Abstract
Icosahedral viral capsids are made of a large number of symmetrically organized protein subunits whose local movements can be essential for infection. In the capsid of the minute virus of mice, events required for infection that involve translocation of peptides through capsid pores are associated with a subtle conformational change. In vitro, this change can be reversibly induced by overcoming the energy barrier through mild heating of the capsid, but little is known about the capsid regions involved in the process. Here, we use hydrogen-deuterium exchange coupled to mass spectrometry to analyze the dynamics of the minute virus of mice capsid at increasing temperatures. Our results indicate that the transition associated with peptide translocation involves the structural rearrangement of regions distant from the capsid pores. These alterations are reflected in an increased dynamics of some secondary-structure elements in the capsid shell from which spikes protrude, and a decreased dynamics in the long intertwined loops that form the large capsid spikes. Thus, the translocation events through capsid pores involve a global conformational rearrangement of the capsid and a complex alteration of its equilibrium dynamics. This study additionally demonstrates the potential of hydrogen-deuterium exchange coupled to mass spectrometry to explore in detail temperature-dependent structural dynamics in large macromolecular protein assemblies. Most importantly, it paves the way for undertaking novel studies of the relationship between structure, dynamics, and biological function in virus particles and other large protein cages.
Introduction
Non-enveloped virus particles include multiple copies of one or a small set of proteins organized in a highly symmetrical capsid that encloses the viral genome. The structures of these large macromolecular assemblies at the atomic level, obtained mainly by x-ray crystallography or cryo-electron microscopy, have contributed enormously to the understanding of these complex biological systems (1, 2, 3). However, the structures depicted by these techniques may be somewhat deceptive. Viruses are not a static ensemble of atoms with a fixed position over time, but highly dynamic structures that constantly fluctuate around an average conformation as a result of thermal energy. These fluctuations are commonly termed “breathing” and represent an important feature in virus dynamics, playing a role in the infection process (1). In addition to these fluctuations, changes in physicochemical conditions or the action of specific biomolecules in vivo can lead to structural transitions between different states separated by an energy barrier (1, 4, 5, 6). Characterization of these and other biologically relevant structural transitions requires specific physicochemical conditions, such as a particular range of temperatures, that frequently fall beyond the technical limits of current high-resolution structural methods.
The minute virus of mice (MVM) represents an excellent model system for studying functionally relevant motions and rearrangements in large protein complexes. MVM is a non-enveloped single-stranded DNA (ssDNA) parvovirus with a T = 1 icosahedral capsid made of 60 equivalent subunits (VP1, VP2, and VP3) that share the same sequence and fold (Fig. 1 A). The only difference between the subunits is the length of their structurally disordered N-terminal segments (Nts) (7). VP2 is the most abundant capsid protein. VP1 is an extended form of VP2 and has a longer Nt, whereas in VP3, the Nt of VP2 has been shortened by proteolytic cleavage during virus entry into a host cell. The Nts of VP2 and VP1, initially buried in the capsid, contain molecular signals that are required during various stages of the infection cycle. They are externalized through capsid pores located at the fivefold symmetry axes (Fig. 1 B) (8, 9, 10, 11, 12, 13, 14). These pores are also involved in packaging and ejection of the viral ssDNA (15, 16).
Figure 1.
MVM capsid and HDX-MS experimental setup. (A) The MVM capsid structure (PDB: 1Z14 (17)) exhibits an icosahedral T = 1 symmetry. The black lines delimit the 60 VP2 subunits. The icosahedral symmetry axes (fivefold, threefold, and twofold) are labeled. (B) Five VP2 subunits (shown as ribbon models) surround a fivefold axis pore (center) through which translocation of peptide segments and viral DNA occurs during viral infection. The capsid contains 12 such pores. (C) Native mass spectrum of the intact MVM VLPs. A well-resolved series of charge states reveals a mass of 3883 kDa, confirming that the capsid consists of 60 copies of the 64 kDa VP2 capsid protein. (D) Schematic representation of the HDX approach used. The uptake of deuterium by the capsid protein backbone is monitored as a function of time at varying incubation temperatures around the transition temperature. To see this figure in color, go online.
