cycle, the late M- (M2) with a deprotonated retinal Schiff base and the consecutive green-absorbing N-state that both equilibrate via reversible reprotonation. In our model, a fraction of the deprotonated M-intermediate of the anti-cycle may be photoconverted—as the L-state—back to its inherent dark state, or to its M-state pendant (M′) of the syn-cycle. The latter reaction pathway requires a C13=C14, C15=N double-isomerization of the retinal chromophore, whereas the intracircular photoconversion of M back to D involves only one C13=C14 double-bond isomerization.
Introduction
Channelrhodopsins (ChRs) are photosensitive ion channels, naturally occurring in the eyes of motile green algae (1, 2, 3, 4). ChRs unify the ability of light perception with a regulated ion conductance in one small protein, which can be coopted to serve as an ideal actuator for optogenetic applications. Hence, ChRs have become wide-spread tools in the field of neuroscience, where they are used to manipulate the membrane voltage of neurons and to noninvasively evoke or inhibit action potentials with high spatial and temporal precision (4, 5, 6, 7, 8, 9). Today, the most widely applied ChR variants are still channelrhodopsin-2 from Chlamydomonas reinhardtii (CrChR2 or simply ChR2) and its gain-of-function mutant H134R (10, 11, 12), which are excited most effectively with blue light of 460 nm. However, due to deeper penetration and reduced scattering in tissues, the use of yellow or even red light would be advantageous. This holds particularly true for in vivo application in rodents, where noninvasive delivery of weakly scattered light is a demanding challenge. Over the last few years, a couple of red-shifted ChRs were discovered, modified, and established as optogenetic tools. In 2008, the native green-absorbing channelrhodopsin-1 from Volvox carteri (VcChR1) with λmax = 535 nm was used to trigger action potentials in neurons as a response to orange light (13). Subsequent mutational and helix shuffling approaches produced variants with improved expression, larger photocurrents in mammalian cells, and action spectra that were red-shifted compared with ChR2 (14, 15). One of the chimeric constructs was the CrChR1/VcChR1 hybrid C1V1ET (E122T) enabling fast AP firing even upon stimulation with 630 nm light (14).
In 2013, the red-activatable ChR (ReaChR) was introduced as an improved VcChR1 variant with the N-terminus of CrChR1 (amino acid 1–94), helix 6 from VcChR2, and the L171I mutation in helix 3 (CrChR2 I131) to reduce desensitization (16) (see Fig. 1 A). In comparison with C1V1ET, ReaChR shows improved expression, faster channel closing rates, and larger stationary currents with an action spectrum that substantially deviates from rhodopsin absorption respective shape and side maximum at 630 nm (16). Additionally, the authors demonstrated deep brain tissue activation through the intact skull of awake mice without removal or surgical thinning of the cranium. In a subsequent study, ReaChR was employed to excite central nervous system neurons of Drosophila melanogaster to study behavior of freely moving flies (17). Remarkably, ReaChR possesses a significant propensity for two-photon absorption most likely due to a high 2P cross section, which was exploited for in-depth excitation of acute brain slices (18) and manipulation of transgenic D. melanogaster with near-infrared light (19). Moreover, Sengupta et al. (20) highlighted the enormous potential of ReaChR to restore vision in blind retinitis pigmentosa mice. Its superior properties mentioned before render ReaChR a versatile and frequently used red-light-activated optogenetic tool for in vitro and in vivo applications.
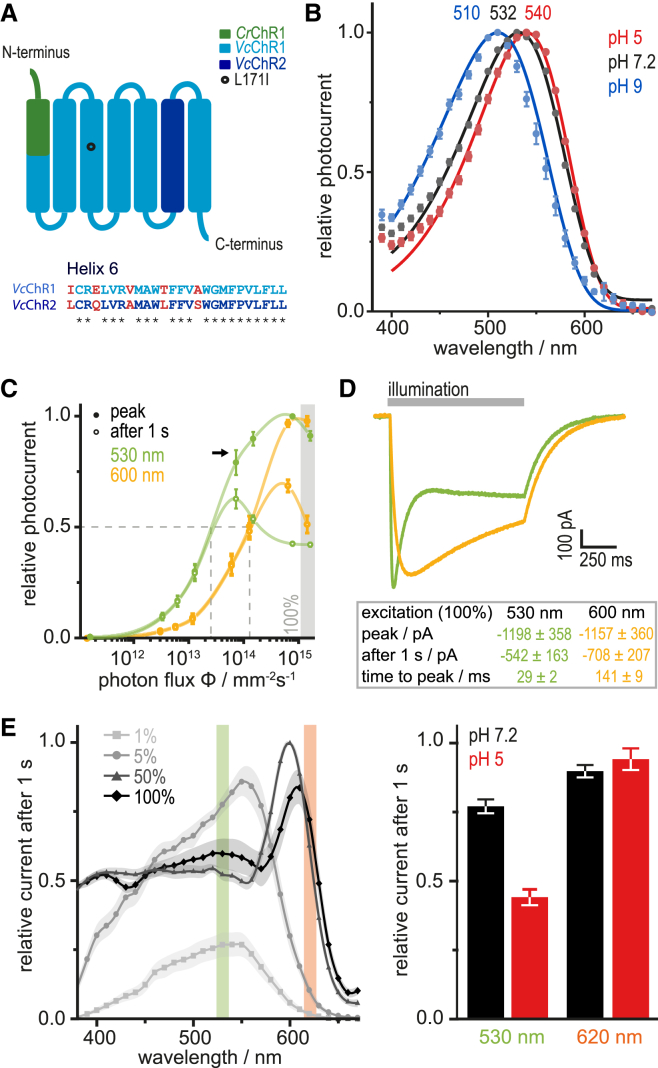
Figure 1.
Photocurrent properties of ReaChR in HEK293 cells. (A) Helix architecture of the chimeric ReaChR consisting of VcChR1, helix 6 (H6) of VcChR2, and the N-terminal sequence of CrChR1. H6 of VcChR1 and VcChR2 only differ in five amino acids. (B) Action spectra (10 ms activation) of ReaChR at different external pH (pHe) values (mean ± SE, n = 7–12). (C) Light titration curve after excitation with green (530 nm; intensities from 6.03 × 10−5 mW mm−2 (0.01%) to 0.61 mW mm−2 (100%)) or orange light (600 nm; 6.65 × 10−5 to 0.47 mW mm−2); photocurrents are normalized to the highest peak response of each cell (mean ± SE, n = 6), lines were added for visual guidance. (D) Comparison of photocurrents evoked with 530 and 600 nm at the same cell (equal photon flux, n = 6). (E) Stationary action spectra at 1, 5, 50 and 100% light intensity (left, mean ± SE, n = 5–6). At pH 5.0 (red bars, right), reduction of the stationary current is more pronounced at 530 nm compared to 620 nm, implying that the photoreactive intermediate is accumulated more at pH 5.0 (mean ± SE, n = 6–8).
