Abstract
While kinases are typically composed of one or two subunits, calcium-calmodulin (CaM)-dependent protein kinase II (CaMKII) is composed of 8–14 subunits arranged as pairs around a central core. It is not clear if the CaMKII holoenzyme functions as an assembly of independent subunits, as catalytic pairs, or as a single unit. One strategy to address this question is to genetically engineer monomeric and dimeric CaMKII and evaluate how their activity compares to the wild-type (WT) holoenzyme. Here a technique that combines fluorescence correlation spectroscopy and homo-FRET analysis was used to characterize assembly mutants of Venus-tagged CaMKIIα to identify a dimeric CaMKII. Spectroscopy was then used to compare how holoenzyme structure and function changes in response to activation with CaM in the dimeric mutant, WT-holoenzyme, and a monomeric CaMKII oligomerization-domain deletion mutant control. CaM triggered an increase in hydrodynamic volume in both WT and dimeric CaMKII without altering subunit stoichiometry or the net homo-FRET between Venus-tagged catalytic domains. Biochemical analysis revealed that the dimeric mutant also functioned like WT holoenzyme in terms of its kinase activity with an exogenous substrate, and for endogenous T286 autophosphorylation. We conclude that the fundamental functional units of CaMKII holoenzyme are paired catalytic-domains.
Introduction
The calcium-calmodulin (CaM)-dependent protein kinase II (CaMKII) is a key modulator of synaptic efficiency (1). Calcium influx into neurons triggers the activation of CaMKII and its translocation from dendrites into postsynaptic spines (2, 3, 4, 5, 6, 7), thus altering kinase substrate specificity (8) and synaptic plasticity. While most kinases are typically composed of one or two subunits, the autoinhibited CaMKII holoenzyme has multiple subunits arranged as 4–7 catalytic-domain pairs distributed around a central oligomerization-domain hub (9, 10). Lateral and transverse protein interactions between adjacent oligomerization-domains allow for the assembly and maintenance of the holoenzyme hub (11, 12), though the mechanistic significance of assembling 8–14 catalytic-domains is poorly understood. Similarly, the purpose of arranging these catalytic-domains into pairs is not known. In response to calcium influx, and subsequent CaM binding, CaMKII is activated by displacing an adjacent pseudo-substrate motif from the kinase catalytic site (13). CaMKII activation is typically transient, occurring while CaM remains bound to the kinase regulatory domain. Using fluorescence correlation spectroscopy (FCS) in conjunction with homo-Förster resonance energy transfer (homo-FRET) measurements to simultaneously monitor CaMKII holoenzyme hydrodynamic volume and catalytic domain pairing, we have previously shown that activation with CaM triggers an increase in the hydrodynamic volume of the holoenzyme, which we have interpreted as an extension of catalytic domains away from the central oligomerization core hub (9). Coincident CaM activation of two neighboring subunits in the holoenzyme can trigger T286 autophosphorylation resulting in a persistent kinase activity even after intracellular calcium concentrations returns to basal levels (14). In our previous study, we have shown that T286 autophosphorylation is required for the extension of catalytic-domain pairs away from the holoenzyme central hub, and that the amount of T286 autophosphorylation triggered by different concentrations of CaM was correlated with the magnitude of the change in hydrodynamic volume (9). Surprisingly, we found that catalytic-domain pairing was maintained through this extension process, although subtle transient changes in homo-FRET were observed (9). Within domain pairs, it is not known if both T286 sites must be autophosphorylated or if only one is sufficient for extension. Additionally, it is unknown if kinase subunits in a catalytic-domain pair can phosphorylate T286 sites on neighboring pairs. The reason for maintaining catalytic domain pairing in the holoenzyme is not understood, but we hypothesize that its purpose is to allow individual catalytic-domain pairs to act as semiautonomous calcium coincidence detectors within the context of the holoenzyme. In this scheme, coincident CaM binding within a catalytic-domain pair allows one catalytic-domain to phosphorylate the T286 site on its paired catalytic-domain to trigger the extension of that catalytic-domain pair and its persistent activation, without intervention from neighboring catalytic-domain pairs. Regarding CaM activation and extension, each pair acts autonomously, but all pairs in a holoenzyme are colocalized within the cell. If this model is correct, T286 autophosphorylation should primarily occur within catalytic-domain pairs, and the kinase activity of an isolated catalytic-domain pair should recapitulate the activity observed in intact holoenzyme.
To investigate the structural and functional significance of CaMKII multimerization, and specifically define the role of catalytic domain-pairs within the holoenzyme structure, we used fluorescent polarization and fluctuation analysis (FPFA) (10) in conjunction with a T286 autophosphorylation assay and a kinase assay using an exogenous CaMKII substrate to characterize and compare assembly mutants of Venus-tagged CaMKIIα holoenzyme in freshly prepared cell lysates.
Materials and Methods
Molecular biology
DNA clones encoding monomeric Venus (Addgene ID: 27794) and V-CaMKIIα (Addgene ID: 29428), are available from Addgene (http://www.addgene.org/Steven_Vogel#). Mutations in V-CaMKIIα were introduced by site-directed mutagenesis PCR as described previously (9) using the PCR primer pairs shown below:
D393A 5′-GGAACCTGGTGGAGGGCCTGGCCTTTCATCGATTCTATTTTGAAAACTTGT GGTC-3′ and antisense 5′-GACCACAAGTTTTCAAAATAGAATCGATGAAAGGCCAGGCCCTCCACCAGGTTCC-3′
F394A 5′-GGAACCTGGTGGAGGGCCTGGACGCTCATCGATTCTATTTTGAAAACTTGTGGTC-3′ and antisense 5′-GACCACAAGTTTTCAAAATAGAATCGATGAGCGTCCAGGCCCTCCACCAGGTTCC-3′
F397A 5′-CTGGTGGAGGGCCTGGACTTTCATCGAGCCTATTTTGAAAACTTGTGGTCCCGGAACAG-3′ and antisense 5′-CTGTTCCGGGACCACAAGTTTTCAAAATAGGCTCGATGAAAGTCCAGGCCCTCCACCAG-3′
Y398A 5′-CTGGTGGAGGGCCTGGACTTTCATCGATTCGCTTTTGAAAACTTGTGGTCCCGGAACAG-3′ and antisense 5′-CTGTTCCGGGACCACAAGTTTTCAAAAGCGAATCGATGAAAGTCCAGGCCCTCCACCAG-3′
N401A 5′-GCCTGGACTTTCATCGATTCTATTTTGAAGCCTTGTGGTCCCGGAACAGCAAGC-3′ and antisense 5′-GCTTGCTGTTCCGGGACCACAAGGCTTCAAAATAGAATCGATGAAAGTCCAGGC-3′
H411A 5′-CCCGGAACAGCAAGCCCGTGGCCACCACCATCCTGAACCC-3′ and antisense 5′-GGGTTCAGGATGGTGGTGGCCACGGGCTTGCTGTTCCGGG-3′
T413A 5′-CGGAACAGCAAGCCCGTGCACACCGCCATCCTGAACCCTCACATCCACCTGATGGG-3′ and antisense 5′-CCCATCAGGTGGATGTGAGGGTTCAGGATGGCGGTGTGCACGGGCTTGCTGTTCCG-3′
L415A 5′-GCCCGTGCACACCACCATCGCGAACCCTCACATCCACCTGATGG-3′ and antisense 5′-CCATCAGGTGGATGTGAGGGTTCGCGATGGTGGTGTGCACGGGC-3′
I434A 5′-GCCTGCATCGCCTACATCCGCGCGACTCAGTACCTGGATGCGGG-3′ and antisense 5′-CCCGCATCCAGGTACTGAGTCGCGCGGATGTAGGCGATGCAGGC-3′
Q436A 5′-GCCTACATCCGCATCACTGCGTACCTGGATGCGGGTGGC-3′ and antisense 5′-GCCACCCGCATCCAGGTACGCAGTGATGCGGATGTAGGC-3′
Constructs with multiple mutations in V-CaMKIIα were prepared using combinations of the above primers. In cases where the mutations were in close proximity, single primer sets containing combinations of the above mutations were used.
