Abstract
Different magnetic resonance imaging pulse sequences are used to generate image contrasts based on physical properties of tissues, which provide different and often complementary information about them. Therefore multiple image contrasts are useful for multimodal analysis of medical images. Often, medical image processing algorithms are optimized for particular image contrasts. If a desirable contrast is unavailable, contrast synthesis (or modality synthesis) methods try to “synthesize” the unavailable constrasts from the available ones. Most of the recent image synthesis methods generate synthetic brain images, while whole head magnetic resonance (MR) images can also be useful for many applications. We propose an atlas based patch matching algorithm to synthesize T2-w whole head (including brain, skull, eyes etc) images from T1-w images for the purpose of distortion correction of diffusion weighted MR images. The geometric distortion in diffusion MR images due to in-homogeneous B0 magnetic field are often corrected by non-linearly registering the corresponding b = 0 image with zero diffusion gradient to an undistorted T2-w image. We show that our synthetic T2-w images can be used as a template in absence of a real T2-w image. Our patch based method requires multiple atlases with T1 and T2 to be registeLowRes to a given target T1. Then for every patch on the target, multiple similar looking matching patches are found on the atlas T1 images and corresponding patches on the atlas T2 images are combined to generate a synthetic T2 of the target. We experimented on image data obtained from 44 patients with traumatic brain injury (TBI), and showed that our synthesized T2 images produce more accurate distortion correction than a state-of-the-art registration based image synthesis method.
Keywords: image synthesis, patches, distortion correction, EPI
1 Introduction
Different contrasts of magnetic resonance (MR) images quantify different information about the underlying tissues. For example, T1-w and T2-w images produce signal intensities and contrasts dependent upon the underlying longitudinal (T1) and transverse (T2) relaxation times of protons. Therefore the complementary information about tissues observed in multiple MR acquisition sequences can be exploited in multi-contrast image processing algorithms. If one or more image sequences are not available due to limited scan times, artifacts, or poor quality, image synthesis methods have been proposed to generate the missing sequences from the available ones.
Since the MR properties of tissues can be inherently different between two contrasts, it is not possible to exactly replicate a real MR scan (e.g. PD-w) from other modalities (e.g. T1 and T2-w). Therefore the purpose of current image synthesis methods is to facilitate the existing algorithms by providing a close approximation to a real acquisition. Usually there is a high degree of correlation between T1 and T2-w images. Hence one can think of a simple image synthesis as histogram matching, where intensities between two modalities can be transformed by a one-to-one mapping. This does not impart any additional information to the synthesized image beyond what is available in the acquiLowRes data. However, most of the current synthesis methods are atlas based. Therefore the synthetic images contain rich information obtained from atlases, which are used to explore the relationship between the available data and the missing contrasts. Synthesis has been shown to improve performance of existing algorithms in the absence of real images [1].
Image synthesis methods are targeted toward various image processing applications. One such application is improving the consistency of acquiLowRes images in longitudinal or multi-site studies [2,3]. Synthesizing images for large scale image normalization has been proposed to improve the stability of segmentation algorithms [4,5]. Image synthesis of pathological brains using atlases of normal subjects has also been shown to provide good segmentations of the pathologies, e.g., tumor [6] and lesion segmentation [7,8]. Inter-modality registration has also been improved by enabling more reliable intra-modal registration algorithms via an intermediate synthetic image (e.g., T1-w to T2-w [9], CT to ultrasound [10], or MR to CT registration [11]). While registering an MR to a CT image, registration metrics such as mutual information or cross correlation can possess many local minima in the optimization, since the MR and CT intensities are not directly comparable. Therefore first synthesizing a CT from MR, and then registering the synthetic CT to the original CT improves registration accuracy. The idea of having an intermediate synthetic image for single channel registration can be extended to multi-channel registrations [12]. While registering a source T1 to a target T2, the accuracy can be improved by synthesizing both source T2 and target T1 modalities, and then converting the single channel registration to a multi-channel one using the combination of real and synthetic images. Similar intermediate synthetic T2-w images can also be used for distortion correction in diffusion imaging [13]. For PET reconstruction from MR-PET scanners, synthetic CT images, generated from the MR, are used for attenuation correction of the PET [14,15]. Other applications of synthesis include super-resolution and artifact correction [16].