Although ReaChR is successfully used in optogenetic experiments, many aspects of its basic photochemistry were not reported in the initial publication and are still unknown, representing the starting point for this investigation. Within the last decade, photochemical reactions as they occur in ChRs have been studied in great detail with different spectroscopic and electrophysiological methods leading to the development of complex photocycle models. Spectroscopic data of ChR2 under single-turnover conditions could be properly described by a single reaction cascade comprising one dark state and four spectrally distinct intermediates (P500, P390, P520, P480) (21, 22, 23). However, explanation of photocurrents under extended illumination demands a more complex two-cycle model with at least two closed and two open states with different conductances and ion selectivities (24, 25, 26, 27). More recently, spectroscopic data of various recombinant ChRs supported the two-cycle-model (28, 29, 30, 31, 32, 33). Bruun et al. (30) reported that both all-trans and 13-cis retinal isomers of the two dark states may directly photoconvert into each other at low temperature without passing through the photocycle. Here, we present a comprehensive electrical and spectroscopic analysis of ReaChR allowing us to explain the unusual action spectrum by secondary photochemical reaction(s).
Materials and Methods
Molecular biology
For spectroscopic experiments, DNA encoding ReaChR (GenBank: KF448069.1, 1–345 amino acids) with a C-terminal 1D4 epitope (T-E-T-S-Q-V-A-P-A) was inserted into the pMT4 expression vector (34) between EcoRI/NotI (Thermo Fisher Scientific, Waltham, MA), while for patch-clamp recordings ReaChR was C-terminally fused to the mCerulean3 fluorophore (35) and cloned into the pEGFP-N1 vector between HindIII/XbaI (Thermo Fisher Scientific). The mutations C168A and C168S were created via site-directed mutagenesis (QuikChange; Agilent Technologies, Santa Clara, CA).
Patch-clamp recordings
HEK293 cells were seeded onto coverslips (1.5 × 105 cells per 40 mm dish; TPP Techno Plastic Products, Trasadingen, Switzerland) and supplemented with 1 μM all-trans retinal (Sigma-Aldrich, Munich, Germany). 48–56 h before recordings cells were transiently transfected with the respective plasmid DNA using FuGENE HD transfection reagent (Promega, Madison, WI). Measurements were performed in whole-cell mode at a holding potential of −60 mV. Signals were amplified with an AxoPatch 200B and digitized using a DigiData 1400 (Molecular Devices, Sunnyvale, CA). Light was delivered by a Polychrome V (TILL Photonics, Planegg, Germany) or a Xenon lamp and regulated with a programmable shutter, while photon densities were adjusted using a neutral density filter wheel and/or neutral density filters. Data was acquired with the Clampex software and evaluated using Clampfit 10.4 (Molecular Devices). If not stated otherwise, pH was intra- and extracellularly adjusted to 7.2 (internal: 110 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 1 mM KCl, 1 mM CsCl, 10 mM HEPES, 10 mM EGTA; external: 140 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 1 mM KCl, 1 mM CsCl, 10 mM HEPES).
Expression and purification
HEK293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (DMEM-S; Biochrom) at 37°C and 5% CO2. For transfection, cells were grown on 100 mm culture dishes (Corning; Sigma-Aldrich) to 70–80% confluency and treated with TurboFect (Thermo Fisher Scientific) according to the manufacture’s protocol. Cells were harvested 36–48 h posttransfection in the presence of protease inhibitor (complete EDTA-free; Roche, Mannheim, Germany). Protein purification was achieved under safelight (>600 nm) at 4°C. During whole-cell treatment with 1% (w/v) n-dodecyl β-D-maltopyranoside (DDM; Glycon, Luckenwalde, Germany) in Dulbecco’s phosphate buffered saline (DPBS), pH 7.4 (Biochrom), and 10 μM all-trans retinal (Sigma-Aldrich) for 4–6 h, the photoreceptors were solubilized and subsequently purified by 1D4 immunoaffinity chromatography. For this purpose, 1D4-antibody-resin was prepared according to Oprian et al. (36). Immobilized protein was eluted with DPBS buffer containing 400 μM 1D4-peptide (T-E-T-S-Q-V-A-P-A, GenScript, NY) and 0.03% DDM, whereas for pH-dependent measurements concentrated ReaChR samples were diluted in 10 mM citric acid pH 5.0, 100 mM NaCl, 0.03% DDM, and 10 mM TRIS pH 9.0, 100 mM NaCl, 0.03% DDM, respectively. For retinal extraction experiments, protein was expressed in Pichia pastoris and purified in buffer containing 10 mM TRIS (pH 7.4), 100 mM NaCl, and 0.03% DDM as described in the literature (23, 37, 38).
Ultraviolet-visible absorption spectroscopy
Absorption spectra and slow kinetic traces were recorded by a Cary 50 Bio spectrophotometer (Varian, Palo Alto, CA) at 22°C. Initial dark state (IDA) spectra were recorded with protein purified under safelight. ReaChR C168S was illuminated with a 530 nm light of 1 × 1020 photons m−2 s−1 (Luxeon LED; Phillips, Amsterdam, the Netherlands) while photon flux was decreased to 5.6 × 1019 photons m−2 s−1 for dual-color experiments. If necessary, brighter blue-light pulses (1 × 1020 photons m−2 s−1) were applied using an LED (400 nm; Thorlabs, Newton, NJ). Flash-photolysis experiments and data processing were performed as described in Wietek et al. (39). For this purpose, samples with an optical density of A280 = 1.0 were excited with green flashes (530 nm, ∼10 ns, 5 mJ/flash). Global analysis is based on a sequential model with four or five spectral components. Absolute spectra of ReaChR wild-type at pH 5.0 were calculated based on the IDA spectrum (see Fig. 2 A) and the corresponding nonnormalized evolutionary associated difference spectra (EADS) (data not shown; compare to Fig. S1). We assumed 10% photoreceptor activation by the laser flash. Cryostatic ultraviolet-visible (UV-vis) measurements were performed using a sample chamber placed inside a cryostat (DN; Oxford Instruments, Abingdon, UK) according to the following protocol: After 2 min of illumination (∼530 nm) and succeeding 30 min of dark adaptation at minimum 20°C, sample was cooled down to the desired temperature. Before the next measuring cycle, the sample was heated up again to a minimum of 20°C. Recorded spectra were scaled by normalizing to the dark state chromophore absorption band.