Construction of V-CaMKIIα(Δ315) was accomplished as described in Thaler et al. (15).
Cell culture, transfection, and homogenate preparation
Experiments were performed using HEK 293 cells (American Type Culture Collection, Manassas, VA) as previously described (9). For live-cell imaging measurements, cells were transfected with DNA encoding V-CaMKIIα (WT), V-CaMKIIα-F394A (F394A), or V-CaMKIIα-Δ315 (Δ315) (typically 1 μg DNA/250,000 cells). Transfected cells were plated on 35 mm glass-bottom dishes and incubated overnight in phenol red-free DMEM media. The following day, DMEM media was replaced with DPBS buffer (1×, with calcium and magnesium; Mediatech, Manassas, VA). Images of the cellular fluorescence were collected by a DCS-120 Confocal FLIM Scanning System (Becker & Hickl, Berlin, Germany). Cell homogenates for FPFA were prepared as previously described (9). In short, 24 h after DNA transfection, cells were washed with calcium-free Dulbecco’s Phosphate Buffered Saline (PBS; Sigma-Aldrich, St. Louis, MO), then harvested and lysed using 1× passive lysis buffer (Promega, Madison, WI) augmented with 1× Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA). Homogenates were centrifuged at 100,000 × g for 30 min to remove membranes and particulate matter. Supernatants were diluted to yield a photon count rate between 20,000 and 100,000 cps. The clarified homogenates were then loaded into 35 mm glass-bottom dishes (MatTek, Ashland, MA) and micro- and macrotimes were measured by FPFA the same day.
CaMKII kinase activity assay
Phosphorylation of TAMRA-Autocamtide-3 (Molecular Devices, Sunnyvale, CA) by Venus-tagged CaMKIIα was measured in freshly prepared homogenates as previously described (9).
Immunocapillary electrophoresis using the WES system
The T286 autophosphorylation status of inactive or activated Venus-tagged CaMKIIα in cell homogenates was determined using Venus fluorescence and immunocapillary electrophoresis. The abundance of Venus-tagged protein was determined by measuring the fluorescence signal of the lysates using a PHERAstar FS luminometer equipped with a 500 nm excitation filter and a 550 nm emission filter (BMG Labtech, Ortenberg, Germany). Lysate concentrations were adjusted accordingly for kinase assay. Immunocapillary electrophoresis was performed using a Wes Immunoprobing System (12–230 kDa Master Kit; ProteinSimple, San Jose, CA). The Wes Immunoprobing System uses capillary electrophoresis coupled with light-based protein immobilization and immunoprobing for size-based immunodetection of proteins. Eight microliters of a reaction mix aliquot were transferred to a tube and denatured with 2 μL of Fluorescent 5× Master Mix as per the manufacturer’s instructions (ProteinSimple). The sample, primary antibodies (1:10,000 dilution of polyclonal rabbit anti-GFP antibody, cat. No. ab6556, and 1:10,000 of dilution mouse monoclonal phospho-T286 CaMKII antibody, cat. No. ab2724; Abcam, Cambridge, UK), respective secondary antibodies, luminol solution, antibody diluent, and wash buffers were added in the appropriate wells of a well plate provided by the manufacturer (all reagents were provided in the manufacturer’s kit; Abcam) as per the manufacturer’s instructions. Protein migration and immunoreactivity rates were analyzed using the manufacturer’s software, Compass (Ver. 2.6.6; ProteinSimple, and http://www.proteinsimple.com/software_compass_simplewestern.html). The output from the Compass software is luminosity (due to peroxidase-luminol interaction) as a function of apparent molecular weight of the proteins from which it is detected. Using the area under the curve of luminosity-molecular mass plots, the total amount of immunoreactivity detected by the antibody between 66 and 116 kDa was quantified. A ratio of total immunoreactivity detected by phospho-T286 antibody and the GFP antibodies was calculated for each sample. The ratio for each sample from three independent experiments was used to generate a bar graph of T286 autophosphorylation.
Optical spectroscopic experimental setup
The FPFA experimental setup and validation were utilized as previously described (10). FCS, time-resolved anisotropy, and fluorescence lifetimes were measured in freshly prepared HEK cell homogenates expressing Venus-tagged CaMKII constructs before and after the addition of saturating calcium-Calmodulin as previously described (9, 10).
Data analysis and calibration
Data analyses of time-resolved anisotropy, lifetime, polarized fluorescence fluctuation analyses, and FPFA calibration have been described elsewhere (9, 10). The integral of the time-dependent fluorescent intensity of parallel and perpendicular polarization channels were used to calculate the steady-state anisotropy (r) of Venus-tagged CaMKII constructs expressed in cellular cytoplasm, as follows:
where I//(t) and I⊥(t) are the fluorescence intensity decay of parallel and perpendicular channels (dark-noise subtracted), respectively; and g is the instrument correction factor, which, for our microscope had a value of 0.99 as estimated by calibration using fluorescein tail fitting (9). The fluorescence lifetime decay was calculated from I//(t) and I⊥(t) using the following equation:
FCS analysis was performed by cross-correlating photons detected in the parallel and perpendicular light paths as previously described (10), and was used instead of autocorrelation to eliminate noncorrelated detector noise such as after-pulsing.
