Abstract
Live imaging of genome has offered important insights into the dynamics of the genome organization and gene expression. The demand to image simultaneously multiple genomic loci has prompted a flurry of exciting advances in multicolor CRISPR imaging, although color-based multiplexing is limited by the need for spectrally distinct fluorophores. Here we introduce an approach to achieve highly multiplexed live recording via correlative CRISPR imaging and sequential DNA fluorescence in situ hybridization (FISH). This approach first performs one-color live imaging of multiple genomic loci and then uses sequential rounds of DNA FISH to determine the loci identity. We have optimized the FISH protocol so that each round is complete in 1 min, demonstrating the identification of seven genomic elements and the capability to sustain reversible staining and washing for up to 20 rounds. We have also developed a correlation-based algorithm to faithfully register live and FISH images. Our approach keeps the rest of the color palette open to image other cellular phenomena of interest, as demonstrated by our simultaneous live imaging of genomic loci together with a cell cycle reporter. Furthermore, the algorithm to register faithfully between live and fixed imaging is directly transferrable to other systems such as multiplex RNA imaging with RNA-FISH and multiplex protein imaging with antibody-staining.
Introduction
Live imaging of genome has offered important insights into the dynamics of the genome organization and gene expression, both at the global nucleus scale (1, 2) and local chromatin scale (3, 4). Recent engineering efforts on DNA-binding protein systems have led to facile imaging of endogenous sequence-specific genomic loci in living cells (5, 6). The demand to image simultaneously multiple genomic loci has prompted a flurry of exciting advances in multicolor imaging methods where intriguing heterogeneous dynamics were observed for different loci. For example, in the CRISPR imaging systems, genomic loci are distinguished by labeling with different fluorescence proteins through Cas9 protein orthologs (7, 8) or modified single-guide RNA (sgRNA) scaffolds that recruit different RNA-binding proteins (9, 10, 11, 12). In all these systems, the number of loci that can be distinguished simultaneously is still limited by the choice of fluorescence proteins that have sufficient color separation. Meanwhile, in fixed systems, highly multiplexed fluorescence in situ hybridization for both RNA (13, 14, 15) and DNA (16) has been reported by sequentially applying and imaging different probes following a prearranged code. Tens or even hundreds of DNA or RNA species can be distinguished in this way.
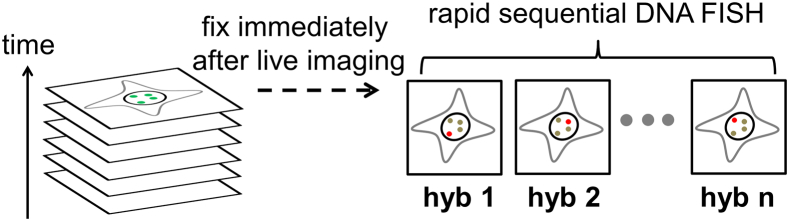
Here we report a correlative imaging method that combines the dynamic tracking capability of CRISPR imaging with the multiplicity of sequential fluorescence in situ hybridization (FISH). This method allows us to perform live-cell CRISPR imaging first to obtain the dynamics of many genomic loci using one Cas9 protein and the corresponding sgRNAs followed by sequential rounds of DNA FISH to decode loci identity (Fig. 1).
Figure 1.
Schematic of correlative CRISPR imaging and sequential DNA FISH. Live cells are first imaged in time-lapse mode to acquire dynamics information. Multiple genomic loci are simultaneously imaged without distinguishing their identities. Cells are fixed immediately after live imaging. Rapid sequential rounds of DNA FISH are performed afterwards. As probes specifically bound to a locus are introduced in each round, the identity of the locus is resolved by comparing the last frame of live image and fixed images. To see this figure in color, go online.
Materials and Methods
Cell culture
Human retinal pigment epithelium (RPE) cells (CRL-4000; American Type Culture Collection, Manassas, VA) were maintained in Dulbecco’s modified Eagle medium/Nutrient Mixture F-12 with GlutaMAX supplement (DMEM/F-12; Gibco/Thermo Fisher Scientific, Waltham, MA) in 10% Tet-system-approved fetal bovine serum from Clontech (Mountain View, CA). The human embryonic kidney (HEK) cell line HEK293T was maintained in DMEM with high glucose (University of California, San Francisco, Cell Culture Facility, San Francisco, CA) in 10% Tet-system-approved fetal bovine serum (Clontech). Cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Lentiviral production and stable expression of dCas9, sgRNA, and Fucci constructs
For viral production, HEK293T cells were seeded onto six-well plates 1 day before transfection. A quantity of 0.1 μg of pMD2.G plasmid, 0.8 μg of pCMV-dR8.91, and 1 μg of the lentiviral vector (Tet-on 3G, dCas9-EGFP, sgRNA, or Fucci) were cotransfected into HEK293T cells using FuGENE (Promega, Madison, WI) following the manufacturer’s recommended protocol. Virus was harvested 48 h posttransfection. For viral transduction, cells were incubated with culture-medium-diluted viral supernatant for 12 h. RPE cell lines stably expressing dCas9-EGFP were generated by coinfecting cells with a lentiviral cocktail containing viruses encoding both dCas9-EGFP and the Tet-on 3G transactivator protein (Clontech). Clonal cell lines expressing dCas9-EGFP were generated by picking a single-cell colony. The clones with low basal level expression of dCas9-EGFP were selected for CRISPR imaging. Clonal RPE cell line expressing dCas9-EGFP were transduced with lentivirus encoding Fucci (Gemini::RFP and Cdt1::mIFP linked by P2A domain) and cells with stable expression of Fucci was sorted using flow cytometry. The Fucci-containing cell line showed normal cell division and cell cycle progression.
