Abstract
In the past decade, microRNAs (miRNAs) have emerged as key regulators of circulating levels of lipoproteins. Specifically, recent work has uncovered the role of miRNAs in controlling the levels of atherogenic low-density lipoprotein LDL (LDL)-cholesterol by post-transcriptionally regulating genes involved in very low-density lipoprotein (VLDL) secretion, cholesterol biosynthesis, and hepatic LDL receptor (LDLR) expression. Interestingly, several of these miRNAs are located in genomic loci associated with abnormal levels of circulating lipids in humans. These findings reinforce the interest of targeting this subset of non-coding RNAs as potential therapeutic avenues for regulating plasma cholesterol and triglyceride (TAG) levels. In this review, we will discuss how these new miRNAs represent potential pre-disposition factors for cardiovascular disease (CVD), and putative therapeutic targets in patients with cardiometabolic disorders. This article is part of a Special Issue, entitled: MicroRNAs and lipid/energy metabolism and related diseases, edited by Carlos Fernández-Hernando and Yajaira Suárez.
INTRODUCTION
Cellular and plasma cholesterol levels are maintained through tightly controlled mechanisms, which regulate the expression and activity of key metabolic genes at both the transcriptional and post-transcriptional level. Alterations in the control of cholesterol and lipid homeostasis can lead to cardiometabolic diseases, including atherosclerosis, a prominent cause of human morbidity and mortality in Western societies [1, 2]. Among the numerous environmental and genetic factors identified to contribute to atherogenesis, increased levels of low-density lipoprotein (LDL)-cholesterol are sufficient to drive the progression of this disease. For this reason, the pathways governing plasma LDL-cholesterol (LDL-C) levels and their translation into effective therapies [e.g. statins and proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors] have been extensively studied. At the forefront of these discoveries are genome-wide association studies (GWAS), which, over the years, have allowed major advances in the identification of novel genetic variants that affect plasma lipoprotein metabolism and increase the risk of developing cardiovascular disease (CVD).
The majority of hypercholesterolemia patients are prescribed statin drugs, which competitively inhibit 3-hydroxy-3methylgutaryl-CoA reductase (HMGCR), the rate-limiting enzyme of cholesterol biosynthesis [3, 4]. The therapeutic benefit of statins is due, at least in part, to the sterol regulatory element-binding protein (SREBP)-mediated increase in the expression of the LDL receptor (LDLR), which promotes the clearance of pro-atherogenic lipoproteins from circulation [5]. Although effective at lowering cholesterol levels and reducing cardiovascular-related deaths, statin therapies are not well tolerated by all individuals and two-thirds of statin patients still experience adverse coronary events [3, 4]. As a result, there has been a significant effort in developing new classes of cholesterol-lowering drugs that can be used alone or in combination with statins, including novel approaches that exploit miRNA-dependent gene silencing [6, 7]. In this review, we summarize the therapeutic potential of manipulating miRNA expression to control plasma LDL-C levels, as well as highlight the most recent findings that uncover the importance of miRNAs in directly regulating LDLR activity in vitro and in vivo. In addition, we focus on several genome-wide association studies (GWAS), which have uncovered a critical role for miRNAs in controlling circulating levels of plasma LDL-C in humans.
miRNAS and LDL-C METABOLISM
It is well-established that miRNAs can have profound effects on cholesterol and lipid homeostasis. Specifically, a great deal of work has identified miRNAs as important regulators of high-density lipoprotein (HDL), increased levels of which are associated with reduced risk for developing CVD [8]. The most well-studied of these miRNAs, miR-33, has been demonstrated to target ATP-binding cassette transporter A1 (ABCA1). ABCA1 is a critical factor for HDL biogenesis and reverse cholesterol transport (RCT), the process through which cells efflux cellular cholesterol for transport to and removal by the liver [9–13]. In addition, miR-33 has been shown to negatively regulate numerous other genes involved in RCT and metabolic function, such as ATP-binding cassette transporter G1 (ABCG1), Niemann-Pick disease, type C1 (NPC1), cholesterol 7α-hydroxylase (CYP7A1), ATP- binding cassette, sub-family B, member 11 (ABCB11), and ATPase, aminophospholipid transporter, class I, type 8B, member 1 (ATP8B1) [9, 14, 15]. Consistent with this, therapeutic inhibition of miR-33 was found to significantly increase circulating levels of HDL-C in mice and non-human primates [9, 10, 13, 16, 17] Moreover, numerous studies have also demonstrated the beneficial effects of miR-33 inhibition on reducing atherosclerotic plaque burden, thus highlighting the therapeutic potential of manipulating miRNA expression to treat CVD [18–22].