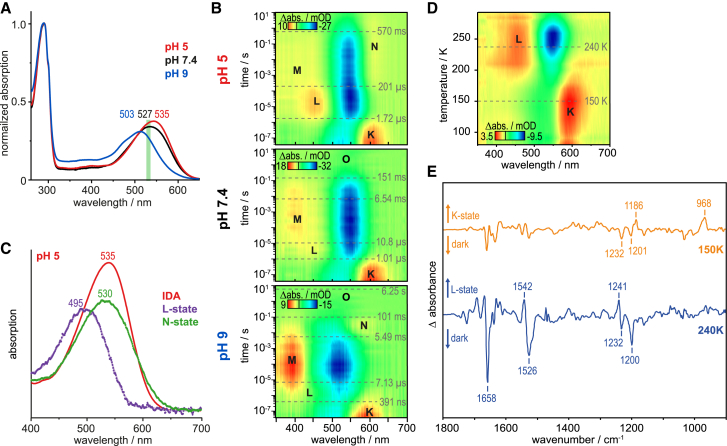
Figure 2.
Spectral properties of recombinant ReaChR. (A) IDA spectra of purified ReaChR at pH 5.0, 7.4, and 9.0 (normalized to 280 nm). (B) Transient absorption changes induced by green laser flashes (10 ns, 530 nm, 5 mJ) were measured in saline detergent (0.03% DDM) solution at pH 5.0 (top, 100 mM NaCl, 10 mM citric acid), 7.4 (middle, DPBS), and 9.0 (bottom, 100 mM NaCl, 10 mM TRIS). Half-life times (t1/2) are highlighted in the contour plots. Five photocycle-intermediates K, L, M, N (not seen at pH 7.4), and O were identified. L and M are enriched at pH 5 and 9, respectively. Acidic pH decelerates photocycle kinetics. (C) Calculated absolute spectra of the L- and N-states of ReaChR at pH 5. Dots resemble raw data points and solid lines were added for visual guidance. (D) Cryostatic UV-vis measurements (light-dark, pH 7.4) of ReaChR under continuous illumination reveal maximal accumulation of the K- and L-states at 150 and 240 K, respectively. (E) FTIR difference (light-dark) of the photostationary states under continuous illumination at 150 (orange) and 240 K (blue) at pH 7.4. Imposing HOOP vibration at 968(+) cm−1 points toward a distorted 13-cis chromophore in the K-state, while cofactor strain is almost completely relieved in the upcoming L-state.
Fourier-transform infrared spectroscopy
Rehydrated samples of ReaChR at pH 7.4 in DPBS were placed between two BaF2 windows and analyzed by an ifs66v/s Fourier-transform infrared (FTIR) spectrometer (Bruker Optics, Karlsruhe, Germany) with a liquid nitrogen-cooled mercury cadmium telluride detector (Kolmar Technologies, Newburyport, MA) and an 1850 cm−1 optical cutoff filter. Cryostatic FTIR measurements were conducted similar to cryo-UV-vis measurements. Corresponding spectra were recorded with a 200 kHz sampling rate and with a spectral resolution of 2 cm−1. Difference spectra were corrected for baseline drifts using a spline algorithm and the baseline-correction mode implemented in the OPUS 6.5 software package (Bruker Optics). The photostationary state was defined as the steady state of the kinetics of the strong absorption band at 1660 cm−1. Spectra were scaled by normalizing to the amide II band in the absolute spectra at ∼1550 cm−1.
Retinal extraction and separation by high-pressure liquid chromatography
Purified protein was illuminated for 10 min with green light (∼530 nm LED) and kept in the dark for 10 min. Chromophore extraction was achieved by addition of ice-cold ethanol (delay time ≤ 2 s). After 2 min, the extracted retinal was dissolved in heptane containing 7% diethyl ether (w/w). After 2 min, phase separation was achieved by centrifugation (1 min, 2000 rpm). All working steps, except sample illumination, were conducted under dim red light and at room temperature. Separation of the retinal isomers in the heptane/diethyl ether mixture was performed using a LC-20AD high-pressure liquid chromatography (HPLC) device (Shimadzu, Kyoto, Japan) equipped with a ReproSil 70 Si, 5 μm column (Dr. Maisch, Ammerbuch-Entringen, Germany). Isomer ratio was determined by peak integration. The chromatogram was corrected for baseline-drifts.
Results
Photocurrent properties in HEK293 cells
In 2013, Lin et al. (16) reported unusual, non-rhodopsin-like action spectra for ReaChR with maximal activity at 590 nm excitation for the peak and even 630 nm for the stationary photocurrent. However, subsequent studies on ReaChR revealed that this ChR is most sensitive to green light (40, 41). Based on this discrepancy, we reinvestigated ReaChR by electrophysiological recordings in HEK293 cells. First, flash-induced photocurrents (10 ms excitation) were recorded at different wavelengths and three extracellular pH-values (pHe = 5.0, 7.2, and 9.0) revealing maximal sensitivity to 532 nm light (pHe 7.2), and small shifts after pHe change. These action spectra were rhodopsin-shaped without any particular unusual characteristics (Fig. 1 B). Upon 1 s illumination, the shape of the photocurrent depends on the intensity of the actinic light as known from CrChR1 (25), CrChR2 (4), and relatives. Titration of the 530 nm activation light (Fig. 1 C, green) revealed that peak photocurrents were half-maximal at a photon irradiance of 2.68 × 1013 mm−2 s−1 (0.010 mW mm−2), while stationary currents saturated at much lower light intensities and were half-maximal already at 0.005 mW mm−2. Interestingly, the curve for the peak current decreased (i.e., kinked) at light intensities already a decade away from saturation (black arrow). Second, the stationary currents decreased again at higher intensities, which was much more pronounced in ReaChR compared with CrChR2 (10). In comparison, the peak current sensitivity at 600 nm (Fig. 1 C, orange) is shifted to four-times higher intensities (I50 = 0.042 mW mm−2; 1.27 × 1014 photons mm−2 s−1), in agreement with a reduced absorption coefficient at 600 nm as seen from the flash-induced action spectrum for pHe 7.2 (Fig. 1 B). However, the reduced slope of the stimulus response curve at intensities 10-fold below saturation is not visible for 600 nm activation (Fig. 1 C). Fig. 1 D shows a direct comparison of photocurrents recorded from the same cell upon activation with 530 and 600 nm light at equal photon irradiance (100%).
In accordance with the unusual shape of the light titration curves at high photon densities, action spectra of the steady-state currents were strongly intensity dependent. At low light intensities of 1 and 5% the spectra peaked at 550 nm, whereas maxima were shifted to 600 nm for 50% and 610 nm for 100% activation light, revealing a complex secondary photochemistry at high photon irradiance (Fig. 1 E).