Curve fitting and statistics
SPCM software (Becker & Hickl) was used to calculate cross-correlation curves from macrotime data as well as time-decay curves from microtime data. The software Igor Pro (Ver. 6.4; WaveMetrics, Lake Oswego, OR, https://www.wavemetrics.com/Products/igorpro/igorpro.htm) was used for fitting of cross-correlation curves, and for calculating time-resolved and steady-state anisotropy. The software Prism 6 (GraphPad, https://www.graphpad.com/) was used for fitting CaMKII activity curves and calculating the Autocamtide-2 Kd and Hill coefficient. Prism 6 was also used to calculate means, standard errors of the mean ± SE, standard deviations (SD), and 95% cumulative index (CI). Prism 6 was also used to calculate ANOVA test using Tukey’s multiple comparison to compare individual values within a data set.
Results
Our strategy for investigating the impact of CaMKII multimerization on enzyme structure and activity was to compare the wild-type (WT) Venus-tagged CaMKII holoenzyme, typically composed of 10 subunits in vitro (10), with monomeric and dimeric mutants. Individual CaMKII subunits are composed of the N-terminal catalytic-domain (amino-acid residues 1–272 in mouse CaMKIIα) that forms pairs in the holoenzyme (9, 10, 15), and the C-terminal oligomerization-domain (residues 336–478), which is attached to the catalytic domain via the regulatory-domain (residues 273–314), and includes the T286 phosphorylation site and the calmodulin binding site and a short linker region (residues 315–335).
X-ray crystallography modeling (11, 16) and immunoelectron microscopy (12, 17) have shown that isolated C-terminal oligomerization-domains assemble to form the central hub, which is composed of two stacked rings (18) (Fig. 1 A). Crystal structures of the CaMKII hub revealed at least two forms, where each of the two rings is composed of either seven (16) or six (11) individual oligomerization-domain subunits to form oligomers of 14 or 12 subunits. Because a holoenzyme composed of 8–10 subunits has also been identified (10, 17), individual rings presumably can also be formed from four to five subunits.
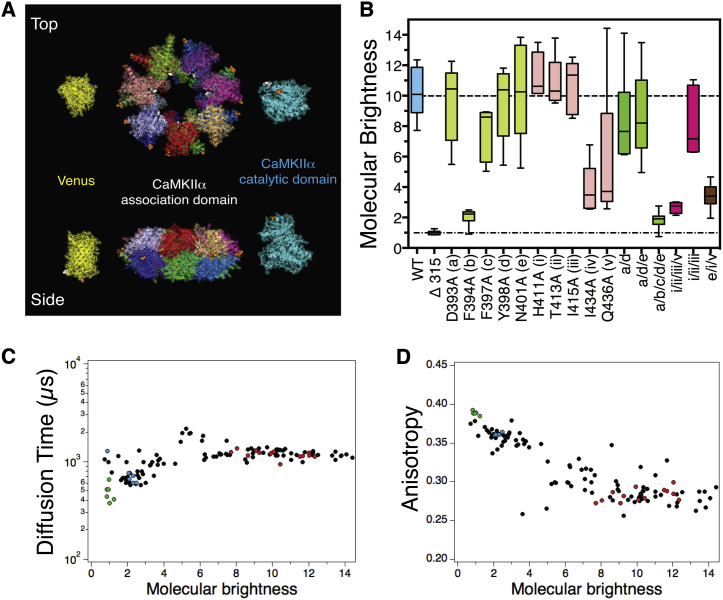
Figure 1.
CaMKIIα association domain mutants. (A) Cartoon depicting the three-dimensional structures of Venus, the CaMKII holoenzyme association domain core structure, and the CaMKII catalytic domain. (White) C-termini; (orange) N-termini. Note that the CaMKII association domains interface with other association domains on three surfaces to form the holoenzyme core structure: two lateral interfaces within each ring structure and one transverse interface between the two ring structures. (B) Mutations were made in the lateral interaction surfaces of the CaMKII association domain and molecular brightness normalized to the molecular brightness of a Venus monomer was plotted for each mutant. Wild-type CaMKII and a monomeric Δ315 mutant are shown for comparison. Parametric plots of diffusion time (C), and steady-state anisotropy (D) as a function of molecular brightness of all individual samples plotted in (B). (Red) WT CaMKII; (green) monomeric Δ315 mutant; (blue) dimeric F394A mutant; (black) all other constructs.
In each case, an oligomerization-domain contacts three others—two laterally, with interaction surfaces on either side, which are oriented head-to-tail (surface area for each is ∼1100 Å2) and propagate around the ring; and one transverse interaction (surface area is ∼2200 Å2), which is oriented head-to-head with an oligomerization-domain on the opposite ring. The lateral head-to-tail orientation runs in opposite directions for each ring. These three interaction surfaces are responsible for the multimeric nature of the holoenzyme, are high affinity (11), and do not require catalytic or regulatory domains to interact (19).
A monomeric CaMKII was engineered by inserting a stop codon to generate a deletion mutant removing the oligomerization-domain (Δ315). Our strategy for generating a dimeric CaMKII holoenzyme involved introducing point mutations in the lateral interaction surfaces while preserving the transverse interaction surface, and then screening these mutants by FPFA (9, 10) to determine if any of these mutations could destabilize the lateral interaction surface to form a dimeric holoenzyme. Fig. 1 B shows molecular brightness analysis of WT Venus-tagged CaMKIIα, the Δ315 mutation, as well as of five single point mutations in the first lateral interaction surface (a–e), five single point mutations in the complimentary lateral interaction surface (i–v), and six other mutants with multiple point mutation. In this experiment the Venus-tagged WT holoenzyme had a molecular brightness of 10.2 ± 0.4 (mean ± SE, n = 13, 10 times as bright as a Venus monomer) with a range of brightness values from 8 to 12 as shown in Nguyen et al. (10). In contrast, the Δ315 mutant had a brightness of 1.0 ± 0.1 (n = 3), as expected for a Venus-tagged CaMKII monomer. Of the 10 single point mutations generated, the F394A mutant (clone b) had a molecular brightness of 2.0 ± 0.2 (n = 3), indicating it assembles into a dimer. In addition to providing information on molecular brightness, FPFA also can be used to simultaneously measure diffusion time (a measure of hydrodynamic volume), and fluorescence anisotropy (a measure of Venus homo-FRET) (20, 21). In Fig. 1 C, diffusion time is plotted as a function of molecular brightness for all constructs generated in this study. Diffusion time had a value slightly larger than 1000 μs for constructs having eight or more subunits. Apparently, the hydrodynamic volume of a holoenzyme with 8–14 subunits does not significantly change. The diffusion time for constructs with fewer than eight subunits had more variance, but in general as the number of subunits decreased, so did diffusion time. These results suggest that the holoenzyme can form its characteristic ring structure when it has at least eight subunits. In Fig. 1 D, the steady-state anisotropy is plotted as a function of molecular brightness for the same samples. Constructs having eight or more subunits typically had steady-state anisotropy values between 0.25 and 0.30. We have previously shown that when Venus is attached to the N-terminus (catalytic-domain) of CaMKIIα, anisotropy primarily reports on catalytic-domain pairing (homo-FRET) (9, 10, 15). For constructs with fewer than eight subunits, anisotropy values increased as the number of subunits decreased. This may indicate that the distance between catalytic-domain pairs increases once a ring structure can no longer form, or the fraction of paired subunits decreases.