Recombinantly expressed VP2 capsid proteins are able to assemble into DNA-free virus-like particles (VLPs) that are structurally indistinguishable from natural empty MVM capsids, except for the absence of the disordered VP1 Nts (17). These VLPs represent an excellent model system to explore the structural determinants of MVM capsid assembly and its physical properties and function (16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26). By excluding the VP1 Nts, these VLPs facilitate specific structural investigation of the biologically required externalization of VP2 Nts. This event is triggered in vivo by DNA encapsidation (14), but can also be mimicked in vitro by mild heating of the VLPs to >∼46°C (18, 19). The transition causes a reversible cooperative conformational rearrangement that involves slight changes in the degree of solvent exposure of some tryptophan residues that can be measured by spectrofluorometry (19). Previous attempts to characterize this conformational rearrangement by conventional methods, however, have been hampered by the required heating conditions.
Hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) (27, 28, 29, 30, 31, 32) has been previously used to probe the dynamics of virus capsids at equilibrium (33, 34, 35, 36, 37, 38, 39). However, to our knowledge, HDX-MS had not been used to probe and structurally characterize conformational transitions in virus particles or other large protein-based complexes. In this work, we apply HDX-MS to study the conformational rearrangement in the MVM capsid and probe temperature-dependent variations in capsid dynamics.
First, we study capsid dynamics at 0°C and show that structural elements important for virus infectivity, such as the pore region or the Nts, present the fastest exchange. Second, we monitor the deuterium uptake at increasing temperatures to determine region-specific temperature-dependent changes in capsid dynamics. Our results show that the cooperative transition required for viral infection and associated translocation events through the capsid pores is not constrained to the vicinity of the pores but involves a global cooperative rearrangement of the capsid.
Materials and Methods
Expression and purification of VLPs of MVM
Bacmid BM-VP2 (19) was used for the production of VLPs of MVM in H-5 insect cells. VLPs were purified as previously described (18), with minor modifications. VLP preparations were extensively dialyzed against phosphate-buffered saline. Capsid purity and integrity were assessed by electrophoresis and electron microscopy.
Spectrofluorometry of VLPs
The heat-induced conformational transition of the VLP associated to Nt externalization was initially probed by following the variation in intrinsic Trp fluorescence in thermal gradients, as previously described (19). Reversibility of the conformational rearrangement was ascertained by repeated fluorescence analysis after heating to a controlled maximum temperature (60°C) and cooling. Capsid dissociation was observed only at high (75°C) temperatures (19).
Native MS
Native MS samples were prepared by exchanging the buffer in which purified VLPs were obtained for aqueous ammonium acetate using multiple concentration and dilution steps with a centrifuge filter (Millipore, Billerica, MA). For the experiments shown in Fig. 1, samples were prepared in 100 mM ammonium acetate (pH 7.4) and kept on ice. For control experiments under HDX conditions, samples were heated at 60°C in water and spectra were acquired at multiple time points. Before injection into the mass spectrometer, low amounts (<25 mM) of triethylammonium acetate were added. MS experiments were performed on a modified QToF II (MS Vision, Waters, Elstree, United Kingdom) (40), operating at 10 mbar source pressure, 1300–1500 V capillary voltage, 100 V cone voltage, and 300 V collision energy with 2 × 10−2 mbar pressure in the collision cell using xenon as the collision gas (41). Tandem MS experiments were acquired by full envelope selection and fragmentation using a 400 V potential over the collision cell. Aliquots of the samples (at ∼2 μM monomer concentration) were introduced in the mass spectrometer through nano-electrospray ionization using gold-coated boro-silicate capillaries produced in house. All data were analyzed using Masslynx 4.1 software (Waters).
Hydrogen-deuterium exchange MS
Fluorometry studies showed that the structural transition is complete for most of the particles within 5 min at >50°C (19). Aliquots of MVM VLPs of 3 mg/mL and D2O (99.99% deuterium content) were heated separately at different temperatures (25, 37, 45, 50, 55, and 60°C) for 5 min before the start of the exchange reaction. The exchange reaction was initiated by addition of the D2O (50-fold dilution, 98% of deuteration) and heating was maintained during the reaction. Reactions were quenched at set time points (10, 30, 60, 600, 1800, 3600, and 14,400 s) by rapid addition of ice-cold quench solution (mixing to a final concentration of 100 mM TCEP and 2 M guanidine HCl (pH 2.5)). Immediately after quenching, the samples were injected into a Waters HDX/nanoAcquity system for digestion on an online pepsin column operating at 100 μL/min and 25°C, followed by separation on a 10 min reverse-phase ultra-performance liquid chromatography gradient at 0.5°C and MS on a Waters Xevo QToF G2. For the chromatography, the aqueous component used was 0.1% (v/v) formic acid in water set to pH 2.5 and the organic component was 0.1% (v/v) formic acid in acetonitrile. Acquisition of HDX of MVM without heating was performed identically, but samples were incubated on ice (at 0°C). All experiments at all temperatures were performed in triplicate. Identification of peptides was performed by dilution in H2O and using MSE data acquisition. Eighty-three peptides covering 80% of the sequence were identified for the experiments performed at 0°C (see the complete VP2 sequence in Fig. S1 in the Supporting Material). For VP2 regions covered by more than one peptide, the criteria were that the shortest peptides would be selected for the analysis, always ensuring that redundant information was consistent. This led to a subset of 51 peptides representing the same 80% sequence coverage. For the experiments performed at increasing temperature, 69 different peptides could be identified (over the entire temperature range), with a sequence coverage of 70%. Using the same selection criteria as stated above, a subset of 35 peptides was used for the analysis (peptides marked with solid circles in Fig. 2 A). Data for peptide identification were processed with ProteinLynx Global Server 2.5 software. Deuterium uptake was calculated compared with the control samples in H2O using Waters DynamX 3.0 software.