Spectral properties of recombinant ReaChR
To determine the absorption properties of the dark state, recombinant ReaChR was purified under safelight (>600 nm) from HEK293T and absorption spectra were recorded (Fig. 2 A). The IDA showed maximal absorption at 535 nm at pH 5.0, 527 nm at pH 7.4, and 503 nm at pH 9.0, the latter with a shoulder at ∼450 nm not seen in the action spectra. The extinction coefficient of chromophore absorption increased gradually upon acidification. Next, recombinant ReaChR was exposed to brief green (530 nm) laser pulses of ∼10 ns and absorption changes between 40 ns and 10 s after flash were monitored (Fig. 2 B). Normalized EADS (Fig. S1) and their half-life times (t1/2) were extracted using a global fit routine. Dissected intermediates were designated according to the bacteriorhodopsin (BR) nomenclature. The dark state (D) is converted into a distinctive red-shifted K-state (seen at ∼600 nm) that rises faster than our time-resolution and decays with t1/2 = 1.01 or 1.72 μs at pH 7.4 and 5.0, respectively. At pH 9.0, K-state decay is 2.5 times faster (t1/2 = 391 ns) compared to pH 7.4. At pH 5.0 this early photoproduct evolves into a blue-shifted L-state with maximal absorption at 460 nm and shows roughly 20 times prolonged lifetime compared to neutral pH. Such an L-state was not reported for ChR2 (22). The subsequent well-known M-intermediate with deprotonated chromophore prevails over several orders of time and exhibits a broad unstructured absorption profile with small amplitude at pH 5.0 and 7.4 but high population at pH 9.0. From previous studies on ChR2, it is known that, as for BR, M is actually composed of an early M1 and a late M2 state (42, 43, 44). The decay of M is biphasic, and accelerated at pH 9.0 (Fig. 2 B, bottom). At pH 5.0 and 9.0, but not at pH 7.4, a late photointermediate appears with positive absorption at ∼600 nm on a millisecond timescale, referred to as N-state. At pH 9.0, the N-intermediate decays with t1/2 = 101 ms to a state with a spectral signature hardly discriminable from the dark state spectrum. This intermediate, designated as O-state, finally recovers to D with t1/2 = 6.25 s at pH 9.0 (Fig. 2 B, bottom). In neutral and acidic conditions these last transitions are either poorly resolved or/and highly decelerated, thus not completed during the observation window of 10 s (Fig. 2 B, middle and top). To disentangle photocycle intermediates, absolute spectra of the L- and N-state at pH 5.0 were determined by adding the appropriate nonnormalized EADS (data not shown; and compare to Fig. S1, top) to the IDA spectrum (Fig. 2 A). The calculated L-state spectrum shows maximal absorption at 495 nm while N-state peaks at 530 nm similar to the dark state but with a broader absorption profile toward longer wavelengths (Fig. 2 C).
To investigate structural features associated with the newly identified L-state, ReaChR was analyzed at cryogenic conditions by UV-vis and FTIR spectroscopy. The temperature-versus-wavelength plot shows that up to 200 K, the K-state is enriched, whereas between 200 K and room temperature, L is the dominant photoproduct (Fig. 2 D). FTIR spectra were recorded at 150 and 240 K (Fig. 2 E). The K-minus-D difference spectrum exhibits relatively small changes with the exception of a typical band pattern within the retinal fingerprint region with two negative bands at 1232 and 1201 cm−1 and a positive band at 1186 cm−1, indicative of a net isomerization of the retinal from all-trans to 13-cis (45). A pronounced hydrogen out-of-plane (HOOP) vibration at 968(+) cm−1 indicates a distorted 13-cis chromophore structure (46, 47) in the early state, which relaxes in the upcoming L state. During transition from K- to L-, the amide I region gains intensity with a strong negative band at 1658 cm−1, suggesting major backbone rearrangements probably associated to channel pregating. The L spectrum possesses a unique band at 1241(+) cm−1, which is caused either by stretching vibrations of C12–C13 of the retinal (48) or N–H in-plane bending of the retinal Schiff base (RSB) (49). The blue-shifted L-state absorption maximum (Fig. 2 C) is reflected in the upshift of the C=C stretching mode from 1526(−) to 1542(+) cm−1 in the FTIR spectrum of the D to L transition (50, 51). The presence of a strong positive ethylenic band hints at a protonated RSB (RSBH+) in L, as only small positive ethylenic bands were observed in states with a deprotonated RSB in channelrhodopsin-1 from Chlamydomonas augustae (CaChR1) (52, 53) and CrChR2 (54).
Photoconversion of intermediate states
To investigate the conducting state(s) and subsequent late photointermediates, we generated slow-cycling mutants of ReaChR by modification of the DC-pair residues C168 and D196 (CrChR2 C128 and D156), which play a crucial role for the channel gating kinetics in CrChR2 (55, 56). We introduced C168A and C168S and confirmed the slowed photocurrent decline by electrical analysis, as illustrated in Fig. S2. Wild-type photocurrent decayed with τoff = 181 ± 22 ms (monoexponential fit), whereas the closing kinetics for C168A and C168S were two orders of magnitude slower with τoff = 19.7 and 28.1 s, respectively (Fig. S2). On average, the C168S photocurrent amplitudes were <20% of the wild-type and C168A currents at similar conditions, thus we used the C168A mutant for further electrical analysis.