Optical setup and image acquisition
Fluorescence images were acquired on an inverted wide-field microscope (Ti-E; Nikon, Melville, NY) with a 100× 1.45 N.A. oil immersion objective. The custom-build epi-illumination optics (X-Cite XLED1; Lumen Dynamics, Mississauga, Ontario, Canada) provided excitation in DAPI, FITC, Cy3, and Cy5 channels. A quad-band dichroic excitation filter (ZT405/488/561/640; Semrock, Rochester, NY) was installed in the excitation path and a quad-band emission filter (FF410/504/582/669; Semrock) in the emission path. Additional emission filters at 525 and 595 nm with 50 nm bandwidth were used for emission in FITC and Cy3 channels, respectively, to further reduce background noise. They were mounted into a motorized filter-wheel (Lambda 10-B; Sutter Instrument, Novato, CA). A motorized microscope stage (Applied Scientific Instrumentation (ASI), Eugene, OR) controls the xy and z translation of the sample. The images were recorded with a sCMOS camera (C11440; Hamamatsu Photonics, Iwata City, Japan) in z-stacks of 6 μm with 0.3 μm steps. The microscope, light source, motorized stage, motorized filter wheel, and camera were controlled through custom configuration in the software Micro-Manager (https://www.micro-manager.org). Live imaging was performed in the FITC channel, and FISH imaging was performed in the Cy5 channel, unless noted otherwise. Each image in Cy5 channel has a corresponding image in the DAPI channel at the exactly same position for use in the two-step image registration algorithm described later. To improve the mIFP signal, a final concentration of 25 μM biliverdin (30891; Sigma-Aldrich) was added to Fucci-containing cells at 12 h before live imaging in the FITC, Cy3, and Cy5 channels.
DNA FISH
The bottom cover-glass surface of an eight-well imaging chamber (Nunc Lab-Tek II; Thermo Fisher Scientific) was coated with a 0.01% poly-L-lysine solution (Sigma-Aldrich, St. Louis, MO) solution for 15 min and rinsed with PBS buffer three times. Cells were allowed to attach to the cover-glass surface for overnight. Cells were fixed with 4% paraformaldehyde solution (Chem Cruz; Santa Cruz Biotechnology, Dallas, TX) at room temperature for 5 min followed by PBS buffer wash for three times. Cell membrane and nucleus membrane were permeabilized by methanol incubation for 5 min followed by PBS buffer wash. Cells were then heated on a hot plate at 80°C for 10 min in 80% formamide (Sigma-Aldrich). Cells were incubated for 1 min in hybridization solution of 200 nM oligo probes in the presence of 50% formamide and 2× SSC followed by PBS buffer wash three times. The conventional hybridization reagents such as dextran sulfate and blocking DNA reagents were not required. Imaging was performed in imaging buffer containing glucose, glucose oxidase, and catalase to prevent photobleaching. After each round of imaging in sequential FISH, the cells were washed with 80% formamide at 50°C for 30 s to remove the bound oligo probes followed by a new round of probe hybridization.
Two-step image registration algorithm
To improve the reproducibility of sample positioning during repetitive mounting and unmounting steps, we designed a three-dimensional (3D)-printed stage adaptor (Fig. S1 in the Supporting Material) that ensures a tight fit of eight-well chambers on the microscope stage. First, to precisely register the acquired live and fixed images, we performed a stage registration step (Fig. S2) that allows the original region to be found after the sample is put back onto the microscope stage. The position of the sample cell in live imaging at the last frame is termed as “original”. The position of the sample cell immediately after it is put back on the stage is considered as “initial”. Image of nuclei at the initial position was convoluted to the image of nuclei in the original position to calculate full-image correlation. The images were downsampled (a fraction of pixels, for example, 1/10, in both x and y) to speed up the registration algorithm for near real-time feedback in sample positioning. The images were then converted to binary format and convoluted to calculate correlation. The peak position in correlation corresponded to the original position in live imaging and the compensatory stage displacement to return to the original position was calculated based on the pixel size. The algorithm takes only a few seconds to process two 1024 × 1024 images. The motorized stage was then translated to the adjusted position based on the input of translation displacement. The spatial precision of this algorithm depends on the downsampling and is usually found within 1 μm, consistent with the precision expectation at this step because the distance between every 10 pixels in the x or y direction is ∼1 μm. Typically, the initial position is within the field of view of the original position. However, if necessary, a much larger imaging region, on a millimeter scale, could be rapidly scanned and tiled using the software Micro-Manager and the software ImageJ’s (National Institutes of Health, Bethesda, MD) stitching plugin to find the original position. The algorithm works well based on similar features between images. For example, when nuclei shape can be acquired in live imaging (e.g., diffusive nuclear EGFP signal in this system), the correlation could be done between the last frame of live imaging and fixed DAPI signal. Alternatively, on the stage, the DAPI staining is performed before the sample is taken off the stage. The algorithm works well for both a single slice and a z-stack of nuclei.