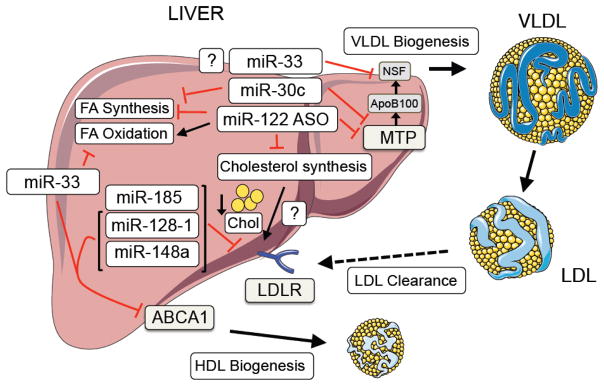
While the role of miRNAs in regulating HDL-C metabolism has been extensively studied, the importance of miRNAs in controlling LDL-C have lagged behind, perhaps due to differences in lipoprotein metabolism between mice and humans. Nevertheless, a number of studies have reported the importance of miRNAs in controlling plasma LDL-C levels by regulating very-low density lipoprotein (VLDL) secretion, cholesterol biosynthesis and LDLR activity (Figure 1).
Figure 1. miRNA regulation of LDL-cholesterol metabolism.
Schematic representation of miRNAS that control circulating levels of LDL-C in vivo.
The liver-restricted miR-122 and miR-30c were the first miRNAs shown to play a role in altering plasma LDL-C in vivo by controlling VLDL secretion and cholesterol biosynthesis. While miR-30c controls plasma cholesterol levels by decreasing lipid synthesis and the secretion of ApoB-containing lipoproteins through direct targeting of the microsomal triglyceride transfer protein (MTP) [23], antisense targeting of miR-122 in mice and non-human primates significantly lowers total plasma cholesterol and triglyceride (TAG) levels [24, 25]. While this highlights the use of miR-122 inhibitors to treat dyslipidemias, germ-line and liver-specific deletion of miR-122 results in increased hepatic steatosis, liver fibrosis and hepatocellular carcinoma [26, 27], thus raising questions about the potential therapeutic value of this treatment. In addition to miR-122 and miR-30c, inhibitors of miR-33 were also shown to modulate VLDL-TAGs in several mouse and non-human primate models, however these results have been controversial. While studies by Goedeke et al and Allen et al showed that long term inhibition of miR-33 resulted in increased levels of VLDL-TAGs in mice fed a high-fat and chow diet, respectively, no adverse effects were observed with long-term inhibition of miR-33 in non-human primates [16, 17, 28, 29]. Indeed, studies by Moore and Temel showed that anti-miR-33 treatment significantly reduced circulating levels of VLDL-TAGs in rhesus monkeys [17]. Additionally, several other miRNAs (e.g. the miR-96/182/183 locus [30]) have been shown to control LDL-C metabolism by regulating the SREBP2-mediated transcription of LDLR. For a comprehensive review of miRNAs that control cholesterol metabolism by affecting VLDL-secretion and/or cholesterol biosynthesis see review by Osborne and Hussain’s groups in this Special Issue of BBA.
A role for miRNAs in contributing to the post-transcriptional regulation of LDLR expression has recently emerged. Notably, miR-27a/b, miR-185, miR-199a, miR-148a, miR-128-1, miR-130b, and miR-301, were shown to directly target the 3′UTR of LDLR and modulate LDLR expression in human and mouse hepatic cells [31–34]. Of these miRNAs, only miR-128-1, miR-148a and miR-185 significantly altered plasma LDL-C in vivo. Specifically, ApoE−/− and APOBTg;Ldlr−/+ mice had markedly less circulating levels of plasma LDL-C when treated with inhibitors of miR-128-1 and miR-148a, respectively (see below for more details [34, 35]). Inhibition of miR-185 expression was also shown to significantly reduce circulating levels of plasma LDL-C in ApoE−/− mice, as well as slow the progression of atherosclerosis [33]. Interestingly, while modulation of miR-27b markedly altered hepatic LDLR expression in wild-type mice, no significant differences in circulating LDL-C levels were observed [32]. As miR-27b plays an extensive role in the processes governing atherosclerosis [36], additional long-term in vivo studies, as well as tissue-specific knockout mice, are needed to further elucidate the therapeutic potential of miR-27b inhibitors.