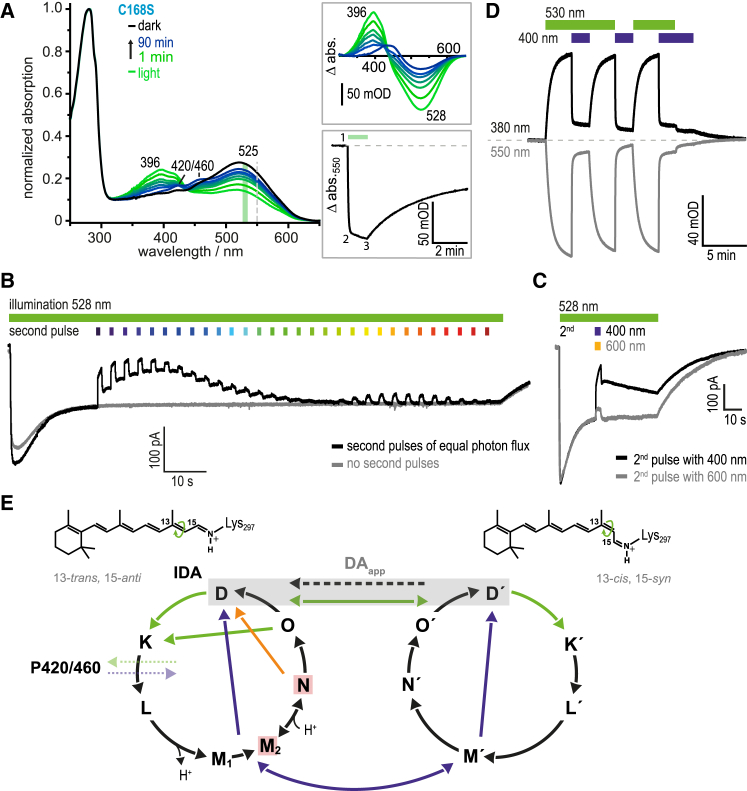
Next, we recorded absorption spectra of purified ReaChR-C168S in the dark and after illumination with 530 nm light (60 s). The IDA spectrum shows maximal absorption at 525 nm, while upon illumination it converts to the UV-absorbing M-state (396 nm) (Fig. 3 A and top inset), which decays biexponentially with time constants in the early to late minute range. Additionally, slowly appearing photoproducts are seen at 420 and 460 nm (P420/460) (Fig. 3 A and top inset), which do not thermally reconvert to the IDA during the entire observation window of 90 min but are fully photoconvertible by UV-light (data not shown). These P420/460 side products are accumulated solely under prolonged illumination (Fig. 3 A, bottom inset, 2→3) and were assigned to inactive side products of the photocycle in analogy to our conclusions drawn for ChR2-C128T (54). Strikingly, even extended green illumination (530 nm light for 60 s) bleaches the absorption at ∼520 nm to maximally ∼55%, suggesting that either a fraction of the protein is not photoactive, or more likely, a green-absorbing photocycle intermediate as the N-state is still prevailing in continuous light.
Figure 3.
Photoconversion of intermediate states and photocycle model. (A) Absorption spectra of ReaChR-C168S recorded for the IDA and after excitation (60 s, 530 nm). Difference spectra were calculated (light-dark, top inset). Upon prolonged illumination, M-state (λmax = 396 nm) is accumulated predominantly, which completely decays within 90 min, but substantial absorption at ∼520 nm (∼55%) indicates the presence of another green-absorbing intermediate (N-state). Chromophore bleaching monitored at 550 nm involves two processes (bottom inset), whereby the faster component is attributed to M-state formation (1→2) and the slower one to formation of minor blue-shifted species seen at 420 and 460 nm (P420/460) (2→3). (B) Photocurrents of HEK cells expressing ReaChR-C168A under continuous illumination with green light and additional stronger light pulses (380–670 nm) inducing current inactivation. The 400 nm light effectively inactivates currents with slow recovery, whereas inactivation at 600 nm is smaller and instantaneously reversible with green light. (C) These current traces show more clearly that application of a single blue-light pulse on top of green background light causes current inactivation and slow recovery, whereas a 600 nm light pulse causes smaller inactivation but fast recovery. (D) Recombinant ReaChR-C168S illuminated with green (530 nm, weak) and/or blue (400 nm, strong) light, and rise and decay of the M- (380 nm) and dark states (550 nm) were monitored by single kinetic traces. Green-light illumination induces formation of M and dark state (D) bleaching in parallel, while blue light reverses this process, indicative for an M→D shortcut. (E) Proposed symmetrical two-cycle model of ReaChR. One cycle is based on the 13-trans, 15-anti retinal configuration (anti-cycle) while the other starts from 13-cis, 15-syn (syn-cycle); the equilibrium of both forms the apparent dark state (DAapp, gray background), while the IDA is supposed to consist of 100% all-trans retinal. Upon illumination with green light, D converts to a red-shifted K-like intermediate, succeeded by the blue-shifted states L and M. The deprotonated M-state decays biexponentially (M1 and M2) and is reprotonated during N-state formation. N decays to an intermediate that is spectrally not distinguishable from the dark state (O-state). (Colored arrows) Proposed photoinduced pathways between the intermediates and both cycles; (black arrows) thermal conversions. The conducting states are assigned to the M2- and N-equilibrium (red box). The syn-cycle consists of the same spectral states marked by a hyphen and undergoes the same protonations; corresponding shortcuts and the M-substates are omitted for clarity. Upon prolonged illumination of slow-cycling mutants, side-products (P420/460) are accumulated, which could be photoconverted by UV-light (data not shown).
With the slow-cycling mutants at hand we electrically analyzed the photoconversion of the conducting state(s) as previously reported for slow-cycling ChR2- and C1V1-variants (15). Continuous illumination of ReaChR-C168A with 528 nm light accumulated the open state(s) after some inactivation and established a steady-state current level. Subsequently, more intense light pulses of different wavelengths but equal photon flux were applied and showed that the steady-state photocurrent can be reduced by UV and orange light. While the inactivation with orange light was smaller and immediately reversed by the green basal illumination, UV or blue-light pulses inactivated the protein to <50% of the steady-state current level (Fig. 3 B) and only a small fraction recovered under green background light within 1 s. The full stationary current level was restored only after a longer time in the minute range under green-light illumination (Fig. 3 C), suggesting that ReaChR was converted into a low- or nonconducting photocycle product by UV and blue light. To elucidate underlying photochemistry under continuous illumination, recombinant ReaChR-C168S was exposed to a series of parallel and sequential light pulses of green (530 nm) and UV (400 nm) light (Fig. 3 D), while M-state rise and decay was monitored by absorbance changes at 380 nm and dark state absorption was monitored at 550 nm. Under permanent green illumination, M was accumulated but was almost completely converted to D when UV-light was interspersed, although the full recovery after cessation of light was slow and occurred on timescales of minutes.
Discussion
Lin et al. (16) described the VcChR1 variant ReaChR that exhibits large photocurrents and shows steady-state current action spectra that substantially differ from rhodopsin absorption spectra. Such spectra are either caused by multiple overlaying photoreceptors—which can be excluded here—or an extended secondary photochemistry. This is the reason why in vivo action spectra that led to the original identification of rhodopsin as the phototaxis photoreceptor in Chlamydomonas were recorded at low light intensities (threshold spectra) (57) or as flash spectra for the correlation between photocurrents and photoreceptor species (58). We confirmed the non-Gaussian-shaped action spectra for ReaChR, but this only holds true for extended illumination periods of a few seconds and high intensities. Low intensity or flash action spectra resemble the absorption spectra of recombinant protein (compare to Figs. 1 B and 2 A) and are similar to the parent VcChR1 (13) and the related C1V1-hybrid (15, 41). The reduced stationary photocurrents under continuous green light can be explained in three ways, namely by photochemical conversion of one or more photocycle intermediates back to the dark state(s), photochemical conversion between intermediates of the syn- and anti-cycle, and conversion of one or more photocycle intermediates into an inactive or less active side product. This interpretation raises the question of the nature of the activated photointermediate and of the secondary photoproducts that are formed more efficiently compared to other ChRs.