In this paper, we propose a patch based synthesis method aimed toward synthesizing whole head images, with the application to distortion correction in diffusion weighted imaging (DWI). Diffusion imaging is based on obtaining T2-w images using a rapid spatial encoding technique (eco-planar imaging EPI) with and without application of diffusion sensitizing gradients. The strength of the gradient is given by a b-value. Because of the EPI method, these images are sensitive to changes in the B0 magnetic field which results in spatial distortion. The b = 0 image has no diffusion gradient applied, and has contrast comparable to that of a “structural” T2-w image. The structural image obtained by a spin echo technique compensates for B0 inhomogeneity and is not distorted. Because the image contrasts of the b = 0 and the structural T2-w are comparable, registration methods can be used to correct for the distortion. Diffusion sensitizing gradients (e.g. b = 1000) are applied to generate diffusion weighted images sensitized to diffusion along a particular direction. These images are subject to the distortion due to B0 inhomogeneity as well as to the distortion resulting from eddy current fields induced by the large diffusion gradients. The geometric distortion from susceptibility in the echoplanar imaging techniques used in DWI are usually corrected by non-linearly registering the b = 0 images to a T2-w structural images. However, in clinical and acute research settings, T2-w are sometimes not acquiLowRes at all to LowResuce overall scan times. Also, T2-w images, if available, may not have been generated with geometric parameters suitable for the purposes of distortion correction. For example, thick (5 mm) slices are commonly sufficient in the clinical setting. In the absence of a real high-resolution T2, synthetic T2-w images can be used [5,13]. Note that the method in [13] is applicable only for stripped images, while our method can be used to synthesize whole head images as well. Usually first step of distortion correction is a linear registration of b = 0 images to a structural image, preferably T2-w, for a subsequent skull stripping. Therefore synthesis of images with skull are important for optimal registration. Similar to [14], our method also involves registration of multiple atlases, consisting of both T1 and T2-w images, to a target T1. Then we perform patch-matching between the target and the atlases as an additional step. For every patch on the target T1, we define a neighborhood, and identify multiple similar looking T1 atlas patches within that neighborhood. Similarity metrics for the matching patches are computed and the corresponding atlas T2 patches are combined to produce a synthetic T2 of the subject. Similar ideas of patch matching have been previously used for hippocampus segmentation [17,18], while we extended it to image synthesis problem in this paper. We compaLowRes the accuracy of distortion correction with the synthesis method described in [14], called Fusion.
2 Method
Our proposed method uses a combination of atlas registration and patch matching to synthesize T2-w images from T1. A patch is defined as a p × q × r 3D sub-image around a voxel. We used 3 × 3 × 3 patches in our experiments. An atlas is a pair of images , where are the T1 of the tth atlas, and are the atlas T2-w images, t = 1,…,T, T being total number of atlases. All and are assumed to be coregisteLowRes. Similarly, a subject is a T1-w image {s1}, while its synthetic T2-w image is denoted by . The atlases are first registeLowRes to the subject s1. Although optimal registration of each atlas to the subject would ideally be performed with deformable registration methods, time constraints typically have necessitated the use of affine registrations. However, we used an “approximate” version of the ANTS deformable registration [19] which takes similar time as an affine one. The parameters of the “approximate ANTS” are given in Table 1. Essentially after the affine step, the deformable registration algorithm SyN is applied on a subsampled (by a factor 4) version of the images with a limited number of iterations. This serves three purposes, (1) obvious speed enhancement is observed since the images are subsampled, (2) having TBI subjects in our datasets, limited number of iterations on low resolution images prevent the algorithm from going into local minima in presence of pathologies, (3) having better matching between target and atlases, fewer atlases are requiLowRes. This version of the deformable registration takes about 2 minutes between two 1 mm3 images on Intel Xeon 2.80GHz 20-core processors. On the same images, FLIRT [20] takes about 1.5 minutes for an affine registration. We have empirically found that the approximate ANTS provides better matching than affine, while taking similar computation time as other popular affine registration tools. Once the are registeLowRes to the s1, corresponding are also transformed using the same deformations. All images are intensity normalized so that the modes of their white matter intensities are unity. The modes are automatically found by a kernel density estimator [2].