miRNAS AND GWAS
Since the advent of high-density single nucleotide polymorphism (SNP) genotyping arrays, GWAS have largely supplanted linkage studies to identify genes involved in various disease etiologies [37]. Numerous GWAS have been completed evaluating genomic linkage to various traits, including anthropometric measures such as weight, height, body mass index, as well as diseases such as metabolic disorders, cancers, and neurodegenerative diseases [38]. Importantly, essentially all GWAS to date have focused on the association of SNPs with nearby protein-coding genes, largely ignoring the presence of regulatory non-coding RNAs such as miRNAs in those genomic regions. Typically, DNA variants are located in intergenic or intronic portions of the genome, not within the coding sequences of genes or non-coding RNAs, requiring further post-GWAS functional characterization in order to determine (i) the causal DNA variant (ii) the gene(s) or non-coding RNAs involved the etiology of the disease, and (iii) the mechanism by which the DNA variant affects the expression of the causative agent.
Given the importance of circulating lipids in the development of cardiometabolic diseases (e.g., atherosclerosis, metabolic syndrome, type 2 diabetes (T2D)), a large number of GWAS have been carried out to characterize new genomic loci associated with circulating levels of lipids and lipoproteins [39]. Recently, a meta-analysis of the previously published GWAS of 188,577 European-ancestry individuals and 7,898 non-European-ancestry individuals performed by The Global Lipids Genetics Consortium led to the identification of 62 new loci within 100 kb of SNPs associated with abnormal circulating lipid [total cholesterol (TC), LDL-C, HDL-C, and TAGs] levels (at p<5×10−8) [40]. In addition to the well-characterized lipid-regulatory genes such as LDLR, ABCA1, and LIPA, new genes not previously known to play a role in lipid homeostasis were found. In particular, SNPs near Sortilin 1 (SORT1), on chromosome 1p13, are strongly associated with LDL-C levels (P=1×10−170) and are also linked to elevated risk of myocardial infarction, the leading cause of death in the Western world [41–44]. SORT1 alters serum levels of LDL-C and very low-density lipoprotein (VLDL) particles by modulating hepatic VLDL secretion [45]. Although the physiological function of SORT1 in regulating circulating lipoprotein homeostasis is still strongly debated, its identification by GWAS highlights the utility of this method to uncover new factors important for the regulation of circulating lipid levels.
To determine whether miRNAs might also be linked to abnormal blood lipids, Wagschal et al re-analyzed the Global Lipids Genetics Consortium GWAS data, identifying 69 miRNAs within 100 kb of signature lipid SNPs (Figure 2) [34]. Several unbiased bioinformatic approaches were then used to further narrow down potential miRNA regulators of circulating blood lipids. Specifically, predicted target genes of each miRNA were extracted using the software prediction program TargetScan [46], and combined with enrichment analyses [functional Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation] using the Database for Annotation, Visualization and Integrated Discovery tool (DAVID: http://david.abcc.ncifcrf.gov). From this analysis miR-130b, miR-301b, miR-148a, and miR-128-1 emerged as potential regulators of circulating blood lipids and metabolic homeostasis.
Figure 2. Identification of miRNAs located in genomic loci enriched for SNPs associated with abnormal circulating total cholesterol, triglycerides, LDL-C and HDL-C levels.