Photochemical back reactions between photocycle intermediates and dark states were previously proposed for wild-type ChR2 (22, 23) and its step-function opsin (SFO) variants (54, 55, 56), although the practical relevance of this secondary photoreaction is low for wild-type ChR2. For ChR2-C128T, side reactions into inactive, slowly recovering species that are not part of the bona fide photocycle were also previously reported (54, 59). Within our study, we identified six states for ReaChR designated as the following: D- (dark), K-, L-, M- (M1 and M2), N-, and O-states that are expected to be consecutive in analogy to BR, with D- and O- having similar spectral properties (Fig. 2 B). Remarkably, a state corresponding to the L-state has not been identified for CrChR2 and CaChR1 (22, 23, 60), but was just recently reported for channelrhodopsin-2 from Platymonas subcordiformis (PsChR2) (32, 33) and the anion-conducting channelrhodopsin-1 from Proteomonas sulcata (PsACR1) (P460) (39) and Guillardia theta (GtACR1) (61). The hypsochromic shift from the K-intermediate absorbing at 600 nm (difference spectrum, Fig. 2 B) to the L-state at 495 nm observed at pH 5.0 (absolute spectrum, Fig. 2 C) is not a result of the protonation state of the RSB but rather from an altered distance between RSBH+ and counterion-complex upon chromophore relaxation. While the 13-cis retinal molecule is twisted and distorted in the early K-state, the cofactor strain is almost relieved in the upcoming L-state, as deduced from the lack of the HOOP band in the L-state IR-spectrum (Fig. 2 E). Most likely, chromophore relaxation is achieved by rotation of the C13=C14 and C15=N bonds, as it occurs in BR (62, 63). As a consequence, the RSBH+ is shifted closer to the water-counterion complex compared to the configurations both in K and D, thus stabilizing the electronic ground state of L and leading to a blue-shifted absorption spectrum (62).
The proposition of the L-state as the main photoreactive intermediate under continuous illumination is further supported by two findings: First, the L-state is strongly accumulated under continuous illumination compared to single-turnover conditions (compare to Fig. 2, B, middle, and D). Second, photochemical inactivation of the photocurrents under continuous green light at pH 5.0 is even more efficient compared to pH 7.2 (Fig. 1 E, bar diagram), in line with an enrichment of L at low pH as seen for the recombinant protein in single flash experiments (Fig. 2 B, top). Because calculated absolute spectra of N- and dark states are almost identical (Fig. 2 C), L-state conversion by blue/green light should be efficient whereas N to D conversion appears less efficient because green light activates N and D simultaneously in the opposite direction.
It might be worthwhile to recapitulate that ReaChR-C168S accumulates mostly M (probably M2) during extended illumination, similar to what was reported for the SFOs ChR2-C128S or D156A (14, 55), but substantial absorption at 520 nm remains (Fig. 3 A); this supports the idea of an equilibrium between M and N, as proposed for ChR2 (P390 and P520) (22, 23). The observed small and fully reversible inactivation with orange light (600 nm) (Fig. 3 C) is explained by the red tail of the N-state (Fig. 2 C). The last question remaining is why 400 nm bleaching is not immediately reversed by green background light. Similar observations were previously reported for the SFO C128T (59) and the stable step-function opsin ChR2-C128S/D156A (14). The slow recovery of the steady state is explained by the two-cycle model based on the two C15=N retinal isomers. Assuming a symmetrical model with similar spectral intermediates in each cycle, photoactivation of the dark state D with 13-trans, 15-anti conformation produces K, L, M, and N intermediates with 13-cis, 15-anti. Thus, we consider this cycle as anti-cycle because the C15=N bond should be anti throughout the whole cycle. Accordingly, the cycle starting from a dark state with 13-cis, 15-syn (D′) is designated syn-cycle. Hence, either a photochemical transition from an M-state of the anti-cycle to an M-state of the syn-cycle occurs or M is converted into an inactive UV-absorbing state, possibly with another cis-isomer than 13-cis, e.g., 9- and 11-cis, which are formed in the ChR2-C128T mutant upon prolonged illumination (28).
Recently, we have reported for the bimodal switchable Histidine Kinase Rhodopsin-1 from C. reinhardtii that UVA-light may not only cause a sole isomerization of the RSB around C13=C14 bond, it can also promote a simultaneous isomerization of both the C13=C14 and the C15=N double bond, preferentially if the RSB chromophore is deprotonated (64). Such a double isomerization would convert the 13-cis, 15-anti M-state into a 13-trans, 15-syn M-state that thermally converts to 13-cis, 15-syn, the expected dark state of the syn-cycle. Because the open state(s) of the syn-cycle is assumed to be less conductive with altered ion selectivity in CrChR2 (27), and both the absorption cross section and the quantum efficiency for a protonated 13-cis, 15-syn chromophore is anticipated to be reduced compared to a 13-trans, 15-anti RSBH+, the slow recovery of the steady-state photocurrent after a UV-light pulse could be explained by the transition from the anti- to the syn-cycle. However, recombinant ReaChR-C168S shows a long-lasting chromophore bleaching and conversion of the dark state(s) into P420/460, which are not representatives of the syn- or anti-cycle. P420/460 recover to the dark state only upon UV-illumination (data not shown). Thus, we consider them as photochemical side products as previously proposed for ChR2-C128T (54).