Second, a further refinement in image registration was applied in image analysis to register between CRISPR loci at the last frame of live-cell imaging and FISH spots (Fig. S3). Based on the nuclei shape, images taken in live EGFP channels at the last frame and fixed DAPI channels were registered and then the same image registration operation was applied to overlay the last frame of live images and FISH images. The registration algorithm is a modified version of image registration (17) that accounts for sample rotation and achieves subpixel precision through upsampled discrete Fourier transforms (DFT) cross correlation. Briefly, the algorithm rotates an image in 1° steps and estimates the two-dimensional translational shift to register with a reference image through calculating the cross-correlation peak by fast Fourier transforming. The highest peak corresponds to not only the two-dimensional translational shift estimate but also the optimal angle of rotation. A refined translational registration with subpixel precision is then achieved through upsampling the DFT in the small neighborhood of that earlier estimate by means of matrix-multiplication DFT. Our algorithm registered the images to a precision within 1/10 of a pixel. Because the algorithm operates on Fourier transform in the frequency domain, it remains robust in image registration when features are more densely distributed (Fig. S4).
Single particle tracking
The positions of the spots in cell nuclei in live images were determined in CellProfiler (cellprofiler.org). The position information at different time points is linked to generate trajectories using custom-written MATLAB (The MathWorks, Natick, MA) codes.
Measurement of loci intensity
Z-stack images were first projected to generate an image using maximum-z projection and intensity measurement is performed on the projected images using custom-written MATLAB codes. The peak intensity of the genomic loci puncta was measured as the peak value in the selected region of interest, subtracting the nuclear background. The nuclear background was calculated as the mean value in nucleus regions lacking detectable puncta.
Target genomic loci
We use the hg19 version of human genome. The regions involved in this study are Chr1: 2581275-2634211; Chr3: 195505721-195515533 (denoted as Chr 3); Chr3: 195199025-195233876 (denoted as Chr3∗); Chr7: 158122661-158135328; Chr13: 112930813-112973591 Chr19: 44720001-44760001 (denoted as NR1); Chr19: 29120001-29160001 (denoted as NR2); ChrX: 30806671-30824818.
sgRNA protospacer sequence to image human genomic loci
sgChr3, 5′ GUGGCGUGACCUGUGGAUGCUG 3′
sgChr7, 5′ GCUCUUAUGGUGAGAGUGU 3′
sgChr13, 5′ GAAGGAAUGGUCCAUGCUUACC 3′
sgChrX, 5′ GGCAAGGCAAGGCAAGGCACA 3′
DNA FISH probe sequence to image human genomic loci
Chr1, 5′ CCAGGTGAGCATCTGACAGCC 3′
Chr3, 5′ CTTCCTGTCACCGAC 3′
Chr3∗, 5′ CCACTGTGATATCATACAGAGG 3′
Chr7, 5′ CCCACACTCTCACCATAAGAGC 3′
Chr13, 5′ GGTAAGCATGGACCATTCCTTC 3′
ChrX, 5′ TTGCCTTGTGCCTTGCCTTGC 3′
Telomere, 5′ CCCTAACCCTAACCCTAA 3′
Centromere, 5′ ATTCGTTGGAAACGGGA 3′
Nonrepetitive probe design and synthesis
Oligo pool library was designed such that seven modules were concatenated. Two sets of index primer pairs were used to amplify the entire oligo pool library or selectively a sublibrary of oligos. A variable region was designed to cover a genomic region of interest. Typically, 200 probes tiling over a 40-kb genomic region led to a detectable FISH signal. The 30 nt variable region was flanked on one side by the T7 promoter used in in vitro transcription and on the other side by a reverse transcription primer sequence shared in the entire library. The sequence in the variable region was first designed in the software OligoArray 2.1 (berry.engin.umich.edu/oligoarray2_1/) using the parameter set -n 20 -l 30 -L 30 -D 1000 -t 70 -T 90 -s 76 -x 72 -p 35 -P 80 -m “GGGG;CCCC;TTTTT;AAAAA”. Sequences with homology of 17 nt or more to the human genome were detected with the tool Blast+ (National Institutes of Health) and removed. Sequences with homology of 14 nt or more due to concatenation between variable region and reverse transcription primer were also removed. The index primers and reverse transcription primers were designed by first truncating sequences to a 20-mer oligo library from a 25-mer random oligo library (18). The oligos with a melting temperature between 75 and 83°C were selected. Sequences with homology of 11 nt or more or 5 nt or more homology to the 3′ end within the 20-mer oligo library were removed. Oligos without a G or C base in the last 2 nt on the 3′ end were removed. Sequences with homology to the T7 promoter sequence were removed. Nonrepetitive probe sequence files for loci NR1 and NR2 are included in the Supporting Material.