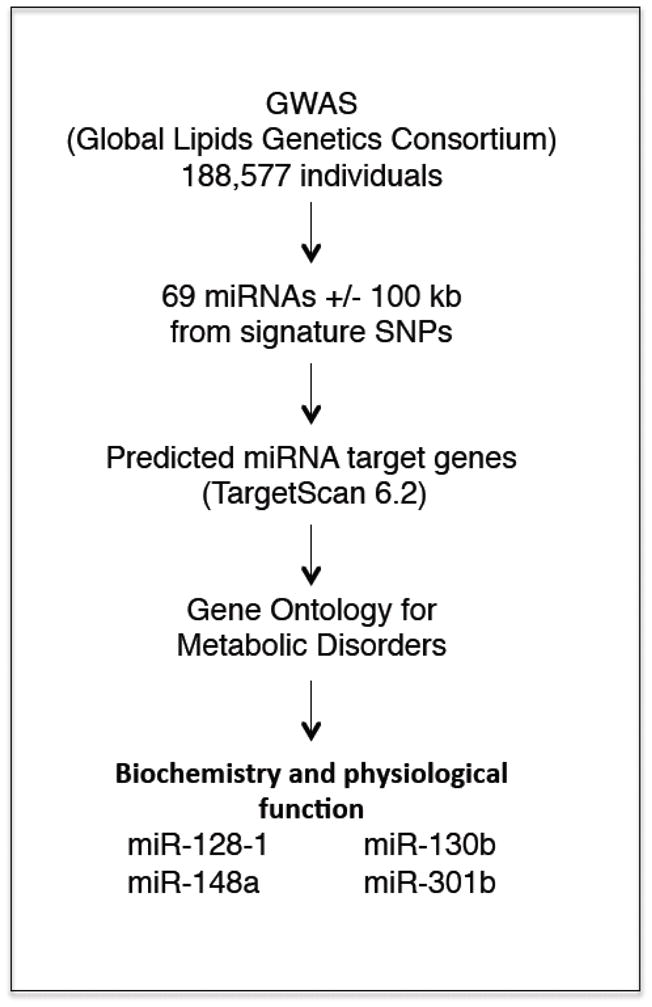
Description of the GWAS meta-analysis method used to identify cholesterol/lipid-regulating miRNAs located in genomic regions associated with abnormal circulating cholesterol/lipids.
miR-130b and miR-301b
miR-130b and miR-301b are located close to one another on human chromosome 22 in a locus associated with abnormal TC and HDL-C levels [34]. These miRNAs share the same seed sequence and are predicted to target the same metabolic factors. For example, both miRNAs directly target the 3′UTR of LDLR and ABCA1, thereby significantly reducing LDL-C uptake and binding and cholesterol efflux to ApoA1 in human hepatoma cells and mouse macrophages, respectively [34]. miR-130b and miR-301b also control the expression of salt-inducible kinase 1 (SIK1), a regulator of hepatic gluconeogenesis and SREBP1-dependent lipogenesis [47], as well as the expression of AMP-activated protein kinase alpha 2 (AMPKα-PRKAA2), a component of the trimeric stress- and energy-sensing kinase AMPK, which monitors and regulates cellular energy status. In addition, miR-130b modulates the expression of PPAR gamma coactivator 1 alpha (PGC-1α, PPARGC1A), a transcriptional coactivator, and insulin-induced gene 1 (INSIG1), a negative regulator of SREBP and HMGCR activities. Thus, these miRNAs may orchestrate tight physiological regulation of lipogenesis through the coordinated control of several upstream regulators of SREBP.
miR-148a
miR-148a is encoded within a gene-poor intergenic region of human chromosome 7 and highly expressed in the adult liver. Interestingly, several labs independently correlated SNPs (rs4722551, rs4719841 and rs6951827) in the promoter region of miR-148a with altered blood levels of TC, LDL-C and TAGs [40, 48, 49]. In particular, a miR-eQTL analysis performed in human livers revealed a strong correlation between SNP status and miR-148a expression [34]. Dietary lipids have also been shown to regulate the expression of miR-148a in the livers of mice and non-human primates, suggesting a conserved link to its metabolic function [35]. Specifically, Goedeke et al identified the presence of several SREBP1 binding sites in the promoter region of miR-148a and demonstrated through a series of in vitro and in vivo experiments that SREBP1c transcriptionally controls the expression of miR-148a in mouse and human hepatocytes [35]. As such, it is likely that these DNA variants in the miR-148a promoter might impact miR-148a transcription levels through altering SREBP1 binding sites and therefore, be considered as predisposition factors for CVD.