ReaChR photocycle model
Based on our study, we conclude that the non-rhodopsin action spectrum for steady-state currents of ReaChR is caused by a complex orchestration of secondary photochemical reactions starting from the newly identified L-state and its subsequent M-states. We propose a symmetrical two-cycle model as recently discussed for CrChR2 (30) with two dark states (D and D′) establishing the apparent dark state (DAapp), and two open states (N and N′) with different conductivities and selectivities, which are in a fast equilibrium with the preceding M2-state (Fig. 3 E), to describe photochemical events as they occur under both single-turnover conditions and continuous illumination. Under continuous green illumination, the initial dark state (IDA) consisting of 100% all-trans retinal is converted to an equilibrium between the two cycles (i.e., conducting states), which is shifted when a second blue- or green-light pulse is applied. Remarkably, blue light drives both intra- and intercircular shortcuts. While the intracircular shortcut is based on C13=C14 isomerization, the latter one is assumed to be based on a C13=C14, C15=N double-isomerization of the retinal chromophore and a conversion between M to M′ or vice versa. However, strictly speaking, a single C15=N anti/syn or syn/anti isomerization cannot be excluded. In such hypothetical cases, isomerization would produce 13-cis, 15-syn or 13-trans, 15-anti retinal Schiff base species that both are expected to be protonated under formation of the O-states. Based on our experiments, it is unclear whether the cycle crossover is purely light-induced or occurs partially thermally. Thermal branching was reported to happen highly efficiently in other microbial rhodopsins such as BR and Anabaena sensory rhodopsin (65), thus could be also possible for ChRs and should be part of further investigations. More evidence for a two-cycle model was given by Szundi et al. (32, 33), who reported that the cation-conducting PsChR2 runs through two cycles in parallel, while 30% of excited protein follow cycle 1 and 70% cycle 2. Retinal extraction and HPLC experiments of ReaChR wild-type in the DAapp-state revealed a proportion of 78:22 of all-trans:13-cis retinal (Fig. S3). This ratio is in line with the composition in CrChR2 (28, 45, 54) and would allow the assignment of a 15-anti cycle with the C15=N bond being always in anti-configuration (previously named “trans-cycle”, according to the 13-trans conformation of the dark state) and a syn-cycle with the C15=N conformation always in syn-conformation. However, an exact assignment would require Raman or NMR experiments, respectively. A better understanding of side reactions into an inactive and only slowly recovering state also requires more extensive studies, which are not trivial due to the heterogeneity of the photoproduct mixture. In general, a deeper knowledge of the photochemistry of ReaChR—a frequently used optogenetic tool—allows for a purposeful implementation within neurophysiological experiments, especially in cases where complex illumination protocols are requested.
Author Contributions
B.S.K. expressed and purified protein, and performed and analyzed UV-vis spectroscopic measurements; C.G. conducted and evaluated electrophysiological experiments; J.C.D.K. performed FTIR and cryo-UV-vis measurements as well as retinal extraction and HPLC experiments; F.S. and T.P.S. provided material and experimental expertise; P.H. and F.J.B. designed and interpreted experiments; and B.S.K. and P.H. wrote the article with further contributions from all authors. All authors reviewed the results and approved the final version of the article.
Acknowledgments
We thank Rolf Hagedorn, Altina Klein, Maila Reh, Manija Kazmi, and He Tian for excellent technical assistance as well as John Lin for providing the plasmid and fruitful discussions. Furthermore, we are grateful for proofreading of the article by Jonas Wietek.
This work was supported by the Deutsche Forschungsgemeinschaft via Sonderforschungsbereich No. 1078, project No. B2 (P.H.) and B5 (F.J.B.), and the Cluster of Excellence 314 “Unifying Concepts in Catalysis” (project No. E4/D4 to P.H.).
Editor: Elsa Yan.
Footnotes
Benjamin S. Krause and Christiane Grimm contributed equally to this work.
Franziska Schneider’s present address is University Heart Centre Freiburg - Bad Krozingen and Medical School, University of Freiburg, Freiburg, Germany.
Three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30155-8.
Contributor Information
Benjamin S. Krause, Email: benjamin.krause@biologie.hu-berlin.de.
Peter Hegemann, Email: hegemann@rz.hu-berlin.de.
Supporting Material
References
- 1.Nagel G., Ollig D., Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 2.Sineshchekov O.A., Jung K.-H., Spudich J.L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T., Yamasaki K., Takahashi T. Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem. Biophys. Res. Commun. 2003;301:711–717. doi: 10.1016/s0006-291x(02)03079-6. [DOI] [PubMed] [Google Scholar]
- 4.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyden E.S., Zhang F., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 6.Li X., Gutierrez D.V., Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishizuka T., Kakuda M., Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Wietek J., Wiegert J.S., Hegemann P. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 9.Berndt A., Lee S.Y., Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J.Y., Lin M.Z., Tsien R.Y. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel G., Brauner M., Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 12.Wietek J., Prigge M. Enhancing channelrhodopsins: an overview. In: Kianianmomeni A., editor. Optogenetics: Methods and Protocols, Methods in Molecular Biology. Springer Science+Business Media; New York: 2016. pp. 141–165. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F., Prigge M., Deisseroth K. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yizhar O., Fenno L.E., Deisseroth K. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prigge M., Schneider F., Hegemann P. Color-tuned channelrhodopsins for multiwavelength optogenetics. J. Biol. Chem. 2012;287:31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin J.Y., Knutsen P.M., Tsien R.Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inagaki H.K., Jung Y., Anderson D.J. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods. 2014;11:325–332. doi: 10.1038/nmeth.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaigneau E., Ronzitti E., Emiliani V. Two-photon holographic stimulation of ReaChR. Front. Cell. Neurosci. 2016;10:234. doi: 10.3389/fncel.2016.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao P.-Y., Tsai C.-L., Chiang A.-S. Non-invasive manipulation of Drosophila behavior by two-photon excited red-activatable channelrhodopsin. Biomed. Opt. Express. 2015;6:4344–4352. doi: 10.1364/BOE.6.004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sengupta A., Chaffiol A., Duebel J. Red-shifted channelrhodopsin stimulation restores light responses in blind mice, macaque retina, and human retina. EMBO Mol. Med. 2016;8:1248–1264. doi: 10.15252/emmm.201505699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst O.P., Sánchez Murcia P.A., Hegemann P. Photoactivation of channelrhodopsin. J. Biol. Chem. 2008;283:1637–1643. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 22.Ritter E., Stehfest K., Bartl F.J. Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J. Biol. Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bamann C., Kirsch T., Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 24.Nikolic K., Grossman N., Degenaar P. Photocycles of channelrhodopsin-2. Photochem. Photobiol. 2009;85:400–411. doi: 10.1111/j.1751-1097.2008.00460.x. [DOI] [PubMed] [Google Scholar]