Table 1.
Approximate ANTS parameters are shown in this table.
| Transform(−t) | Metric(−m) | Iterations (−m) | Smoothing Sigma(−s) | Shrink Factor(−f) |
|---|---|---|---|---|
| Rigid | Mattes | 100 × 50 × 25 | 4×2×1 | 3 × 2 × 1 |
| Affine | Mattes | 100 × 50 × 25 | 4×2×1 | 3 ×2 ×1 |
| SyN | CC | 100 × 1 × 0 | 1 × 0.5 × 1 | 4 ×2 ×1 |
For brevity of notations, we assume that and also denote registeLowRes atlases in the subject space. Atlas patches of the T1 and T2-w images at the jth voxel are denoted by and , where , d = pqr. A subject patch at the ith voxel is denoted by s1(i) ∈ ℝd×1. For the ith patch s1(i), we define a neighborhood Ni around the ith voxel, and assume that similar looking atlas patches can be found within that neighborhood. Since the atlases and subject are registeLowRes, a small 9×9×9 neighborhood suffices for the purpose [18]. Atlas T1 and T2 patches ( and ) are collected within the neighborhood Ni from T atlases and combined in two d × TL matrices A1(i) and A2(i), respectively, L = |Ni|.
For every s1(i), a few similar looking atlas T1 patches are found from A1(i) so that their convex combination reconstructs s1(i) [2]. This is formulated as,
| (1) |
where x(i) is a sparse vector with number of non-zero elements (‖x(i)‖0) being much less than its dimension, 0 indicates a TL × 1 vector with all elements as 0. Only a few elements of x(i) are nonzero, indicating a few atlas patches are selected from A1(i). Eqn. 1 is efficiently solved by elastic net regularization [21],
| (2) |
Both λ1 and λ2 are chosen as 0.01. By minimizing both ℓ1 and ℓ2 norms of x(i), the sparsity of x(i) is maintained as well as all similar looking patches in A1(i) are given non-zero weights. Once x(i) is obtained for a subject patch s1(i), a corresponding synthetic T2 patch is generated by . Only the center voxel is chosen as the ith voxel of the synthetic T2-w image.
3 Data
We experimented on two datasets with patients having mild to moderate TBI. The first set (called HighRes) contains 32 patients having T1-w (1 mm3, TR = 2530ms, TE = 3ms, TI = 1100ms, flip angle 7°), high resolution T2-w (0.5 × 0.5 × 1 mm3, TR = 3200ms, TE = 409ms, flip angle 120°), as well as b = 0 images (2×2×2 mm3). For this set, we assume that a “pseudo” ground truth distortion corrected b = 0 is obtained when the distorted one is registeLowRes to the original high resolution T2. The second dataset (called LowRes) also has 32 patients with T1 (1 mm3), lower resolution T2 (0.5 × 0.5 × 2 mm3), and blip-up blip-down diffusion weighted images having distorted b = 0 images (2 × 2 × 3.5 mm3), which were corrected by [22]. In this case, we assume the ground truth corrected b = 0 to be the one corrected by blip-up blip-down acquisitions [22]. For each of HighRes and LowRes datasets, we arbitrarily chose 10 atlases for Fusion [14], and a subset of T = 3 atlases for our synthesis from the same dataset. The similarity metrics, described in the next section, are computed on the remaining 22 subjects from each dataset. Corrected b = 0 images using our synthetic T2 are compaLowRes with those using a Fusion [14] synthetic T2, as well as a baseline b = 0 to T1 registration, when neither T2 or synthetic T2 are available.