The oligo pool library is synthesized by the software CustomArray (Bothell, WA) and amplified by limited cycle PCR. In vitro transcription (E2040S; New England Biolabs, Ipswich, MA) was performed and dsDNA was converted to RNA with an effective amplification of 200+-fold. The RNA was then converted back to ssDNA in a reverse transcription reaction (EP0751; Thermo Fisher Scientific).
Results
Fast FISH staining of genomic DNA
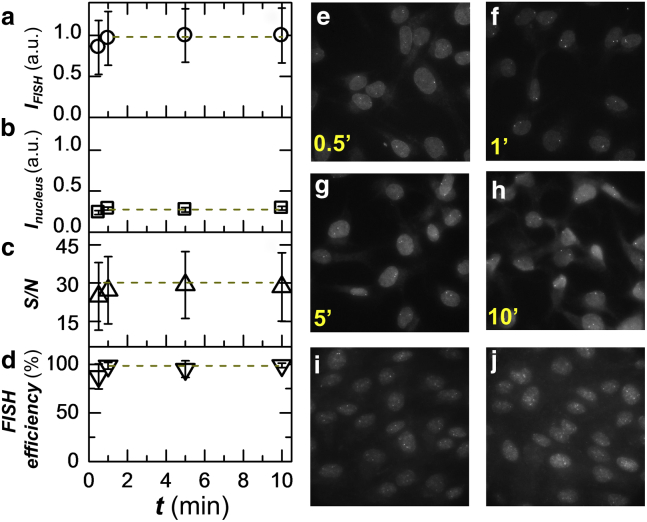
To practically and efficiently perform multiple rounds of DNA FISH on the same sample, we sought to address the technical challenge that DNA FISH often requires many hours or overnight for probe hybridization, which is much longer compared to that needed for RNA FISH because of the double-stranded nature of DNA. As recent reports suggest, oligo DNA probes can rapidly label RNA or a single-stranded DNA motif on the timescale of 5–30 min (15, 16, 19, 20, 21); therefore, we speculate that a rapid binding of oligo probes directly to genomic DNA is possible after sufficient DNA denaturation to separate duplex strands. To simplify the initial test, we started with oligo DNA probes targeting the tandemly repetitive sequence in the human genome (Fig. S5) so only one FISH probe is needed (22). Fig. 2, a–d shows the kinetics of FISH staining targeting a tandemly repetitive sequence region. Under optimized conditions, staining is essentially complete in 1 min and an acceptable signal-to-noise ratio is even achieved in as short as 0.5 min, in drastic contrast to the common practice of overnight incubation. The representative images of cells stained for different durations show that the nuclear background is consistently low throughout staining. The efficiency of FISH staining, calculated as the ratio of observed to expected number of FISH puncta, reaches almost 100% by 1 min (Figs. 2 d and S6). The signal-to-noise ratio reaches ∼30-fold for the ∼800 copies and we thus estimate a lower detection limit to be ∼40 probes (Fig. S7), a number consistent with those reported in recent RNA FISH studies. The signal-to-noise, nuclear background, and FISH efficiency remain constant after 1 min so a wide time window of hybridization works well. To test whether the optimized FISH protocol could also be widely applied to label nonrepetitive genomic sequences, we designed oligo DNA probe pools that tile nonrepetitive genomic regions (23) and observed similarly efficient staining of these regions (Fig. 2, i and j). Typically, we observe two puncta in each cell nucleus as expected for the diploid cells. Thus, this method can rapidly detect aneuploidy in interphase cells and potentially report copy number variations with probes targeting specific genes of interest.
Figure 2.
Rapid staining can be achieved with oligo DNA probes under optimized DNA FISH protocol. (a) Kinetics of the rapid DNA FISH staining. Intensity of FISH puncta for a tandemly repetitive sequence in the human genome is shown. Intensity is averaged over 300+ puncta and error bars denote SD. (b) Intensity of nuclear background staining at different time points. (c) The signal-to-noise ratio of FISH puncta. (d) The efficiency of FISH staining, calculated as the ratio of observed to expected number of puncta. (e–h) Representative images at different time points of staining. Maximum intensity projection is shown for each 3D z-stack. (i–j) Representative images of two genomic regions denoted as NR1 (i) and NR2 (j) of nonrepetitive sequence labeled by tiling ∼200 probes over a 40-kb region with a staining time of 2 min. See Materials and Methods for details on probe sequence. Maximum intensity projection is shown for each 3D z-stack. To see this figure in color, go online.