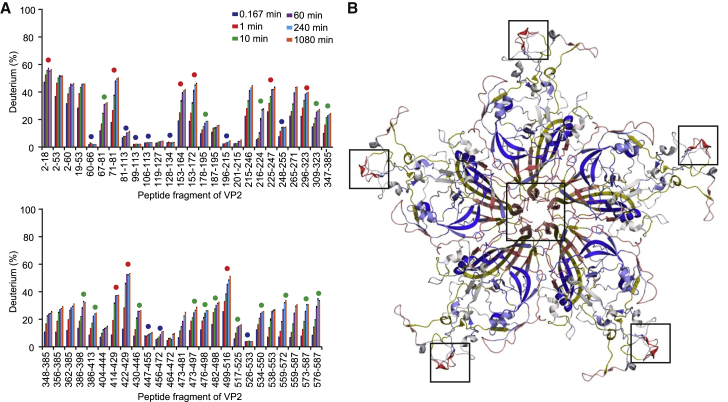
Figure 2.
Deuterium uptake by the MVM capsid at 0°C. (A) Uptake of deuterium for a selection of peptides at different time points expressed as a fraction of the maximal uptake. Circles indicate the peptides used for the analysis at increasing temperature. The circle color indicates the level of uptake after 1 h: red (high uptake), green (medium uptake), and blue (low uptake). (B) Mapping of the peptide uptake levels after 1 h on the capsid structure. For simplicity, only five subunits surrounding a capsid pore (center) are represented. The levels of exposure from the highest to the lowest percentage are colored red, white, and blue, respectively. Capsid regions that could not be probed are shown in yellow. Squares indicate some of the most dynamic regions of the capsid, with peptide 153–164 located around the pore (central square) and peptide 422–429 at the threefold axis spikes (peripheral squares). To see this figure in color, go online.
MS under denaturing conditions
To acquire spectra of denatured VP2, samples were introduced in the Waters HDX/nanoAcquity system without pepsin digestion. Spectra were deconvoluted using the MaxEnt algorithm in Masslynx 4.1 software (Waters).
Uptake plots
The total number of exchangeable backbone amide hydrogens was calculated by subtracting the Nt and the number of prolines from the peptide length and multiplying it by 0.98 (percentage of deuterium in the solvent). Profiles were fitted without considering back exchange or artifactual in-exchange (burst phase), as previously done by Monroe et al. (39). Back-exchange levels appeared to be grossly independent of the incubation temperature of the samples, as we verified using standard peptide mixtures (see Fig. S2).
Fitting of the uptake curves to the two-exponential equation (D = N − A1exp(−k1t) − A2exp(−k2t)) was not successful, since for some of the peptides, the values from the fitting (k1, k2, A1, and A2) did not converge and significant data could not be obtained. Therefore, we decided to base our analysis on the different deuteration levels that were reached over time at increasing temperatures. The number of exchanged amide hydrogens at 4 h was normalized to the total exchangeable amides (N4h/NTOTAL) and plotted against temperature. We limited our analysis to 4 h because data at 18 h time points (data not shown) showed great variation among the technical replicates. Nonetheless, native MS at 60°C for >5 h confirmed the stability of the particles under HDX-MS conditions. Capsid dissociation occurs only at a temperature of 70–80°C (18, 19). All uptake plots can be found in the Supporting Material (Figs. S6–S8).