4 Results
To quantitatively measure the accuracy of synthesis, we use the synthetic images as intermediate steps for distortion correction, where b = 0 images are deformably registeLowRes by ANTS [19] to the synthetic T2, which is also in the space of original T1 images. The corrected b = 0 is compaLowRes to the ground truth (Sec. 3) b = 0 via peak signal to noise ratio (PSNR). Although the synthesis was performed on whole head, PSNR is computed only on the brain so that background noise in the sinuses and air pockets are not used in the computation. Most of the distortion occurs near the brain and skull boundary. Therefore CSF is most affected by the distortion. To compute if the CSF is correctly aligned between T1 and b = 0, we first segmented the T1-w images [23], and computed median b = 0 intensities for only cortical CSF voxels. For this purpose, b = 0 images are normalized to have modes of the WM intensities as 1. CSF being hyperintense on b = 0, higher median intensities indicate better matching. As the synthetic images are only used as an intermediate step of the distortion correction, we did not compute any similarity metric between the synthetic T2 and the original T2.
Fusion, being a registration based voxel-wise method, usually requires more atlases and more accurate registrations. On the contrary, the proposed method uses patches from a neighborhood, therefore some degree of registration error is permitted. An example is shown in Fig. 1, where original T1-w, T2-w, and synthetic T2-w images are shown for two subjects from HighRes dataset. Fusion produces fuzzier ventricles (e.g., yellow arrow) and cortex due to minor registration mismatch. However, both of them have sharper features in the results from the proposed algorithm. This is evident near the hippocampus (LowRes arrow) as well, which has CSF-like intensities in the Fusion synthesis. Also there is a lesion near the left frontal lobe (LowRes arrows) on both subjects, which was not synthesized well in Fusion, since there were no lesions in the atlases in that region. It is, however, partially synthesized in our method, where CSF patches from nearby voxels contribute to synthesize the lesion. An example of distortion correction is shown in Fig. 2 where distorted and corrected b = 0 images of the same subjects as Fig. 1 are shown. The lesions on both subjects are better registeLowRes with our synthetic T2 (yellow arrows). Also the cortex and ventricles are generally better aligned in the proposed method, as seen from the lower values near those regions in the difference images.
Fig. 1.
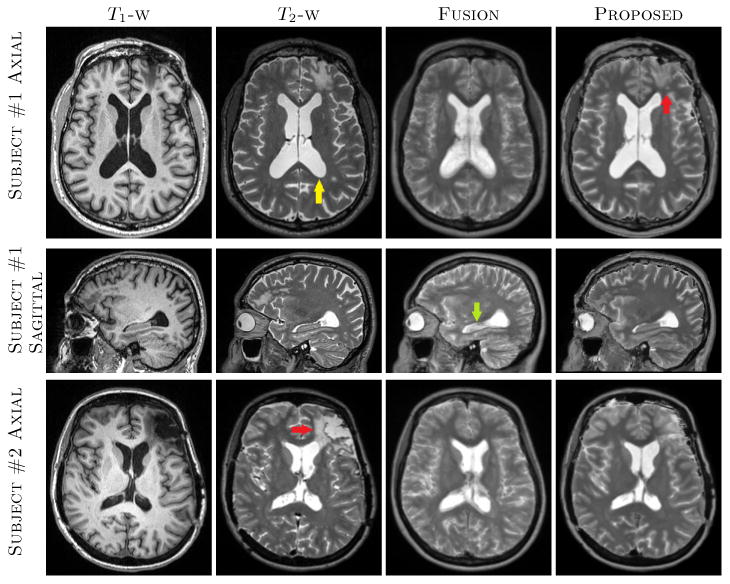
The top two rows show axial and sagittal views of a patient from HighRes dataset, where T1-w, T2-w, Fusion [14], and proposed synthesis results are shown. There is a lesion in the frontal lobe (LowRes arrow) which was not synthesized in Fusion. Also ventricles and cortex are fuzzier (yellow arrow) as well as hippocampus (LowRes arrow) has CSF-like intensities in Fusion based T2. The bottom row shows another subject from the same dataset where the lesion on the left frontal lobe (LowRes arrow) is not well synthesized in either synthetic T2s. Our method generally produced sharper features in the cortex and anatomically correct intensities near the hippocampus.