Three aspects could have contributed to the fast staining: 1) gentle fixation by cross linking and permeabilization by an alcohol wash dissolves most lipids in cell membrane and nuclear membrane; 2) an extended heating step ensures thorough denaturation between double strands of genomic DNA—we find that the genome accessibility directly correlates to the heating denaturation (Fig. S8); and 3) oligo DNA allows extremely fast diffusion through cellular and nuclear structures. Note that this FISH protocol is also greatly simplified in procedure, requiring no crowding reagents such as dextran sulfate to boost probe diffusion or blocking DNA reagents such as salmon sperm DNA and Cot-1 DNA to prevent nonspecific nuclear staining.
Multiple sequential rounds of DNA FISH in multiplex imaging
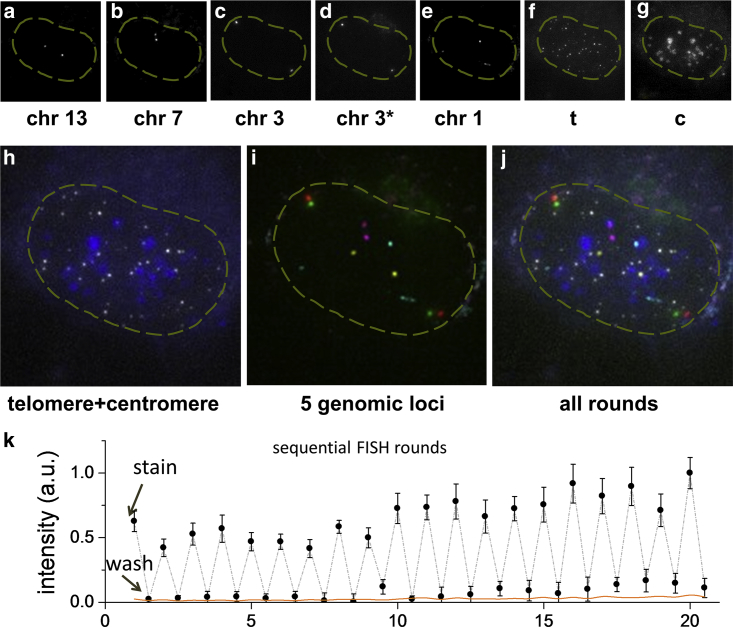
With the ease of performing rapid FISH staining, we then tested whether this technique can be applied to multiple sequential rounds of hybridization. A technical challenge here is to minimize the interference of bound probes with the next round of imaging. In previous studies, this issue was addressed either by DNase enzymatic reactions to degrade DNA probes bound to RNA targets (13) or by photobleaching the dyes on bound probes using powerful lasers and then adding new probes targeting other vacant binding sites (15). Here, to explore a simpler protocol, we apply a stringent wash step using a concentrated formamide solution at an elevated temperature to strip the bound oligo DNA probes after each round of imaging and detection. Fig. 3, a–g, shows an example of sequential FISH rounds where five distinct genomic regions and the telomere and centromere are sequentially detected through six rounds of staining and washing. The images show effective staining of specific sequences without interfering loci signals between sequential rounds. We find that the wash step is efficient, and that probes are effectively removed within 0.5 min. In Fig. 3, we have also demonstrated two-color FISH (Cy3 and Cy5 channels) to distinguish two loci as close as 300 kb away on chromosome 3 in one FISH round, suggesting the multiplex imaging capacity of the method could be further expanded through combining multicolor approaches.
Figure 3.
Multiple genomic loci can be resolved through multiple sequential rounds of DNA FISH. (a–g) Chr13, Chr7, Chr3, Chr3∗, Chr1, telomere, and centromere are sequentially resolved in six rounds of staining of 1 min and wash of 0.5 min. Two sites 300 kb apart on chromosome 3 (denoted as Chr3 and Chr3∗) are costained and distinguished in one round with two different colors. Maximum intensity projection is shown for each 3D z-stack. (Dashed yellow line) Nucleus contour. (h) False-color overlay of telomere (white) and centromere (blue) staining. (i) False-color overlay of Chr13 (yellow), Chr7 (magenta), Chr3 (red), Chr3∗ (green), and Chr1 (cyan). (j) Overlay of all loci determined in the sequential DNA FISH. Color is the same as in (h) and (i). Image area in (a)–(i) is 22 × 22 μm. (k) Peak intensity value of 20 rounds of repeated staining of 1 min and washing of 0.5 min with nuclear background subtracted, averaged over eight loci. Error bars denote SD. The nuclear background (orange line) is calculated as the average intensity within a nuclear region minus the average intensity from the open region without cells.
Recent studies show that multiple rounds can quickly expand the multiplex capacity to thousands or more through various encoding strategies (13, 15, 16). To further test the potential of the method for multiplex imaging, we measured intensities of a specific genomic region through 20 rounds of alternating staining and washing. Fig. 3 k shows that the loci can be reversibly and consistently stained for at least 20 rounds without visible sample deterioration. High contrast is reproducibly seen with consistently high intensity after staining and almost negligible signal after wash. In fact, the residual signal after washing is so dim that it is often undetectable from the nuclear background, especially in earlier rounds. The consistent and reversible staining and washing in multiple DNA FISH rounds suggests the multiplex potential of the method.