Data fitting to a two-state transition
The transition temperature was determined by fitting the data to a two-state transition (19). Thermodynamic parameters (TM and ΔHǂTm) were obtained by nonlinear fitting of the experimental number of deuterons or fluorescence intensity, as previously done by Carreira et al. (19) (see Note S1 in the Supporting Material). For HDX-MS experiments, all peptides showing a linear increase in uptake were fitted, and the ones showing r2 values of 0.99 or higher (11 peptides) were used for calculation of the average Tm. The values of the transition enthalpy (ΔH™) were subjected to large errors that hampered an accurate estimation.
Circular dichroism (CD) spectroscopy
CD measurements were carried out using a Jasco-600 spectropolarimeter equipped with a Neslab RTE-100 computer-operated temperature control unit. Purified MVM VLP preparations at a protein (monomer) concentration of 2.6 μM in phosphate-buffered saline were used. Thermal experiments were carried out using the temperature scan mode and measuring the ellipticity in the far-ultraviolet range at 215 nm. A temperature scan rate of 20°C/h, a response time of 2 s, and a band width of 1 nm were used. Temperature was monitored by a thermocouple in the cuvette holder block.
Results and Discussion
The general workflow of the temperature-dependent HDX experiments described in this article is shown in Fig. 1 D. Overall, the approach is similar to most other HDX experiments but includes additional sample preparation and data analysis steps. Before HDX analysis, we verified both the composition and stoichiometry of the purified VLPs using native MS (Fig. 1 C). In these spectra, a well-resolved series of charge states was detected around 30,000 m/z, which was assigned to a mass of 3883.0 ± 0.8 kDa, corresponding well with the expected mass of 60 copies of the monomeric VP2 protein, 3877.9 kDa. The mass of the VP2 monomer was determined by liquid chromatography MS under denaturing conditions (Fig. S3 B). The oligomeric state was additionally confirmed using tandem MS experiments (Fig. S3 A). Having verified the integrity and purity of the VLPs, we performed HDX analysis (Fig. 1 D). Samples stored in protiated buffer were heated in parallel with a volume of D2O for 5 min. The solutions were subsequently mixed at a ratio that reached 98% deuterium content and continuously heated over different periods of time during which backbone amide hydrogens could exchange with deuterium from the buffer. After stopping the reaction with a quench solution, proteins were denatured, reduced, and digested and this mixture was loaded onto a high-performance liquid chromatography system. The peptides that resulted from the digestion were separated and analyzed by the coupled mass spectrometer, after which the deuterium uptake time courses were measured.
Breathing of the MVM capsid
In previous structure-function studies, mutant MVM particles were compared with wild-type particles using spectrofluorometry, atomic force microscopy (AFM), and infectivity assays (20, 25). Mutation of residues located around the base of the fivefold axis pores abolished viral infectivity, impaired the transition observed by fluorometry, and showed a highly increased mechanical stiffness at the pore regions (20, 25). Other studies showed that viral ssDNA segments bound to the capsid inner wall in the native MVM virion confer a higher resistance to thermal inactivation of its infectivity by increasing the mechanical stiffness of the capsid around the twofold and threefold axes (relative to mutant virions in which some of the capsid-DNA interactions had been removed) (21, 22, 23, 26). This biologically advantageous stabilization of the virion was achieved while preserving the limited mechanical stiffness of the capsid around the fivefold axis pores, which is essential for virus survival. These results indicated that the distribution of mechanical stiffness and variations in capsid conformational dynamics are essential for virus survival. However, the structural elements involved in functionally relevant conformational transitions had not been investigated.
To directly investigate the equilibrium dynamics (“breathing”) of different regions of the MVM capsid in a basal state (before the conformational transition), we performed HDX-MS experiments at 0°C without prior heating (Fig. 2). The results provided a high-resolution map of the level of exposure to solvent of different capsid regions over time, revealing different kinetics in the regions represented by different peptides (Fig. 2 A). We classified the capsid peptides into three different groups according to their level of deuterium uptake after 1 h: 0–15% (blue circles), 16–34% (green circles), and 35–100% (red circles). Fig. 2 B shows the deuterium uptake level after 1 h, color-coded on the crystal structure of the VLP (PDB: 1Z14) (17). The most dynamic capsid regions correspond to the Nts of the capsid protein subunits (located at the capsid interior and not visible in the crystal structure), some highly exposed loops at the tips of the threefold spikes and, interestingly, the structural elements forming the capsid pores at the fivefold axis. These results provide direct proof for conformational fluctuations at equilibrium (breathing) in the capsid pore regions that are involved in translocation events during MVM infection.