To identify structural motifs associated with catalytic-domain pairing, FPFA was used to compare the WT Venus-tagged CaMKIIα (V-CaMKIIα) holoenzyme with the monomeric deletion mutant (V-CaMKIIαΔ315) and the dimeric oligomerization-domain mutant (V-CaMKIIαF394A). FPFA involves collecting time-resolved microtimes (the time delay between a laser excitation pulse and when individual fluorescent photons are emitted) and macrotimes (the time delay from the start of data collection and when individual photons are detected). The orientation of each detected photon is parameterized by using two photon-counting detectors in orthogonal emission light paths designed to isolate parallel and perpendicular polarization relative to the excitation-light electric-field orientation (21). Microtimes are used to calculate fluorescence polarization decays, a measure of homo-FRET between catalytic-domains and molecular rotation (10), while macrotimes are used to calculate cross-correlation curves, a measure of molecular brightness and lateral diffusion (10).
FCS of Venus fluorescence from V-CaMKIIα, V-CaMKIIαF394A, and V-CaMKIIαΔ315 generated cross-correlation curves with dramatically altered amplitudes for samples with the same fluorescence count-rate (Fig. 2 A). The amplitude of the cross-correlation curves is inversely proportional to the number of fluctuating fluorescent molecules in the excitation volume, and is indicative of the number of Venus-tagged subunits per fluctuating holoenzyme. The autocorrelation amplitude was greatest in the WT holoenzyme, then the F394A mutant, and the least in the Δ315 deletion mutant.
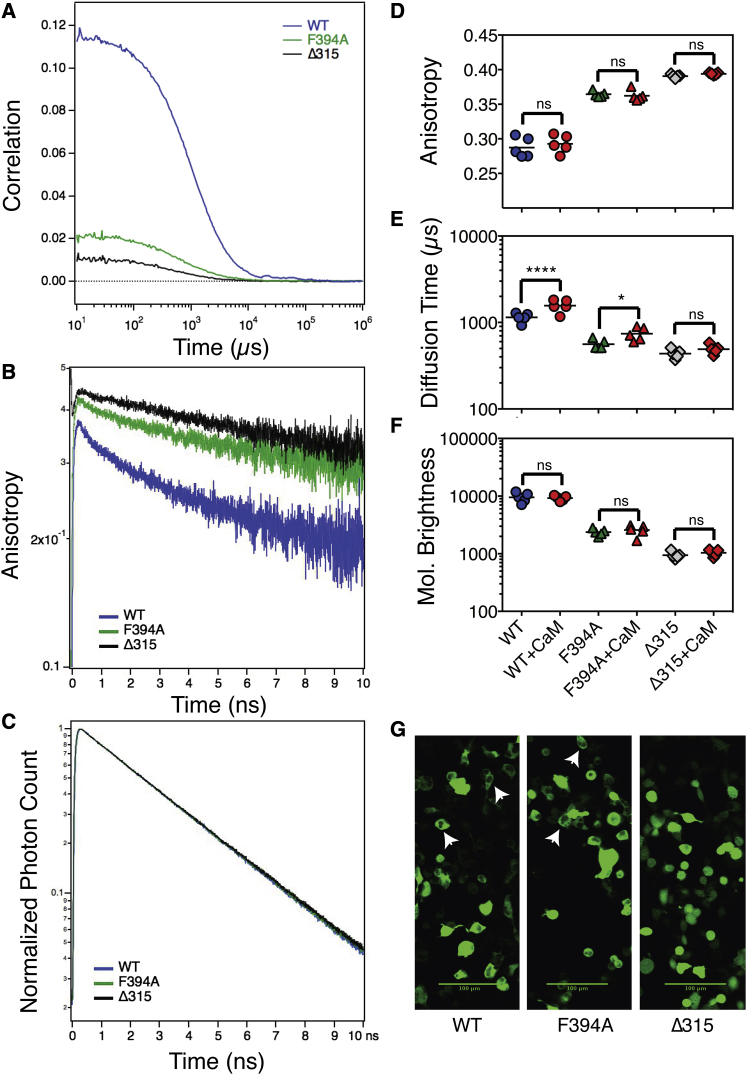
Figure 2.
Biophysical comparison of WT, dimeric (F394A), and monomeric (Δ315) V-CaMKII. (A) Representative cross-correlation curves from FCS analysis of WT (blue), F394A (green), and Δ315 (black) matched for similar counts/seconds in cell homogenates. Representative time-resolved anisotropy (B) and normalized fluorescence lifetime decay (C) of WT, F394A, and Δ315. FPFA analysis was used to compare the steady-state anisotropy (D), diffusion time (E), and molecular brightness (F) of WT (circles), F394A (triangles), and Δ315 (diamonds) before (blue) and after (red) activation with CaM. When expressed in HEK cells, WT CaMKII and F394A CaMKII are occluded from the nucleus (G).
Time-resolved anisotropy analysis revealed that V-CaMKIIα had a large fast-decay component observed within the first 2 ns, consistent with a paired organization of catalytic-domains, as previously described (9, 10, 15) (Fig. 2 B). In contrast, V-CaMKIIαΔ315 anisotropy decayed as a single exponential, indicating that CaMKIIαΔ315 cannot multimerize, and so remains a monomer without homo-FRET. Like V-CaMKIIα, the V-CaMKIIαF394A anisotropy also decayed as a double exponential, but with an attenuated fast decay component, indicating it also has a paired, albeit altered, organization.
Fluorescence lifetime decays analysis of the samples depicted in Fig. 2 B are shown in Fig. 2 C. Despite the dramatic differences observed using time-resolved anisotropy decay (Fig. 2 B), the lifetime decay of these three CaMKII constructs were indistinguishable (Fig. 2 C). Thus, as demonstrated in Nguyen et al. (10) and Koushik and Vogel (22), we show that changes in homo-FRET do not alter the lifetime of Venus fluorophores. The fluorescence lifetime of Venus, when attached to CaMKIIα, CaMKIIαF394A, and CaMKIIαΔ315, decayed for each as a single exponential, with a lifetime of 3.1 ± 0.0 ns (note an error of 0.0 indicates an error of <50 ps).