Sequential DNA FISH after live imaging resolves loci identity
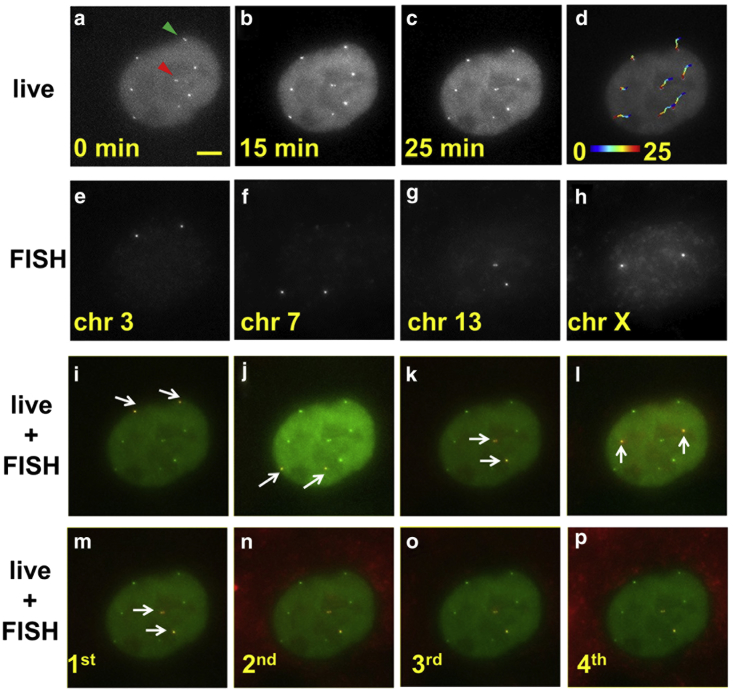
Fig. 4, a–d, shows representative time-lapse images of a cell with simultaneous labeling of multiple genomic loci. Here, four sgRNAs with different protospacer sequences are expressed in the cell nucleus at the presence of dCas9::EGFP fusion protein, resulting in efficient labeling of the corresponding genomic loci and eight bright spots in the nucleus as homologous chromosomes are simultaneously labeled in the diploid cell. As these sgRNAs share the same dCas9 binding motif, one cannot directly distinguish the loci identities based on the live images. Nonetheless, genome dynamics can be extracted regardless of the loci identities. Comparison of images at different time points (Fig. 4, a–c) suggests that the relative positions of these loci are essentially stable, consistent with earlier FRAP measurement on cell nuclei (1, 2). The trajectory over 25 min overlaid on live images reveals a global motion of the nucleus (Fig. 4 d). The adjusted chromosome dynamics after subtracting the contribution of global nucleus movement show a more randomized trajectory direction (Fig. S9). Furthermore, we show in Fig. S10 that the global movement of the nucleus is quite commonly seen, more so on the longer observation timescale. We find that this global scale of nucleus movement is not caused by stage drift as cells in the same field of view have a randomized trajectory direction with respect to one another. The live images also capture the dynamic vibrations between sister chromatids (arrowheads in Fig. 4 a), which are sometimes distinguishable when the distance exceeds the optical diffraction limit.
Figure 4.
Sequential rounds of DNA FISH resolve identities of loci imaged in live-cell mode. (a–c) Representative micrographs of a cell from time-lapse images. Eight spots corresponding to four genomic regions in a diploid RPE cell are seen. (Green arrowhead) Pair of sister chromatids that are distinguished at the beginning of the live imaging, and that came closer than the diffraction limit at a later time. (Red arrowhead) Another pair of sister chromatids that are distinct throughout the image acquisition with fluctuating separation. (d) Overlay of the cell image at the end of the live observation with the corresponding loci trajectories. (Color) Time in minutes. (e–h) FISH images in four sequential FISH rounds reveal the identities of the loci. (i–l) Overlay of live images at 25 min with DNA FISH images from each round shows faithful registration and negligible nuclear deformation. (White arrows) FISH spots in each round. (m–p) Four sequential DNA FISH rounds staining Chr13 show consistent image registration between rounds. All micrographs here show the same image area and scale bar in (a) as 5 μm.
The cells are fixed at the end of the live observation and prepared for sequential DNA FISH. To correlate live imaging of genomic loci with sequential FISH, it is desirable to register the same area with submicrometer precision to correlate images between live condition and fixed FISH rounds. This task is challenging, especially because the denaturation of DNA duplex strands during DNA FISH preparation requires elevated temperatures; therefore, the sample has to be taken off the microscope stage. To address this issue, we use a combination of three measures to achieve faithful image registration. First, a 3D printed microscope stage adapter ensures consistent sample orientation by restricting sample rotation (Fig. S1). Second, we employ a rapid correlation-based algorithm to find the same cell observed in live imaging. Third, and finally, a more sophisticated registration algorithm (17) that accounts for both translation and rotation is applied to register the live and fixed images based on the nucleus shape.