Although the precise mechanism by which these SNPs contribute to altered plasma lipids remains unknown, recent studies provide mechanistic insight into how miR-148a variants may modify circulating levels of LDL-C in humans. In particular, miR-148a was demonstrated to directly target the 3′UTR of LDLR, as well as several other genes involved in lipid metabolism, including ABCA1, SIK1, PGC1a, AMPK and INSIG1 [34, 35]. Importantly, overexpression and inhibition of miR-148a significantly decreases and increases, respectively, hepatic LDLR expression in mice, suggesting a physiological role for miR-148a in controlling LDLR activity in vivo. Indeed, inhibition of miR-148a markedly lowered circulating levels of LDL-C in two different mouse models of hypercholesterolemia [34, 35]. While the effects of long-term inhibition of miR-148a on serum LDL kinetics, hepatic and serum lipids, and the distribution of ApoB and other lipoproteins should be addressed, collectively these studies underscore the therapeutic potential of modulating miR-148a expression to treat dyslipidemias. Moreover, given that miR-148a regulates ABCA1 expression in hepatocytes and macrophages and in the liver in vivo [34, 35], as well as modulating HDL-C levels in both normal mice and ApoE−/− mice fed a high fat diet [34, 35], therapies directed against miR-148a represent viable options to simultaneously increase circulating levels of HDL-C and lower LDL-C. Future assessment of atherosclerotic CVD in miR-148a knockout mice, as well as in mice treated with miR-148a antisense inhibitors, are needed to determine the role of miR-148a during the progression and regression of atherosclerosis.
In addition to altered LDL-C, SNPs in the miR-148a promoter region (rs4719841 and rs6951827) have also been linked to altered levels of circulating TAGs [34, 40, 49]. Interestingly, the rs472251 variant was also found to be associated with altered TAGs, however this association did not reach statistical significance [48]. It is thus plausible that in addition to regulating cholesterol homeostasis, miR-148a may also contribute to the regulation of fatty acid and TAG metabolism. Indeed, miR-148a is predicted to target numerous SREBP1-responsive genes. Furthermore, miR-148a is also highly expressed in adipose tissue. Taken together, these observations necessitate further investigation into the role of miR-148a in fine-tuning SREBP-mediated processes, such as fatty acid/TAG synthesis. While no appreciable differences in circulating TAGs were observed in APOBTg;Ldlr−/+ and ApoE−/− mice treated with inhibitors of miR-148a, future studies using additional mouse and non-human primate models may uncover new roles of miR-148a in regulation of whole-body lipid metabolism.
miR-128-1
Wagschal et al uncovered miR-128-1 as strongly associated with altered levels of circulating TC and LDL-C [34]. A miR-eQTL analysis of 424 liver samples from gastric bypass surgery patients showed a correlation between the SNPs associated with the circulating levels of TC and LDL-C and the SNPs associated with miR-128-1 expression [34]. While this suggests that certain SNPs may affect DNA regulatory sequences linked to miR-128-1 expression, more work is needed to determine how these SNPs regulate miR-128-1 expression and whether they represent predisposition factors for cardiovascular risk.
miR-128-1 is located within an intron of R3HDM1, a gene with unknown function. Biochemical and physiological studies showed that miR-128-1 plays an important role in lipid and carbohydrate metabolism, suggesting that the association of SNPs in this genomic locus with abnormal blood lipids is indeed due to altered miR-128-1 expression [34]. In particular, long-term inhibition of miR-128-1 in hyperlipidemic ApoE-deficient mice leads to (i) a ~35% decrease in circulating TC, (ii) a ~25% decrease in TAGs associated with VLDL (triglyceridemia), (iii) a decrease in hepatic TAGs (steatosis) and (iv) improved glucose tolerance and insulin sensitivity [34]. Although the precise mechanisms responsible for these complex phenotypes are not yet fully understood, the involvement of miR-128-1 in the regulation of lipoprotein transport and insulin signaling highlight the crucial role that miR-128-1 plays in these observed phenotypes.