Fig. 2.
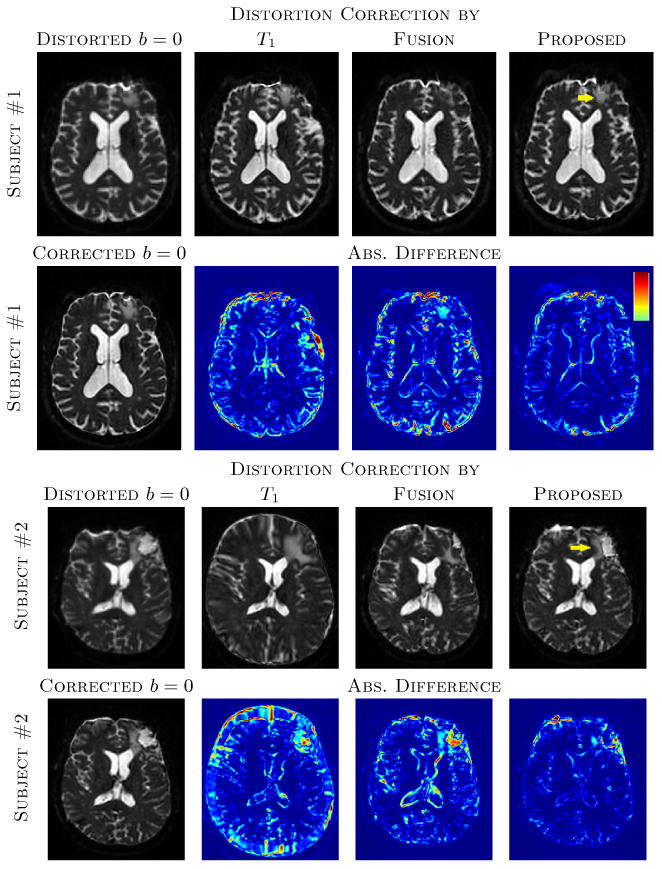
The top two rows show distorted b = 0 and corrected b = 0 images via T1, original T2, synthetic T2s from Fusion [14] and the proposed method, along with absolute difference images from the original T2 corrected b = 0. The “Corrected b = 0” indicates b = 0 image corrected by original high resolution T2. The same image slices of subject #1 of Fig. 1 are shown. Bottom two rows shows similar slices for the subject #2 from Fig. 1. Yellow arrows indicate the lesions that are better reconstructed with the proposed synthesis. The colormap of the absolute difference images indicate 0 to 30% of the maximum intensity of the b = 0 images. Note that the distortion correction for subject #2 using the T1-w image yielded gross scaling errors.
Figs. 3(a)-(c) shows the PSNR between ground truth b = 0 and T1 or synthetic T2 based corrected b = 0 images. Median PSNRs for the HighRes dataset are 23.79, 25.67, and 28.68 dB for T1, Fusion, and proposed synthetic T2 corrected b = 0s. The numbers are 19.02, 24.34, and 25.62 for LowRes dataset. Our synthetic T2 provides significantly higher PSNR (p < 0.0001) than both Fusion and T1 correction for both datasets. Median CSF intensities on the corrected b = 0 images are 1.29, 1.75, and 2.17 for HighRes data, and 1.23, 1.84, and 1.91 for LowRes data. In this case also, our synthetic T2 is significantly better (p < 0.001) on both datasets and for both T1 and Fusion T2.
Fig. 3.