The identities of the four genomic loci are resolved in four rounds of DNA FISH as probes specific to each locus are sequentially introduced in each round (Fig. 4, e–h). Furthermore, overlay of the live-cell images and fixed-cell images demonstrate consistent loci position between live condition and fixed condition across various loci with a root mean square error of ∼52 nm in loci registration (Figs. 4, i–l and S11). Similarly, we find images between sequential FISH rounds for a given probe register well with a root mean square error of ∼43 nm (Figs. 4, m–p, and S12). The successful image registration and principle component analysis on nuclear morphology (Fig. S13) also confirms that negligible deformation is introduced during the FISH preparation steps. A closer inspection of Fig. 4, m–p, shows that positions of sister chromatids and the distances between them are faithfully maintained in repeated sequential rounds and registered well with the live image, further confirming the method maintains almost intact nuclear morphology at both global and local scale and is compatible with high-resolution imaging to resolve fine chromatin structure.
Multicolor live imaging enabled by correlative CRISPR imaging and sequential DNA FISH
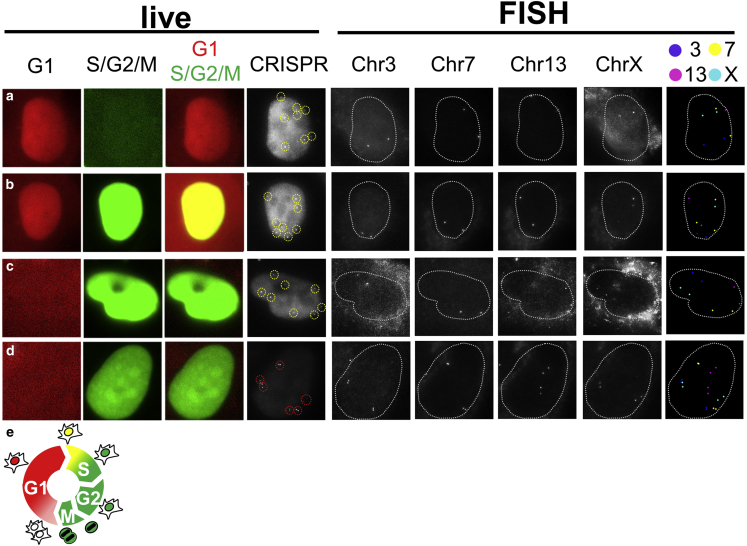
Because the correlative CRISPR imaging and sequential DNA FISH uses a single color channel in live imaging to track multiple genomic regions, it opens up other color imaging channels in live cell imaging to extract information otherwise difficult to obtain. Here we demonstrated this capability by performing CRISPR imaging of four loci in cells expressing the Fucci probe, a widely used cell-cycle tracker that uses two colors to mark G1 or S/G2/M cell phases, respectively (24). With the help from the Fucci probe, we were able to distinguish G1 phase cells and early S phase cells (Fig. 5, a–c), both of which display singlet spots for each locus in CRISPR imaging results, while late S/G2 phase cells could be identified by doublet spots corresponding to replicated sister chromatids (Fig. 5 d). Because the Fucci reporter itself already occupies two of the three color channels (green, orange, and far red) so that multicolor live cell imaging can be easily performed, this experiment is challenging for other multicolor CRISPR imaging methods. Potentially, with Fucci reporting the onset of the S phase and CRISPR imaging detecting the replication of given genomic loci (e.g., based on intensity change of the labeled spot), we can profile replication timing and test how it affects the spatial organization of genome and the formation of topologically associated domains formation (as recently proposed based on Hi-C results (25)). In addition, with other color channels opened up, our approach can also to be used to monitor the interaction of genomic loci with other nuclear components such as lamin, nuclear pore complex, and various nuclear bodies (26), many of which are known to be active genome organizers.
Figure 5.
Simultaneous live CRISPR imaging of multiple genomic elements and cell cycle tracking. (a–d) Correlative imaging of CRISPR imaging and sequential DNA-FISH is performed on cells with a Fucci cell cycle tracker at the G1, onset of S, early S, and G2 phases, respectively. The first three columns in the live panel correspond to signals from Fucci cell tracker and the fourth column in the live panel shows CRISPR imaging where puncta are highlighted (circle). The first four columns in the FISH panel are FISH images corresponding to four different genomic loci; and the last column in the FISH panel (false color) shows all FISH puncta identified in sequential rounds. (Dotted line) Nuclei. (e) Schematic of Fucci cell cycle tracker.