miR-128-1 controls circulating lipoprotein metabolism by directly targeting the 3′UTR of LDLR and ABCA1. In accordance with the function of ABCA1, miR-128-1 regulates cellular cholesterol efflux to ApoA1 in human hepatoma cells and mouse macrophages [34]. Furthermore, overexpression of miR-128-1 (using viral particles encoding the miR-128-1 precursor) decreased hepatic ABCA1 expression and circulating levels of HDL-C in C57BL/6J mice. While other miRNAs have been shown to regulate ABCA1 expression and HDL-C levels (e.g. miR-33), miR-128-1 also regulates hepatic LDLR expression, similar to miR-148a. Specifically, modulation of miR-128-1 was shown to strongly affect LDL binding in human hepatoma cells and LDL-C clearance in ApoE−/− mice. Indeed, acute (5 days) inhibition of miR-128-1 was sufficient to significantly alter lipoprotein metabolism, resulting in decreased plasma VLDL-C and LDL-C levels. Based on the ability of miR-128-1 inhibitors to affect both HDL-C and LDL-C, miR-128-1 antisense treatment may decrease atherosclerosis by improving the LDL-C/HDL-C ratio.
In addition to controlling cholesterol metabolism, miR-128-1 also regulates hepatic insulin signaling. In particular, miR-128-1 directly controls the expression of the insulin receptor (INSR), insulin receptor substrate 1 (IRS-1), and the downstream phosphorylation levels of Akt [34, 50]. Importantly, antisense inhibition of miR-128-1 improved both glucose tolerance and insulin sensitivity in ApoE−/− mice fed a Western-type diet [34], indicating that miR-128-1 plays a critical physiological role in regulating insulin signaling and glucose homeostasis in vivo. Given the key role of insulin in controlling carbohydrate and lipid metabolism in skeletal muscle, further studies on miR-128-1 are needed to shed light on its insulin-dependent functions in other metabolic tissues.
In the liver of patients with T2D, abnormal activation of the insulin-signaling pathway leads to a deregulation of lipid metabolism [51]. Indeed, insulin suppresses hepatic ApoB secretion and promotes the clearance of ApoB-containing circulating lipoproteins via several receptors, including the LDLR, LDLR-related protein 1 (LRP1), and heparan sulfate proteoglycans [52]. Among the most common lipoprotein abnormalities, patients with T2D frequently present low circulating HDL-C and high TAGs, consequently leading to elevated TAG-rich remnant lipoproteins, in both fasting and postprandial states [52]. As such, the role of miR-128-1 in suppressing hepatic insulin signaling could contribute to the decreased lipoprotein levels observed in mice treated with anti-miR-128-1, specifically with respect to altered TAGs and VLDL.
In addition to the effect of miR-128-1 on hepatic insulin signaling, miR-128-1 was also found to control synthesis of free fatty acids through the regulation of fatty acid synthase (FASN) expression [34]. Interestingly, miR-128-1 was shown to negatively regulate the expression of SIRT1, an NAD+-dependent energy sensor and lysine deacetylase that can directly deacetylate and inactivate SREBP1 and thus modulate SREBP1-dependent lipogenesis [53]. It is thus tempting to speculate that the effects of anti-miR-128-1 on FASN and hepatic steatosis may at least partially be due to the miR-128-1-mediated repression of SIRT1, however, additional studies are needed to reveal the precise function of miR-128-1 in regulating lipogenesis and the potential role of SIRT1 in this process.
CONCLUSIONS AND FUTURE MIRNA-BASED THERAPEUTICS
The recent discoveries of miRNAs that control LDLR expression and activity have greatly expanded our understanding of the molecular mechanisms that govern plasma LDL-C. This miRNA-centered regulatory network continues to grow at a rapid pace, and will no doubt expand to include other miRNAs that regulate LDL-C levels, both directly, by regulating VLDL secretion and LDL catabolism, and indirectly, by controlling hepatic cholesterol biosynthesis. While the regulatory function of these miRNAs in health and disease states, as well as how these networks fit into the already well-established transcriptional and post-transcriptional pathways that control hepatic LDLR expression, remain to be fully elucidated, future studies in humanized mouse models and non-human primates will likely help determine the relative contribution of each of these miRNAs in controlling LDLR activity and LDL-C trafficking and cholesterol homeostasis.