Time-resolved anisotropy curves were transformed into steady-state anisotropy values to facilitate statistical comparison of the impact of kinase activation on catalytic-domain homo-FRET. A net change in homo-FRET between catalytic-domain-attached Venus molecules was not observed when WT, F394A, and Δ315 CaMKII were activated with CaM (Fig. 2 D). Similarly, the brightness (the number of Venus-tagged subunits) of these samples was not statistically altered by activation as determined by ANOVA (Fig. 2 F). In contrast, the diffusion time for WT and F394A (but not Δ315) increased with activation (ANOVA multiple comparison test, p < 0.0001 and p = 0.05, respectively) (Fig. 2 E). These results indicate that the dimeric V-CaMKIIαF394A holoenzyme increases its hydrodynamic volume upon activation without altering its subunit stoichiometry or the separation distance between catalytic domains. A similar observation has been made for WT V-CaMKII holoenzyme (9), suggesting that they share the same activation mechanism. Visual examination of HEK cells expressing WT, F394A, and Δ315 revealed that Venus fluorescence was attenuated in nuclei in cells expressing WT or F394A (Fig. 2 G), indicating again that the dimeric and WT holoenzyme behave similarly to each other but distinct from the monomeric enzyme.
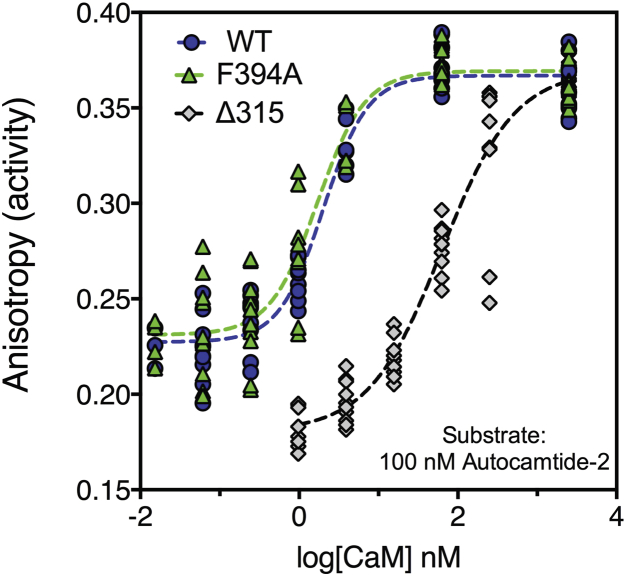
The enzyme kinetics for activation with CaM was next compared for the WT, the F394A holoenzyme, and the monomeric Δ315 mutant using autocamtide-2 as an exogenous substrate (Fig. 3). WT holoenzyme had a KD for CaM of 2.0 nM (95% CI, 1.6–2.5 nM) with a Hill coefficient of 1.6 (95% CI, 1.1–2.0). The dimeric F394A mutant holoenzyme was statistically indistinguishable with a KD for CaM of 1.7 nM (95% CI, 1.2–2.4 nM) with a Hill coefficient of 1.4 (95% CI, 0.8–2.1). In contrast, the monomeric Δ315 mutant had a KD for CaM of 62 nM (95% CI, 45–87 nM) with a Hill coefficient of 1.0 (95% CI, 0.6–1.2). The Vmax of all three enzymes was similar, but the monomeric deletion mutant had a reduced basal activity. This experiment suggests that the mild cooperativity for CaM observed for activation in the WT holoenzyme results from catalytic-domain pairing.
Figure 3.
Enzymatic comparison of WT, dimeric, and monomeric V-CaMKII. Equivalent concentrations of WT, F394A, and Δ315 were assayed for phosphorylation activity of the exogenous substrate autocamtide-2 using an anisotropy-based assay. (Dashed lines) Nonlinear fits to the Hill equation.
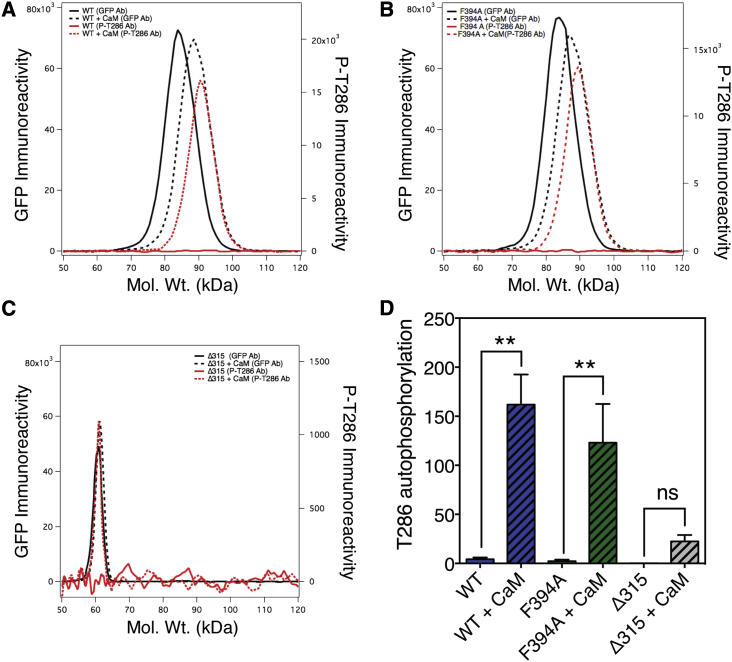
The mechanism facilitating an increased hydrodynamic-volume of the holoenzyme with activation is not known, but appears to require T286 autophosphorylation (9). T286 autophosphorylation requires two activated CaMKII subunits—one to catalyze the phosphorylation reaction and the other to present its regulatory domain T286 site as substrate (14). It is not clear if T286 autophosphorylation involves the catalytic domains within a pair, or if catalytic domains in adjacent pairs interact. Furthermore, if subunits within a pair mediate T286 autophosphorylation, it is possible that each subunit can act sequentially as enzyme and substrate for the other. Alternatively, hysteresis may occur after a random initial phosphorylation event that then locks each subunit into acting as either enzyme or substrate. To investigate this possibility, we performed quantitative immunocapillary electrophoresis on homogenates prepared from HEK cells expressing V-CaMKIIα, V-CaMKIIαF394A, or V-CaMKIIαΔ315 (as a negative control), before and after the addition of saturating concentrations of CaM (Fig. 4). Two antibodies were used: a GFP specific-antibody that cross-reacts with Venus adducts, and a phospho-T286-specific antibody that reacts only with phosphorylated T286 sites. The addition of CaM caused a decrease in migration (increased apparent molecular weight) under denaturing conditions. This was observed for both V-CaMKIIα and V-CaMKIIαF394A (black solid and dashed traces in Fig. 4, A and B). Because there was no appreciable phospho-T286-specific immunoreactivity in any of the samples before incubation with CaM (solid red traces, Fig. 4, A–C), we conclude that the species detected by the GFP-specific antibody (solid black traces) represents unphosphorylated CaMKII subunits. After CaM incubation, a phospho-T286 immunoreactive component appeared in all three samples (red dotted trace), but as expected its abundance was highly attenuated in the monomeric V-CaMKIIαΔ315 (Fig. 4 C). For both V-CaMKIIα and V-CaMKIIαF394A, the species detected after CaM incubation migrated more slowly than the activated component identified with the GFP-specific antibody Fig. 4, A and B. These traces represent the migration of CaMKII subunits that have been phosphorylated at T286. Because the phosopho-T286-specific antibody only reacts with phosphorylated subunits while the GFP-specific antibody can detect Venus-tagged subunits regardless of their phosphorylation state, we conclude that the intermediate extent of migration observed with the GFP-specific antibody after CaM activation (black dashed traces) indicates that for both WT and F394A, some subunits were phosphorylated (migrating slower), and some were not. Interestingly, the extent of T286 autophosphorylation for V-CaMKIIα and V-CaMKIIαF394A was statistically indistinguishable (as determined by ANOVA; Fig. 4 D).