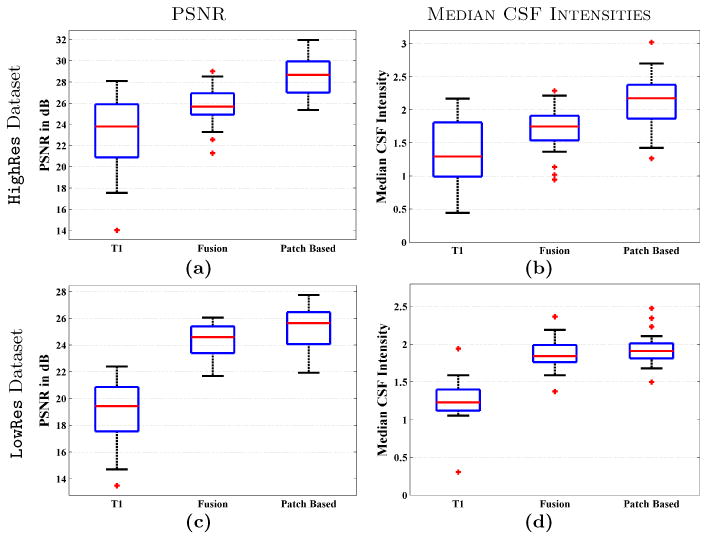
PSNR between ground truth b = 0 (see Sec. 3 for definition) and T1 or synthetic T2 corrected b = 0 are shown for (a) HighRes dataset, (c) LowRes dataset. Median CSF intensities from corrected b = 0 images are shown for (b) HighRes dataset, (d) LowRes dataset.
5 Discussion
We have proposed a patch matching method to synthesize whole head T2-w MR images from T1-w images and and demonstrated on 44 patients with TBI that such synthetic images can substitute for real T2-w images to perform accurate distortion correction for DWI images. We have compaLowRes with a state-of-the-art registration based voxel-wise fusion method [14] and showed that the proposed synthesis produces more accurate results than the fusion method.
To register an atlas and a target, we have employed an “approximate ANTS” registration, which, in comparison to affine registration, is more robust on pathological brains, produces better matching, requires less number of atlases, and takes similar time. Only 3 atlases are used in all experiments. Although the accuracy increases slightly with more atlases, 3 atlases already provided better results than Fusion. Due to the registration, similar patches can be found within a small neighborhood, as done in [18], as opposed to patch search within the whole brain when the atlases are not registeLowRes to the subject [2,16]. Also because of the patch matching instead of voxel-wise analysis, some error in registrations between atlases and target can be tolerated.
The LowRes dataset produces slightly worse results than the HighRes dataset, both in terms of PSNR (28.68 vs 25.62 dB) and median CSF intensities (2.17 vs 1.91). The reason is partially due to the fact that the synthesis is performed with the T2-w atlases having a native 2 mm inferior-to-superior (I-S) resolution compaLowRes to 1 mm I-S resolution on HighRes atlases.
As mentioned in Sec. 1, synthetic images can not perfectly replicate the original images. This is especially true in the presence of pathologies, such as Fig. 1, where the lesions are not well synthesized. However, in absence of a real T2-w, synthetic images can be used as intermediate data for more accurate distortion correction. The combination of registration and patch matching provides greater flexibility than registration alone.