- 25.Hegemann P., Ehlenbeck S., Gradmann D. Multiple photocycles of channelrhodopsin. Biophys. J. 2005;89:3911–3918. doi: 10.1529/biophysj.105.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berndt A., Prigge M., Hegemann P. Two open states with progressive proton selectivities in the branched channelrhodopsin-2 photocycle. Biophys. J. 2010;98:753–761. doi: 10.1016/j.bpj.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider F., Gradmann D., Hegemann P. Ion selectivity and competition in channelrhodopsins. Biophys. J. 2013;105:91–100. doi: 10.1016/j.bpj.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritter E., Piwowarski P., Bartl F.J. Light-dark adaptation of channelrhodopsin C128T mutant. J. Biol. Chem. 2013;288:10451–10458. doi: 10.1074/jbc.M112.446427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lórenz-Fonfría V.A., Schultz B.-J., Heberle J. Pre-gating conformational changes in the ChETA variant of channelrhodopsin-2 monitored by nanosecond IR spectroscopy. J. Am. Chem. Soc. 2015;137:1850–1861. doi: 10.1021/ja5108595. [DOI] [PubMed] [Google Scholar]
- 30.Bruun S., Stoeppler D., Stehfest K. Light-dark adaptation of channelrhodopsin involves photoconversion between the all-trans and 13-cis retinal isomers. Biochemistry. 2015;54:5389–5400. doi: 10.1021/acs.biochem.5b00597. [DOI] [PubMed] [Google Scholar]
- 31.Becker-Baldus J., Bamann C., Glaubitz C. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2015;112:9896–9901. doi: 10.1073/pnas.1507713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szundi I., Bogomolni R., Kliger D.S. Platymonas subcordiformis channelrhodopsin-2 (PsChR2) function: II. Relationship of the photochemical reaction cycle to channel currents. J. Biol. Chem. 2015;290:16585–16594. doi: 10.1074/jbc.M115.653071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szundi I., Li H., Kliger D.S. Platymonas subcordiformis channelrhodopsin-2 function: I. The photochemical reaction cycle. J. Biol. Chem. 2015;290:16573–16584. doi: 10.1074/jbc.M114.631614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karnik S.S., Sakmar T.P., Khorana H.G. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc. Natl. Acad. Sci. USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markwardt M.L., Kremers G.-J., Rizzo M.A. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oprian D.D., Molday R.S., Khorana H.G. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc. Natl. Acad. Sci. USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luck M., Mathes T., Hegemann P. A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J. Biol. Chem. 2012;287:40083–40090. doi: 10.1074/jbc.M112.401604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruun S., Naumann H., Hildebrandt P. The chromophore structure of the long-lived intermediate of the C128T channelrhodopsin-2 variant. FEBS Lett. 2011;585:3998–4001. doi: 10.1016/j.febslet.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Wietek J., Broser M., Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J. Biol. Chem. 2016;291:4121–4127. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawydow A., Gueta R., Kittel R.J. Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications. Proc. Natl. Acad. Sci. USA. 2014;111:13972–13977. doi: 10.1073/pnas.1408269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider F., Grimm C., Hegemann P. Biophysics of channelrhodopsin. Annu. Rev. Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 42.Lórenz-Fonfría V.A., Heberle J. Channelrhodopsin unchained: structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Spudich J.L., Sineshchekov O.A., Govorunova E.G. Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta. 2014;1837:546–552. doi: 10.1016/j.bbabio.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lórenz-Fonfría V.A., Resler T., Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nack M., Radu I., Heberle J. The retinal structure of channelrhodopsin-2 assessed by resonance Raman spectroscopy. FEBS Lett. 2009;583:3676–3680. doi: 10.1016/j.febslet.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 46.Smith S.O., Lugtenburg J., Mathies R.A. Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J. Membr. Biol. 1985;85:95–109. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- 47.Eyring G., Curry B., Mathies R. Assignment and interpretation of hydrogen out-of-plane vibrations in the resonance Raman spectra of rhodopsin and bathorhodopsin. Biochemistry. 1982;21:384–393. doi: 10.1021/bi00531a028. [DOI] [PubMed] [Google Scholar]
- 48.Smith S., Pardoen J.A., Mathies R.A. Vibrational analysis of the 13-cis-retinal chromophore in dark-adapted bacteriorhodopsin. J. Phys. Chem. 1987;91:804–819. [Google Scholar]
- 49.Hashimoto K., Choi A.R., Kandori H. Low-temperature FTIR study of Gloeobacter rhodopsin: presence of strongly hydrogen-bonded water and long-range structural protein perturbation upon retinal photoisomerization. Biochemistry. 2010;49:3343–3350. doi: 10.1021/bi100184k. [DOI] [PubMed] [Google Scholar]
- 50.Aton B., Doukas A.G., Ebrey T.G. Resonance Raman studies of the purple membrane. Biochemistry. 1977;16:2995–2999. doi: 10.1021/bi00632a029. [DOI] [PubMed] [Google Scholar]
- 51.van den Berg R., Du-Jeon-Jang H.C., El-Sayed M.A. Subpicosecond resonance Raman spectra of the early intermediates in the photocycle of bacteriorhodopsin. Biophys. J. 1990;58:135–141. doi: 10.1016/S0006-3495(90)82359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogren J.I., Yi A., Rothschild K.J. Proton transfers in a channelrhodopsin-1 studied by Fourier transform infrared (FTIR) difference spectroscopy and site-directed mutagenesis. J. Biol. Chem. 2015;290:12719–12730. doi: 10.1074/jbc.M114.634840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muders V., Kerruth S., Schlesinger R. Resonance Raman and FTIR spectroscopic characterization of the closed and open states of channelrhodopsin-1. FEBS Lett. 2014;588:2301–2306. doi: 10.1016/j.febslet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 54.Stehfest K., Ritter E., Hegemann P. The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J. Mol. Biol. 2010;398:690–702. doi: 10.1016/j.jmb.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 55.Bamann C., Gueta R., Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 56.Berndt A., Yizhar O., Deisseroth K. Bi-stable neural state switches. Nat. Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 57.Foster K.W., Saranak J., Nakanishi K. A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature. 1984;311:756–759. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- 58.Harz H., Hegemann P. Rhodopsin-regulated calcium currents in Chlamydomonas. Nature. 1991;351:489–491. [Google Scholar]
- 59.Schoenenberger P., Gerosa D., Oertner T.G. Temporal control of immediate early gene induction by light. PLoS One. 2009;4:e8185. doi: 10.1371/journal.pone.0008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sineshchekov O.A., Govorunova E.G., Spudich J.L. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sineshchekov O.A., Li H., Spudich J.L. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc. Natl. Acad. Sci. USA. 2016;113:E1993–E2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanyi J.K., Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M1, M2, and M2′ intermediates of the photocycle. J. Mol. Biol. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- 63.Schobert B., Cupp-Vickery J., Lanyi J. Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal. J. Mol. Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 64.Luck M., Bruun S., Hildebrandt P. Photochemical chromophore isomerization in histidine kinase rhodopsin HKR1. FEBS Lett. 2015;589:1067–1071. doi: 10.1016/j.febslet.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 65.Kawanabe A., Furutani Y., Kandori H. Photochromism of Anabaena sensory rhodopsin. J. Am. Chem. Soc. 2007;129:8644–8649. doi: 10.1021/ja072085a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.