Figure 4.
T-286 autophosphorylation comparison of WT, dimeric, and monomeric V-CaMKII. The mobility of WT (A), dimeric (B), and monomeric (C) CaMKII was assayed before (solid traces) and after (dashed traces) activation with CaM using immunocapillary electrophoresis with a GFP-specific antibody (black traces) or a phospho-T286-specific antibody (red traces). Note the predicted molecular mass of V-CaMKIIα and V-CaMKIIαF394A based on their amino-acid sequence is ∼82 kDa while the predicted molecular mass of V-CaMKIIαΔ315 deletion mutant is ∼64 kDa. (D) The amount of phospho-T286 immunoreactivity normalized to the abundance (GFP immunoreactivity) of the V-CaMKII sample before and after activation is plotted.
Discussion
The use of molecular brightness analysis for measuring the extent of oligomerization of fluorescent protein-tagged constructs is well established (23, 24, 25, 26, 27). Nonetheless, aberrant molecular brightness can be caused by factors that alter the fluorescence lifetime of fluorophores, and by factors that alter the stoichiometry of fluorescent and nonfluorescent subunits composing a complex. Because both types of factors can result in erroneous molecular brightness values, it is prudent to consider and control for their influence. When fluorescent proteins are used to tag proteins of interest for brightness analysis, four major factors can alter the lifetime of a fluorophore: hetero-FRET (28, 29), homo-FRET (22, 30, 31), pH (32), and refractive index (33). Fortunately, the impact of these factors on molecular brightness can be directly monitored by measuring the fluorescent lifetimes of fluorophore-tagged samples, as we have done here for Venus-tagged wild-type CaMKII, and the F394A and Δ315 CaMKII mutants (Fig. 2 C). Because all three of these constructs had the same Venus fluorescent lifetime, it is unlikely that any of the factors mentioned above were responsible for the large changes in molecular brightness observed for these mutants (Fig. 2, A and F). It is worth noting that different fluorophores can have different susceptibilities to many of these factors, and so experimental errors introduced by these factors can often be mitigated by the judicious selection of fluorophores. For example, some fluorescent proteins are more susceptible to pH changes than others (34), so pH-sensitive fluorophores should be avoided in molecular brightness experiments where pH is expected to change. Alternatively, while it is known that Venus fluorescence is mildly sensitive to pH (pKa of 6.0) (34, 35), in the in vitro studies described here, pH-induced changes in Venus fluorescence were readily controlled by pH buffering of our solutions. Another example is fluorophore sensitivity to homo-FRET. While the lifetimes of CFP (30) and Cerulean (22) can shorten if they transfer energy by homo-FRET, the Venus lifetime does not (10, 22). Clearly, if oligomerization of a protein is thought to occur, concomitant homo-FRET may also occur. Thus, the use of Venus to tag the protein, rather than CFP or Cerulean, might mitigate the odds of observing anomalous changes in molecular brightness.
The second category of issues that can complicate the interpretation of molecular brightness measurements are mechanisms that alter the stoichiometry of fluorescent and nonfluorescent subunits composing a complex. This includes photobleaching (36, 37, 38), incomplete fluorescent protein maturation (38), dark states (37, 38, 39), the incorporation of nontagged endogenous subunits in molecular complexes, and proteolysis. The impact of these types of factors can typically be evaluated by a combination of tests. By measuring the molecular brightness of covalent fluorescent protein oligomers (monomers, dimers, trimers, etc.) expressed in the same types of cells, and measuring under the same experimental conditions (excitation power, one-photon versus two-photon excitations, etc.), negative impact from bleaching, slow fluorophore maturation, and dark states will be observed as a nonlinear response. To illustrate this, consider that a fluorescent protein dimer should theoretically have a molecular brightness two times the brightness of a monomer (37). If, however, 50% of the fluorescent proteins are in a dark state, then the molecular brightness of a dimer will appear on average to be only 1.33 times the brightness of a monomer. This nonintuitive behavior arises because only fluorescent monomers contribute to molecular brightness measurements. In contrast, a population of dimers in which 50% are in a dark state will consist of a mixture where 25% of dimers have two normal fluorophores, 50% will have one dark and one normal, and 25% will have both dark. Again, dimers with two dark fluorophores will not influence brightness measurements; the dimers contributing to the molecular brightness measurement will be 33% normal dimers and 67% dimers with one dark fluorophore. In fact, such anomalous behavior has been observed with EGFP oligomers. A comparison of the molecular brightness of EGFP monomers, dimers, and trimers revealed a nonlinear relationship between fluorophore number and molecular brightness (38). This anomalous behavior was attributed to a pH-dependent change in the abundance of EGFP molecules in dark states. In contrast, our lab has shown that the molecular brightness of Venus monomers, dimers, trimers, tetramers, pentamers, and hexamers were linear with the number of Venus molecules in each oligomer (10). Based on the literature, we can only speculate about the reasons for this apparent discrepancy. The two most obvious differences in these studies are: 1) They used different fluorescent proteins, EGFP versus Venus, with each having their own unique photophysical behaviors; and 2) The EGFP study used one-photon excitation for brightness analysis while the Venus study used two-photon excitation. We note that intersystem crossing to intensity-dependent dark states are less likely with two-photon excitation (39), so this might explain the discrepancy. Regardless, because Venus monomers through hexamers had a linear increase in their molecular brightness (10), it is unlikely that photobleaching, incomplete fluorescent protein maturation, or dark states influence the apparent molecular brightness of Venus-tagged CaMKII holoenzyme reported here.