The recent findings that manipulating miRNA expression may reduce circulating LDL-C opens new avenues for the treatment of dyslipidemias, especially for those patients who are resistant to statin treatment. Indeed, clinical trials using antisense inhibitors targeting miR-122 are currently underway [54]. Furthermore, multiple groups have demonstrated that inhibition of miR-33 results in a significant increase in plasma HDL-C in mouse and non-human primates and protection against atherosclerosis in hyperlipidemic mouse models [9, 10, 13, 16–22]. In addition, a report from Hussain’s group showed that prolonged delivery of miR-30c mimics markedly reduced plasma LDL-C levels and atherogenesis in mice [23].
The recent GWAS discussed in this review also highlights the therapeutic potential of inhibiting miR-128-1 and miR-148a expression to simultaneously lower and increase circulating levels of LDL-C and HDL-C, respectively. As inhibitors of these miRNAs promote cholesterol efflux in macrophages, as well as HDL biogenesis in the liver, antisense treatments would be expected to not only increase HDL-C levels, but increase HDL functionality as well, and thus be of greater clinical relevance. Moreover, given the role of miR-128-1 in regulating insulin signaling and lipoprotein metabolism, inhibition of this miRNA also represents an attractive therapeutic strategy to combat metabolic syndrome and T2D-associated dyslipidemia, such as increased VLDL-TAGs and decreased HDL-C.
Highlights.
Discussion of the role of miRNAs in regulation of LDL-cholesterol homeostasis
Unbiased high-throughput and GWAS approaches identify lipid regulatory miRNAs
Highlights miRNAs as potential therapeutic targets for cardiometabolic diseases
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01DK094184 and U54HL119145 to AMN and R01HL107953 and R01HL106063 to CF-H) and The Leducq Transatlantic Network in Vascular microRNAs (MIRVAD) to CF-H. Figure 1 was generated using the Servier Medical Art illustration resources (http://www.servier.com). We apologize to those whose work could not be cited due to space limitations.
Footnotes
CONFLICTS OF INTEREST
AMN has patents issued and pending on the use of miR-128-1 inhibitors, AMN and AW have patents pending on the use of miR-148a inhibitors, AMN and CF-H have patents issued and pending on the use of miR-33 inhibitors, and CF-H and LG have patents pending on the use of miR-27b and miR-148a inhibitors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 2.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg D. The statins in preventive cardiology. N Engl J Med. 2008;359:1426–1427. doi: 10.1056/NEJMp0806479. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg D, Glass CK, Witztum JL. Evidence mandating earlier and more aggressive treatment of hypercholesterolemia. Circulation. 2008;118:672–677. doi: 10.1161/CIRCULATIONAHA.107.753152. [DOI] [PubMed] [Google Scholar]
- 5.Brown MS, Goldstein JL. Receptor-mediated control of cholesterol metabolism. Science. 1976;191:150–154. doi: 10.1126/science.174194. [DOI] [PubMed] [Google Scholar]
- 6.Grundy SM. Promise of low-density lipoprotein-lowering therapy for primary and secondary prevention. Circulation. 2008;117:569–573. doi: 10.1161/CIRCULATIONAHA.107.720300. discussion 573. [DOI] [PubMed] [Google Scholar]
- 7.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ P Coordinating Committee of the National Cholesterol Education. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. J Am Coll Cardiol. 2004;44:720–732. doi: 10.1016/j.jacc.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Rader DJ. Illuminating HDL--is it still a viable therapeutic target? N Engl J Med. 2007;357:2180–2183. doi: 10.1056/NEJMe0707210. [DOI] [PubMed] [Google Scholar]
- 9.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, Macdougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010 doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013;58:1111–1121. doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Naar AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horie T, Baba O, Kuwabara Y, Chujo Y, Watanabe S, Kinoshita M, Horiguchi M, Nakamura T, Chonabayashi K, Hishizawa M, Hasegawa K, Kume N, Yokode M, Kita T, Kimura T, Ono K. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE−/− mice. J Am Heart Assoc. 2012;1:e003376. doi: 10.1161/JAHA.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotllan N, Ramirez CM, Aryal B, Esau CC, Fernandez-Hernando C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice--brief report. Arterioscler Thromb Vasc Biol. 2013;33:1973–1977. doi: 10.1161/ATVBAHA.113.301732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karunakaran D, Thrush AB, Nguyen MA, Richards L, Geoffrion M, Singaravelu R, Ramphos E, Shangari P, Ouimet M, Pezacki JP, Moore KJ, Perisic L, Maegdefessel L, Hedin U, Harper ME, Rayner KJ. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ Res. 2015;117:266–278. doi: 10.1161/CIRCRESAHA.117.305624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, Rinehold K, van Solingen C, Fullerton MD, Cecchini K, Rayner KJ, Steinberg GR, Zamore PD, Fisher EA, Loke P, Moore KJ. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;2015 doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goedeke L, Salerno A, Ramirez CM, Guo L, Allen RM, Yin X, Langley SR, Esau C, Wanschel A, Fisher EA, Suarez Y, Baldan A, Mayr M, Fernandez-Hernando C. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med. 2014;6:1133–1141. doi: 10.15252/emmm.201404046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen RM, Marquart TJ, Jesse JJ, Baldan A. Control of very low-density lipoprotein secretion by N-ethylmaleimide-sensitive factor and miR-33. Circ Res. 2014;115:10–22. doi: 10.1161/CIRCRESAHA.115.303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, Osborne TF. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez ML, Khosroheidari M, Eddy E, Done SC. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis. 2015;242:595–604. doi: 10.1016/j.atherosclerosis.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goedeke L, Rotllan N, Ramirez CM, Aranda JF, Canfran-Duque A, Araldi E, Fernandez-Hernando A, Langhi C, de Cabo R, Baldan A, Suarez Y, Fernandez-Hernando C. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis. 2015;243:499–509. doi: 10.1016/j.atherosclerosis.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H, Zhang J, Du Y, Jia X, Yang F, Si S, Wang L, Hong B. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243:523–532. doi: 10.1016/j.atherosclerosis.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 34.Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, Hatoum I, Shah N, Lu Y, Kristo F, Psychogios N, Vrbanac V, Lu YC, Hla T, de Cabo R, Tsang JS, Schadt E, Sabeti PC, Kathiresan S, Cohen DE, Whetstine J, Chung RT, Fernandez-Hernando C, Kaplan LM, Bernards A, Gerszten RE, Naar AM. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med. 2015;21:1290–1297. doi: 10.1038/nm.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, Horton JD, Lasuncion MA, Naar AM, Suarez Y, Fernandez-Hernando C. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med. 2015;21:1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WJ, Yin K, Zhao GJ, Fu YC, Tang CK. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis. 2012;222:314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. American journal of human genetics. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atanasovska B, Kumar V, Fu J, Wijmenga C, Hofker MH. GWAS as a Driver of Gene Discovery in Cardiometabolic Diseases. Trends Endocrinol Metab. 2015;26:722–732. doi: 10.1016/j.tem.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 40.C. Global Lipids Genetics. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, Wtccc C the Cardiogenics. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.C. Myocardial Infarction Genetics; Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O’Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O’Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O’Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D Wellcome Trust Case Control C. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strong A, Patel K, Rader DJ. Sortilin and lipoprotein metabolism: making sense out of complexity. Current opinion in lipidology. 2014;25:350–357. doi: 10.1097/MOL.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon YS, Seo WY, Lee MW, Kim ST, Koo SH. Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J Biol Chem. 2009;284:10446–10452. doi: 10.1074/jbc.M900096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O’Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Altshuler D, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Mohlke KL, Ingelsson E, Abecasis GR, Daly MJ, Neale BM, Kathiresan S. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huan T, Rong J, Liu C, Zhang X, Tanriverdi K, Joehanes R, Chen BH, Murabito JM, Yao C, Courchesne P, Munson PJ, O’Donnell CJ, Cox N, Johnson AD, Larson MG, Levy D, Freedman JE. Genome-wide identification of microRNA expression quantitative trait loci. Nat Commun. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126:2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sparks JD, Sparks CE, Adeli K. Selective hepatic insulin resistance, VLDL overproduction, and hypertriglyceridemia. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:2104–2112. doi: 10.1161/ATVBAHA.111.241463. [DOI] [PubMed] [Google Scholar]
- 52.Haas ME, Attie AD, Biddinger SB. The regulation of ApoB metabolism by insulin. Trends Endocrinol Metab. 2013;24:391–397. doi: 10.1016/j.tem.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]