Changes in MVM capsid dynamics during biologically relevant translocation events
Unlike techniques such as fluorescence spectroscopy, which do not provide any detailed information about the structural elements and residues involved in the transition, HDX-MS offers peptide resolution that should permit the identification of structural elements involved in structural transitions in very large protein complexes, even viral particles. To prove this, we compared the equilibrium dynamics of different regions of the MVM capsid at different temperatures below and above the transition temperature, TM, of the conformational rearrangement. Purified VLP aliquots were incubated for between 10 s and 4 h at six different temperatures (25, 37, 45, 50, 55, and 60°C). An initial examination of the HDX time-course plots at different temperatures revealed differences between peptides that suggest region-dependent variations in dynamics. A representative set of these distinct behaviors is shown in Fig. 3 A. In the left column, the time courses of four different peptides are plotted at each different temperature. In the right column, the relative uptake after 4 h of incubation (N4h/NTOTAL) is plotted as a function of temperature. We found that some peptides were characterized by a rapid exchange rate that was not dependent on temperature (e.g., 153–164), whereas others presented increasing deuteration levels at increasing temperatures (e.g., 196–215, 178–195, and 248–255). It should be noted that for some peptides (e.g., 178–195 and 248–255), the deuteration level changed at temperatures close to the reported TM of the capsid conformational change detected by fluorescence and associated with Nt externalization through the pores. To test whether those changes in dynamics could be associated to this transition, we fitted our HDX data to a unimolecular two-state transition in the same way that it had been done in the fluorescence spectroscopy experiments (Fig. 3 C and Note S1 in the Supporting Material) (19). We found that the TM values obtained by fluorescence spectroscopy (47.2 ± 0.3°C, Fig. 3 B) and HDX-MS experiments (TM = 48 ± 1°C, Fig. 3 C) were indistinguishable, suggesting that changes in dynamics around the TM can be used as markers to identify capsid regions that are structurally altered during the transition.
Figure 3.
Probing the heat-induced transition of the MVM capsid by fluorescence and HDX-MS. (A) HDX time-course plots of four representative peptides (left) and plots of N4h/NTOTAL as a function of temperature (right). (B) Tryptophan-specific fluorescence of the MVM VLPs as a function of temperature. Data (circles) were fitted to a two-state transition (solid line). (C) HDX uptake plots of the number of deuterons exchanged at t = 4 h as a function of the temperature. Data (circles) were fitted to a two-state transition (solid lines). To see this figure in color, go online.
To systematically analyze the different behaviors caused by the conformational change, we calculated the slope of the relative uptake plots before (m25–45°C) and after (m50–60°C) the transition temperature, TM. In this way, we could classify the peptides in three different groups: 1) peptides presenting the same slope before and after the transition temperature, corresponding to peptides that expose most of their amide sites at low temperature or do not undergo a change in dynamics (e.g., 153–164 and 196–215); 2) peptides that decrease the slope after the transition, corresponding to peptides that abruptly reach their fully deuterated level or decrease their dynamics (e.g., 178–195); and 3) peptides that increase the slope after the transition, corresponding to peptides whose amide hydrogens become exposed at higher temperatures (e.g., 248–255). In principle, the observed increase in amide exposure of this last group could be a fingerprint of higher dynamics, but it could also arise because of a loss of capsid integrity at higher temperatures. To verify that the capsids would not fall apart during heating in the HDX buffer, we acquired native MS spectra of MVM capsids under identical conditions. Even over an extended period of time (>5 h) at 60°C, the spectrum does not appear different from the spectrum acquired at 0°C. This corroborated that the integrity of the particles was not compromised (Fig. S3 C). It was also important to ascertain that the conformational transition probed was reversible. Fluorescence assays showed that after heating and cooling the sample from 27°C to 61°C (waiting 30 min at the highest temperature), particles not only maintained their integrity but also kept their ability to undergo the temperature-induced conformational change (results not shown), which corroborates the reversibility of the process.