The impact of the incorporation of nontagged endogenous subunits in molecular complexes and/or proteolysis on molecular brightness can be evaluated by immunoblotting or immunocapillary electrophoresis. For example, in this study, immunocapillary electrophoresis using a phospho-T286-specific antibody detected our exogenous Venus-tagged autophosphorylated protein (∼90 KDa; see red dotted trace in Fig. 4 A), but failed to detect any endogenous CaMKII subunits (lacking the 28 KDa Venus moiety) that are expected to migrate between 50 and 65 KDa. Thus, in this study, it is unlikely that the incorporation of nontagged endogenous subunits into CaMKII holoenzymes perturb our molecular brightness measurements. Similarly, proteolytic degradation products were not observed with both the GFP-specific antibody or the phospho-T286 specific antibody (Fig. 4). Thus, it is unlikely that proteolytic cleavage of Venus or the linker that attaches Venus to CaMKII, removed fluorophores from CaMKII subunits to perturb our molecular brightness measurements.
We have previously shown that brightness analysis can be used to measure the number of subunits in CaMKII holoenzyme, and these values were in agreement with biochemical means of estimating CaMKII holoenzyme subunit stoichiometry (9, 10). In this study, brightness analysis was used to monitor the impact of mutations in the CaMKII oligomerization-domain on subunit stoichiometry (Fig. 1 B). Based on previous studies (9, 10), the brightness of WT Venus-tagged CaMKII holoenzyme was used as a positive assembly control and was found to be 10 times as bright as a Venus monomer, our baseline brightness value. Brightness values were normalized to the molecular brightness of a Venus monomer having the Zacharias mutation to prevent fluorescent protein dimerization (40). The monomeric nature of Venus was confirmed by time-resolved anisotropy analysis of Venus that decayed as a single exponential, indicating the absence of homo-FRET (41). FCS measurements of molecular brightness indicates that a single point mutation (F394A) on the αD helix of the lateral interaction surface between CaMKIIα oligomerization-domains results in a Venus-tagged holoenzyme that has a molecular brightness twice as bright as a Venus monomer and one-fifth the brightness of the WT Venus-tagged CaMKIIα holoenzyme (Fig. 1 B). Single point mutations (F397A, I434A, and Q436A) in the lateral interaction surface (αD helix, β3- and β4-sheets) destabilized the holoenzyme structure, but were composed of more than two subunits. Others seemed to stabilize the holoenzyme based on an increase in the number of subunits (H411A, T413A, and I415A), while D393A, Y398A, and N401A had no obvious effect on the holoenzyme stoichiometry. The oligomerization-domain Δ315 deletion mutant, in contrast to F394A, had a brightness of 1.0 ± 0.1, indicating that it is a monomer. V-CaMKIIαF394A has a fast anisotropy decay component (Fig. 2 A), indicative of homo-FRET; thus, it must have at least two Venus-tagged subunits in close proximity, while V-CaMKIIαΔ315 decayed as a single exponential and therefore did not have any observable homo-FRET. The V-CaMKIIαF394A diffusion time (560.5 ± 71.4 μs; mean ± SD, n = 5) was only slightly larger, and not significantly different (paired Student t-test, p = 0.0715) from the diffusion time of monomeric V-CaMKIIαΔ315 (436.4 ± 52.7 μs, n = 5), suggesting that its mass and hydrodynamic volume was only slightly larger than that of the monomeric mutant. Conventional two-photon FCS measurements can only be made using low concentrations of sample, typically 10–100 nM. Thus, the oligomerization state of CaMKII holoenzyme detected using brightness analysis also indicates that a high-affinity interaction between subunits must persist at these low concentrations. Thus, based on molecular brightness analysis and corroborating data from fluorescence anisotropy measurements and diffusion time analysis, we conclude that when V-CaMKIIαF394A is expressed in HEK cells, it self-assembles with high affinity to form a dimer while V-CaMKIIαΔ315, lacking an oligomerization domain, remains a monomer.
A comparison of structural changes that occur upon activation with CaM revealed that activation does not alter the stoichiometry of the holoenzyme for WT-, dimeric-, or monomeric-CaMKII (Fig. 2 D). The molecular brightness of each CaMKII construct remained the same before and after activation, indicating that the net number of subunits was not altered. Similarly, a net change in the Venus anisotropy signal with activation was not observed for all three constructs (Fig. 2 C), indicating that activation with CaM did not alter the homo-FRET signal from these constructs. Because the time-resolved anisotropy signal of WT V-CaMKIIα and V-CaMKIIαF394A (Fig. 2 A) each had both a slow decay component associated with the constructs’ molecular rotation as well as a fast decay component associated with homo-FRET (20), WT V-CaMKIIα and V-CaMKIIαF394A each must have at least two Venus-tagged subunits. In contrast, V-CaMKIIαΔ315 anisotropy decayed with a single slow decay component, which is consistent with it being a monomer. Thus, structurally CaMKII holoenzyme activation is associated with an increase in hydrodynamic volume (Fig. 2 E), previously interpreted as catalytic-domain pair extension (9), without perturbing the pairing of catalytic domains as measured by homo-FRET. These structural changes in the WT holoenzyme were recapitulated by the V-CaMKIIαF394A dimeric mutant.
It is worth noting that while both WT and CaMKIIαF394A each had anisotropy values consistent with a dimeric structure (Fig. 2 A), homo-FRET values were larger in the WT holoenzyme. This suggests that some aspect of catalytic-domain pairing is altered in V-CaMKIIαF394A. One possible explanation for this is that normal catalytic-domain pairing requires the ring structure of the holoenzyme oligomerization domain core. Consistent with this interpretation, we saw a progressive increase in anisotropy (decrease in homo-FRET) for Venus-tagged holoenzyme composed of fewer than eight subunits (Fig. 1 D).
The kinase activity of V-CaMKIIα and V-CaMKIIαF394A for an exogenous substrate was indistinguishable in terms of CaM KD, Vmax, Hill coefficient, and basal activity (Fig. 3). In contrast, the KD of V-CaMKIIαΔ315 exceeded V-CaMKIIα by a factor of 30 to achieve the same activity. As expected for a monomer, cooperativity was not observed for V-CaMKIIαΔ315, even though it shared the same CaM Vmax as WT and F394A. V-CaMKIIαΔ315 also differed from both WT and V-CaMKIIαF394A by having a lower basal kinase activity, suggesting that catalytic-domain pairing increases the basal kinase activity.