Discussion
This method thus combines the advantage of acquiring dynamics in live-cell imaging and multiplex imaging capacity in sequential FISH. It frees up other in vivo color channels for imaging applications such as RNA expression and processing (4, 27), protein expression (28, 29), and for imaging various nuclear components that are closely related to the spatial organization and dynamics of the genome. Moreover, a simpler system of live imaging with more uniform expression and assembly of Cas9 protein and sgRNA could potentially reduce system variability and facilitate further quantitative analysis. As DNA FISH has demonstrated the power to probe genome organization in high-throughput fashion such as HIP-map (30) and with superresolution imaging using the tool Oligopaints (http://genetics.med.harvard.edu/oligopaints/) (31, 32), this work could potentially add a new dimension of dynamic information. As the method of image registration between live and fixed conditions is directly transferrable to other systems, the concept of multiplex imaging through correlative imaging between live and fixed cells could be similarly applied to RNA imaging resolved by sequential RNA-FISH and protein imaging by sequential antibody-staining.
Conclusion
In summary, we introduce a correlative imaging method that combines the dynamic tracking capability of CRISPR imaging and the multiplicity of sequential FISH. After live imaging to obtain dynamics information of multiple genomic loci using a one-color CRISPR system of one Cas9 protein and multiple sgRNAs, we perform rapid sequential rounds of DNA FISH to resolve loci identities. We also demonstrate a greatly simplified DNA FISH protocol that effectively stains genomic DNA in as short as 30 s in contrast to the common practice of overnight incubation. Our correlation-based algorithm to faithfully register between live images and fixed images can be readily adapted for other multiplex imaging applications.
Author Contributions
J.G. and B.H. designed research; J.G., H.L., and S.F. performed research; J.G., X.S., and H.L. analyzed data; and J.G. and B.H. wrote the article.
Acknowledgments
This work is supported by a W.M. Keck Foundation Medical Research grant, the National Institutes of Health (No. R21EB021453 and the Single Cell Analysis Program No. R33EB019784). H.L. receives the National Science Foundation Graduate Research Fellowship.
Editor: Tamar Schlick.
Footnotes
Supporting Materials and Methods, thirteen figures, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30162-5.
Supporting Material
The last frame shows the tracked trajectories.
References
- 1.Strickfaden H., Zunhammer A., Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 2010;1:284–297. doi: 10.4161/nucl.1.3.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlich D., Beaudouin J., Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 3.Belmont A.S. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr. Opin. Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masui O., Bonnet I., Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B., Gilbert L.A., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B., Guan J., Huang B. Imaging specific genomic DNA in living cells. Annu. Rev. Biophys. 2016;45:1–23. doi: 10.1146/annurev-biophys-062215-010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B., Hu J., Huang B. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016;44:e75. doi: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H., Naseri A., Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl. Acad. Sci. USA. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y., Rocha P.P., Skok J.A. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat. Commun. 2016;7:11707. doi: 10.1038/ncomms11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao S., Zhang W., Sun Y. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 2016;44:e86. doi: 10.1093/nar/gkw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., Su J.H., Zhuang X. An RNA-aptamer-based two-color CRISPR labeling system. Sci. Rep. 2016;6:26857. doi: 10.1038/srep26857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma H., Tu L.C., Pederson T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016;34:528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubeck E., Coskun A.F., Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coskun A.F., Cai L. Dense transcript profiling in single cells by image correlation decoding. Nat. Methods. 2016;13:657–660. doi: 10.1038/nmeth.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K.H., Boettiger A.N., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Su J.H., Zhuang X. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353:598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guizar-Sicairos M., Thurman S.T., Fienup J.R. Efficient subpixel image registration algorithms. Opt. Lett. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q., Schlabach M.R., Elledge S.J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. USA. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agerholm I.E., Ziebe S., Kølvraa S. Sequential FISH analysis using competitive displacement of labelled peptide nucleic acid probes for eight chromosomes in human blastomeres. Hum. Reprod. 2005;20:1072–1077. doi: 10.1093/humrep/deh735. [DOI] [PubMed] [Google Scholar]
- 20.Shaffer S.M., Wu M.T., Raj A. Turbo FISH: a method for rapid single molecule RNA FISH. PLoS One. 2013;8:e75120. doi: 10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng W., Shi X., Singer R.H. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. USA. 2015;112:11870–11875. doi: 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson G. Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beliveau B.J., Joyce E.F., Wu C.T. Versatile design and synthesis platform for visualizing genomes with Oligopaints FISH probes. Proc. Natl. Acad. Sci. USA. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaue-Sawano A., Kurokawa H., Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 25.Pope B.D., Ryba T., Gilbert D.M. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., Sawyer I.A., Dundr M. Cajal bodies are linked to genome conformation. Nat. Commun. 2016;7:10966. doi: 10.1038/ncomms10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levesque M.J., Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods. 2013;10:246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clowney E.J., LeGros M.A., Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood A.M., van Bortle K., Corces V.G. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol. Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shachar S., Voss T.C., Misteli T. Identification of gene positioning factors using high-throughput imaging mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beliveau B.J., Boettiger A.N., Wu C.T. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaints FISH probes. Nat. Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boettiger A.N., Bintu B., Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The last frame shows the tracked trajectories.