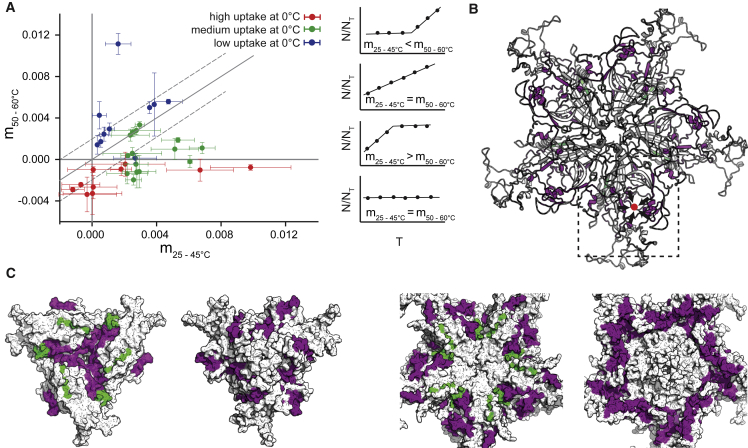
To identify which regions undergo the most abrupt changes in deuterium incorporation after the cooperative transition, we plotted for each peptide the slope before the transition (m25–45°C) versus the slope after the transition (m50–60°C) (Figs. 4 A and S4). With this approach, peptides showing a similar uptake before and after the transition would fall close to the region represented by slope m = 1 (Fig. 4 A, solid line); peptides that show an increase in dynamics after the transition fall above this region, and peptides that show reduced dynamics or reach a fully deuterated state after the transition fall below it. Peptides presenting rapid exchange at 0°C (Fig. 3 A, red circles) had slope values close to 0. Any transition-associated increase in the equilibrium dynamics of these peptides, which include the disordered Nts within the capsid and the five long β-hairpins that delimit each capsid pore, was beyond the range of our experiments, because exchange was nearly complete even at the shortest times that could be reliably tested. Peptides 414–429 and 422–429 also belong to this fast-exchange group but present an abrupt change in slope. They correspond to an exposed loop situated close to the tip of each threefold, spike and their slope decreases 10-fold after the transition. However, two-state fittings for these two peptides suggest that the HDX does not occur at the transition temperature, indicating that a structural change in this loop is not associated with the global transition. Blue circles correspond to peptides with low uptake at 0°C and green circles to peptides with moderate uptake at 0°C. Not surprisingly, peptides presenting an increased dynamics after the transition fall above the line with slope m = 1 and are blue, and peptides with a decreased dynamics fall below m = 1 and are green. For example, peptides 248–255 (green) and 178–195 (blue) are the ones falling the furthest away from the line m = 1, and they represent the most notorious cases of each change in dynamics. To identify the peptides presenting the most significant changes in dynamics, we selected a region around m = 1 (accounting for 25% of the area covered by all the peptides) and only included the peptides that fell beyond that region in our classification (Table S1). Mapping of these selected peptides in the capsid structure revealed local conformational changes in multiple capsid regions that can be collectively described as follows (Fig. 4, B and C):
-
1)
Capsid regions with transition-associated conformational rearrangements leading to reduced dynamics or abrupt fully deuterated states after the transition. These include substantial portions of several long, convoluted loops of each capsid subunit that generally define parts of the thick walls of the spikes centered at the capsid threefold axes, away from the pores at the capsid fivefold axes. Peptides defining these structural elements involve as many as one-fourth of the capsid amino acid residues.
-
2)
Capsid regions with transition-associated conformational rearrangements leading to an abrupt increase in local dynamics correspond to a β-strand that belongs to the β-sandwich fold of each capsid protein subunit from which long peptides protrude to form the capsid spikes.
Figure 4.
Peptides that present an abrupt change in dynamics around the heat-induced conformational transition of the MVM capsid. (A) Plot showing the slope change of each peptide before (m25–45°C) and after (m50–60°C) the transition (defined by the transition temperature, TM). The colors indicate the dynamics of the peptides at 0°C. (B) Peptides presenting an abrupt change in deuterium uptake are mapped in the MVM capsid structure. For simplicity, only five capsid subunits surrounding a pore are represented. In green are shown peptides that expose more residues after the transition, and in purple are peptides that present a plateau after the transition. The inset square delimits a region of a capsid subunit close to the threefold axes where there is a substantial reduction in dynamics after the TM. Peptides 178–195 and 347–385, which present the most significant reduction, are located in the inner core of this cavity. (C) Detailed views of a 3- and a fivefold symmetry axis. From left to right are a threefold axis inside view, a threefold axis outside view, a fivefold axis inside view, and a fivefold axis outside view. The peptides that become more exposed after the TM (green) are located in the inner part of the capsid. To see this figure in color, go online.
To ensure that these changes in dynamics were due to a global conformational change and not to a loss of secondary structure, we performed far-ultraviolet CD experiments at increasing temperatures to see whether heating causes changes in the β-sheet content of the MVM capsid protein (Fig. S5). We could not detect any effect of heating, which excludes the possibility that a loss of secondary structure becomes convoluted with the conformational changes around the transition temperature. Overall, our results provide a detailed description based on HDX-MS analysis of structural elements involved in a functionally relevant conformational transition in a large macromolecular complex, and they show that a global conformational rearrangement of a viral capsid is involved in externalization of biological signals through capsid pores.