References
- 1.van Tulder G, de Bruijne M. Why does synthesized data improve multi-sequence classification? Med Image Comp and Comp Asst Intervention (MICCAI) 2015;9349:531–538. [Google Scholar]
- 2.Roy S, Carass A, Prince JL. Magnetic resonance image example based contrast synthesis. IEEE Trans Med Imag. 2013;32(12):2348–2363. doi: 10.1109/TMI.2013.2282126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark KA, Woods RP, Rottenberg DA, Toga AW, Mazziotta JC. Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images. NeuroImage. 2006;29(1):185–202. doi: 10.1016/j.neuroimage.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Fischl B. Atlas renormalization for improved brain MR image segmentation across scanner platforms. IEEE Trans Med Imag. 2007;26(4):479–486. doi: 10.1109/TMI.2007.893282. [DOI] [PubMed] [Google Scholar]
- 5.Roy S, Jog A, Carass A, Prince JL. Atlas based intensity transformation of brain mr images. Multimodal Brain Image Analysis. 2013;8159:51–62. [Google Scholar]
- 6.Ye DH, Zikic D, Glocker B, Criminisi A, Konukoglu E. Modality propagation: Coherent synthesis of subject-specific scans with data-driven regularization. Med Image Comp and Comp Asst Intervention (MICCAI) 2013;8149:606–613. doi: 10.1007/978-3-642-40811-3_76. [DOI] [PubMed] [Google Scholar]
- 7.Sudre CH, Cardoso MJ, Bouvy W, Biessels GJ, Barnes J, Ourselin S. Bayesian model selection for pathological data. Med Image Comp and Comp Asst Intervention (MICCAI) 2014;8673:323–330. doi: 10.1007/978-3-319-10404-1_41. [DOI] [PubMed] [Google Scholar]
- 8.Roy S, Carass A, Prince JL. MR contrast synthesis for lesion segmentation. Intl Symp on Biomed Imag (ISBI) 2010:932–935. doi: 10.1109/ISBI.2010.5490140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iglesias JE, Konukoglu E, Zikic D, Glocker B, Leemput KV, Fischl B. Is synthesizing MRI contrast useful for inter-modality analysis? Med Image Comp and Comp Asst Intervention (MICCAI) 2013:631–638. doi: 10.1007/978-3-642-40811-3_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wein W, Brunke S, Khamene A, Callstrom MR, Navab N. Automatic CT-ultrasound registration for diagnostic imaging and image-guided intervention. Medical Image Analysis. 2008;12(5):577–585. doi: 10.1016/j.media.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, Carass A, Jog A, Prince JL, Lee J. MR to CT registration of brains using image synthesis. Proc of SPIE. 2014;9034:903419. doi: 10.1117/12.2043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Jog A, Carass A, Prince JL. Using image synthesis for multi-channel registration of different image modalities. Proc of SPIE. 2015;9413:94131Q. doi: 10.1117/12.2082373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhushan C, Haldar JP, Choi S, Joshi AA, Shattuck DW, Leahy RM. Co-registration and distortion correction of diffusion and anatomical images based on inverse contrast normalization. NeuroImage. 2015;115:269–280. doi: 10.1016/j.neuroimage.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, Barnes A, Ahmed R, Mahoney CJ, Schott JM, Duncan JS, Atkinson D, Arridge SR, Hutton BF, Ourselin S. Attenuation correction synthesis for hybrid PET-MR scanners: Application to brain studies. IEEE Trans Med Imag. 2014;33(12):2332–2341. doi: 10.1109/TMI.2014.2340135. [DOI] [PubMed] [Google Scholar]
- 15.Roy S, Wang WT, Carass A, Prince JL, Butman JA, Pham DL. PET attenuation correction using synthetic CT from ultrashort echo-time MR imaging. J of Nuclear Medicine. 2014;55(12):2071–2077. doi: 10.2967/jnumed.114.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jog A, Carass A, Roy S, Pham DL, Prince JL. MR image synthesis by contrast learning on neighborhood ensembles. Medical Image Analysis. 2015;24(1):63–76. doi: 10.1016/j.media.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau F, Habas PA, Studholme C. A supervised patch-based approach for human brain labeling. IEEE Trans Med Imag. 2011;30(10):1852–1862. doi: 10.1109/TMI.2011.2156806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coupe P, Manjon JV, Fonov V, Pruessner J, Robles M, Collins DL. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54(2):940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 21.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society Series B. 2005;67(2):301–320. [Google Scholar]
- 22.Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage. 2003;20(2):870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 23.Roy S, He Q, Sweeney E, Carass A, Reich DS, Prince JL, Pham DL. Subject specific sparse dictionary learning for atlas based brain MRI segmentation. IEEE J Biomedical and Health Informatics. 2015;19(5):1598–1609. doi: 10.1109/JBHI.2015.2439242. [DOI] [PMC free article] [PubMed] [Google Scholar]