Conclusions
In this work, we have used HDX-MS at various incubation temperatures to identify, with high spatial resolution, structural changes in the MVM capsid that are required for viral infection. Transition-associated, abrupt changes in local dynamics of peptide segments in the capsid served as markers to identify structural elements that modified their conformation during the transition. The results revealed that translocation events through capsid pores that occur during the infection cycle involve a global structural rearrangement of the capsid. Many of the elements showing altered conformations were located far from the pores and included loops with abruptly decreased dynamics as well as regular secondary-structure elements that showed a marked increase in dynamics. This study provides, to our knowledge, a novel demonstration of the potential of HDX-MS to structurally analyze temperature-dependent structural transitions, even in mega-Dalton protein complexes. Particularly, it enables further investigation of the links between the viral infection process and the conformational landscape that is sampled by a virus capsid. This paves the way for more comprehensive studies where MVM and other viruses are functionally investigated at the (bio)chemical, mechanical, and thermodynamic levels. These studies can have important outcomes in virology and nanotechnology, for instance for the design of antiviral drugs and development of functional nanoparticles or gene-therapy vehicles.
Author Contributions
M.v.d.W., A.L., J.S., P.J.d.P., M.G.M., and A.J.R.H. conceived and coordinated the study and wrote the article. M.v.d.W., A.L., and J.S. performed and analyzed mass spectrometry experiments. A.V., A.R.-H., M.A.F., and M.G.M. provided MVM samples and performed tryptophan fluorescence and CD studies. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
M.v.d.W., J.S., and A.J.R.H. are supported by Proteins@Work (project no. 184.032.201), funded by The Netherlands Organisation for Scientific Research (NWO). M.v.d.W. and A.J.R.H. are additionally supported by a project ruimte grant (12PR3303-2) from Fundamenteel Onderzoek der Materie(FOM). A.V., A.R.-H., M.A.F., and M.G.M. are funded by grants BIO2012-37649 and BIO2015-69928-R from the Ministerio de Economía, Industria y Competitividad (MINECO/EU) and by an institutional grant from Fundación Ramón Areces. A.L. and P.J.d.P. are funded by grant FIS2011-29493 from MINECO/EU. A.V. is the recipient of a postdoctoral contract by MINECO/EU. M.G.M. is an associate member of the Institute of Biocomputation and Physics of Complex Systems, Zaragoza, Spain.
Editor: Elizabeth Komives.
Footnotes
Michiel van de Waterbeemd and Aida Llauró contributed equally to this work.
Eight figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30158-3.
Supporting Material
References
- 1.Bothner B., Hilmer J.K. Viral vectors for gene delivery. In: Agbandje-McKenna M., McKenna R., editors. Structural Virology. RSC Publishing; Cambridge, United Kingdom: 2011. pp. 41–61. [Google Scholar]
- 2.Rossmann M.G., Rao V.B., editors. Viral Molecular Machines. Springer; New York: 2012. [Google Scholar]
- 3.Mateu M.G., editor. Structure and Physics of Viruses. Springer; Dordrecht, the Netherlands: 2013. [Google Scholar]
- 4.Johnson J.E. Advances in Protein Chemistry, Volume 64. Viral Structure. Academic Press; San Diego, CA: 2003. Virus particle dynamics; pp. 197–218. [Google Scholar]
- 5.Johnson J.E. Virus particle maturation: insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veesler D., Johnson J.E. Virus maturation. Annu. Rev. Biophys. 2012;41:473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agbandje-McKenna M., Llamas-Saiz A.L., Rossmann M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore S.F., D’Abramo A.M., Jr., Tattersall P. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology. 1999;254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- 9.Lombardo E., Ramírez J.C., Almendral J.M. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 2002;76:7049–7059. doi: 10.1128/JVI.76.14.7049-7059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maroto B., Valle N., Almendral J.M. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 2004;78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valle N., Riolobos L., Almendral J.M. Synthesis, post-translational modification and trafficking of the parvovirus structural polypeptides. In: Kerr J.R., Cotmore S.F., Bloom M.E., Linden R.M., editors. Parvoviruses. Edward Arnold; London, United Kingdom: 2006. [Google Scholar]
- 12.Cotmore S.F., Tattersall P. Mutations at the base of the icosahedral five-fold cylinders of minute virus of mice induce 3′-to-5′ genome uncoating and critically impair entry functions. J. Virol. 2012;86:69–80. doi: 10.1128/JVI.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez-Martínez C., Grueso E., Almendral J.M. Essential role of the unordered VP2 n-terminal domain of the parvovirus MVM capsid in nuclear assembly and endosomal enlargement of the virion fivefold channel for cell entry. Virology. 2012;432:45–56. doi: 10.1016/j.virol.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 14.Cotmore S.F., Tattersall P. Parvoviruses: small does not mean simple. Annu. Rev. Virol. 2014;1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore S.F., Hafenstein S., Tattersall P. Depletion of virion-associated divalent cations induces parvovirus minute virus of mice to eject its genome in a 3′-to-5′ direction from an otherwise intact viral particle. J. Virol. 2010;84:1945–1956. doi: 10.1128/JVI.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plevka P., Hafenstein S., Tattersall P. Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis. J. Virol. 2011;85:4822–4827. doi: 10.1128/JVI.02598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontou M., Govindasamy L., Agbandje-McKenna M. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernando E., Llamas-Saiz A.L., Almendral J.M. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology. 2000;267:299–309. doi: 10.1006/viro.1999.0123. [DOI] [PubMed] [Google Scholar]
- 19.Carreira A., Menéndez M., Mateu M.G. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Biol. Chem. 2004;279:6517–6525. doi: 10.1074/jbc.M307662200. [DOI] [PubMed] [Google Scholar]
- 20.Reguera J., Carreira A., Mateu M.G. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reguera J., Grueso E., Mateu M.G. Functional relevance of amino acid residues involved in interactions with ordered nucleic acid in a spherical virus. J. Biol. Chem. 2005;280:17969–17977. doi: 10.1074/jbc.M500867200. [DOI] [PubMed] [Google Scholar]
- 22.Carrasco C., Carreira A., de Pablo P.J. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. USA. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrasco C., Castellanos M., Mateu M.G. Manipulation of the mechanical properties of a virus by protein engineering. Proc. Natl. Acad. Sci. USA. 2008;105:4150–4155. doi: 10.1073/pnas.0708017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellanos M., Pérez R., Mateu M.G. Mechanical disassembly of single virus particles reveals kinetic intermediates predicted by theory. Biophys. J. 2012;102:2615–2624. doi: 10.1016/j.bpj.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellanos M., Pérez R., Mateu M.G. Mechanical elasticity as a physical signature of conformational dynamics in a virus particle. Proc. Natl. Acad. Sci. USA. 2012;109:12028–12033. doi: 10.1073/pnas.1207437109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellanos M., Carrillo P.J.P., Mateu M.G. Quantitatively probing propensity for structural transitions in engineered virus nanoparticles by single-molecule mechanical analysis. Nanoscale. 2015;7:5654–5664. doi: 10.1039/c4nr07046a. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z., Smith D.L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konermann L., Pan J., Liu Y.-H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 29.Hamuro Y., Coales S.J., Griffin P.R. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J. Biomol. Tech. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- 30.Englander S.W., Kallenbach N.R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 31.Englander J.J., Del Mar C., Woods V.L., Jr. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engen J.R. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanman J., Lam T.T., Prevelige P.E., Jr. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 34.Tuma R., Coward L.U., Prevelige P.E., Jr. Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J. Mol. Biol. 2001;306:389–396. doi: 10.1006/jmbi.2000.4383. [DOI] [PubMed] [Google Scholar]
- 35.Gertsman I., Gan L., Johnson J.E. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domitrovic T., Movahed N., Johnson J.E. Virus assembly and maturation: auto-regulation through allosteric molecular switches. J. Mol. Biol. 2013;425:1488–1496. doi: 10.1016/j.jmb.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Smith D.L. Capsid structure and dynamics of a human rhinovirus probed by hydrogen exchange mass spectrometry. Protein Sci. 2005;14:1661–1672. doi: 10.1110/ps.051390405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bereszczak J.Z., Rose R.J., Heck A.J.R. Epitope-distal effects accompany the binding of two distinct antibodies to hepatitis B virus capsids. J. Am. Chem. Soc. 2013;135:6504–6512. doi: 10.1021/ja402023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroe E.B., Kang S., Prevelige P.E. Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure. 2010;18:1483–1491. doi: 10.1016/j.str.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Heuvel R.H.H., van Duijn E., Heck A.J.R. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 41.Tahallah N., Pinkse M., Heck A.J.R. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 2001;15:596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.