Saturating concentrations of CaM (where maximal T286 autophosphorylation was observed) triggered comparable amounts of T286 autophosphorylation levels for both WT and F394A (Fig. 4 D). Monomeric Δ315 achieved lower levels of T286 phosphorylation, presumably because T286 autophosphorylation requires one subunit to act as kinase and a second to act as substrate. We speculate that the limited Δ315 T286 phosphorylation observed resulted from intermolecular reactions between freely diffusing subunits, while the T286 phosphorylation observed in WT and F394A arose primarily from intramolecular reactions within catalytic-domain pairs. We found that T286 autophosphorylation triggered a comparable shift in the migration of individual CaMKII subunits (on capillary electrophoresis) for both WT V-CaMKIIα and V-CaMKIIαF394A (Fig. 4, A and B). Predicated on the assumption that under saturating conditions all T286 sites will be autophosphorylated, we initially hypothesized that the distribution and shift in migration observed when probed with the P-286-specific antibody should be identical to the distribution and shift observed with the GFP-specific antibody that recognizes the Venus adduct. We found instead for both WT and F394A that the shift observed using the P-T286-specific antibody was larger than the shift observed using the GFP antibody (Fig. 4, A and B; compare red and black dashed traces). Similarly, the width of the distribution detected with the P-286-specific antibody was narrower than the width of the distribution detected with the GFP antibody. This discrepancy in observed migration suggests that only a fraction of T286 sites are autophosphorylated. We hypothesize that after kinase activation the GFP antibody recognizes a peak composed of both WT and the shifted autophosphorylated T286 subunits. Assuming the shift in migration caused by T286 autophosphorylation is below the resolution of our instrumentation, it will appear to migrate as a broad peak to an intermediate distance between the two pure species. In contrast, because the P-286 antibody will only bind autophosphorylated T286 subunits, it can only recognize the subfraction that is autophosphorylated. Thus, the component detected using the P-286 antibody should be shifted to a greater extent, and present with a narrower distribution. Thus, our experiments suggest that even under conditions of maximal T286 phosphorylation, substantial amounts of T286 remain unphosphorylated. While we cannot rule out the possibility that limited T286 phosphorylation is indicative of a heterogeneous population in which some catalytic-domain pairs have no autophosphorylation while other pairs are autophosphorylated at both T286 sites, a more plausible explanation is that only one subunit in a catalytic-domain pair can be phosphorylated at its T286 site.
It is known that CaM activation of CaMKII triggers the extension of catalytic domain pairs away from the holoenzyme central core, and that this reaction requires T286 autophosphorylation (9). Here we show, using CaMKII assembly mutants and capillary electrophoresis, that only a fraction of CaMKII subunits in the holoenzyme get phosphorylated at T286 (Fig. 4 A). We hypothesize that submaximal T286 autophosphorylation arises as a consequence of the paired catalytic-domain structure of CaMKII. Accordingly, our data may provide insight into the rules dictating which subunits within a holoenzyme can act as the kinase and which as the substrate. The observation that T286 phosphorylation occurs in the dimeric V-CaMKIIαF394A construct (Fig. 4 B) argues that intracatalytic-domain pair T286 phosphorylation can occur. Because the amount of V-CaMKIIαF394A T286 autophosphorylation was statistically indistinguishable from the autophosphorylation observed in WT V-CaMKIIα (Fig. 4 D), and both were submaximal (Fig. 4, A and B), our data suggests that if any intercatalytic-domain pair T286 phosphorylation occurs in the WT holoenzyme, the amount is limited. Because autoinhibited catalytic-domain pairs are thought to be symmetrical (18), it is reasonable to hypothesize that when intracatalytic-domain pair T286 phosphorylation occurs, each subunit within a pair could act sequentially as either kinase or T286 substrate. Alternatively, intracatalytic-domain pair T286 phosphorylation might generate an asymmetrical structure that prevents any subsequent T286 phosphorylation event from occurring within that catalytic-domain pair. Our observation of submaximal T286 autophosphorylation under saturating conditions supports this alternative hypothesis, as does the observation of an asymmetrical catalytic-domain pair structure by x-ray crystallography (11).
Deciphering how complex molecular machines function often requires differentiating between different types of protein interactions. Some interfaces within molecular machines are involved in maintaining the integrity of the complex. These interactions are usually static over a timescale of minutes to hours. More-dynamic protein interactions typically mediate ligand binding or transient interactions with exogenous proteins, while others dynamic changes may reflect changes in the conformation of the complex associated with its function. One approach to differentiate between multiple types of interactions in a complex is by monitoring several molecular interactions simultaneously. Here we used a method for studying protein interactions in CaMKIIα assemblies that simultaneously monitor homo-FRET (catalytic-domain pairing), molecular brightness (holoenzyme subunit stoichiometry and assembly), and the hydrodynamic volume of the complex (catalytic-domain extension from the oligomerization-domain core). The power and utility of multimodal monitoring of protein complexes arises from the ability of one modality to act as a positive or negative control for the other modalities. For example, it is well known that the absence of a change in FRET does not mean that a conformational change has not occurred. FRET is sensitive to changes in proximity between donor and acceptor (42, 43), but FRET is also sensitive to the relative dipole orientation of donors and acceptors (44, 45) and to the number of donors and acceptors in a complex (46). Thus, while the absence of a change in FRET could indicate that there was no conformational change in a complex, it could also result from structural changes that do not alter donor-acceptor proximity, or even from changes in proximity that were compensated for by changes in dipole orientation or stoichiometry. In this study, a net change in homo-FRET was not observed when Venus-tagged WT CaMKIIα holoenzyme was activated with CaM (Fig. 2 C). Activation did not alter the subunit stoichiometry of the holoenzyme either (Fig. 2 D), so the absence of a change in homo-FRET cannot be attributed to a compensating change in stoichiometry. Activation did change the hydrodynamic volume of the holoenzyme (Fig. 2 E), indicating that a large conformational change occurred, and this is consistent with the changes in kinase activity observed for both exogenous (Fig. 3) and endogenous (Fig. 4 A) substrates. Because a flexible linker was used to attach Venus to CaMKII in these experiments, it is unlikely that a change in dipole orientation could compensate for a change in Venus separation. Presumably the conformational change that occurs with CaMKII holoenzyme activation does not cause a net change in the separation between Venus-tagged catalytic domains (9), but does cause an increase in the holoenzyme hydrodynamic volume. CaMKII activation is a clear example where the absence of a net change in FRET does not mean the absence of a conformational change in the complex. This negative homo-FRET data for both WT and dimeric V-CaMKIIα before and after CaM activation can be more rigorously interpreted because of our multimodal approach.
The picture that emerges for the activation of CaMKII holoenzyme from this and previous studies is that both dimeric V-CaMKIIαF394A and WT holoenzyme undergoes a conformational change when activated by CaM that increases hydrodynamic volume without altering catalytic-domain separation, or subunit stoichiometry. CaM activation requires T286 autophosphorylation (9) of at least one, and is most likely limited to only one subunit per catalytic-domain pair. The concomitant extension of catalytic-domain pairs away from its oligomerization-domain core is responsible for the increase in holoenzyme hydrodynamic volume. Finally, the catalytic activity of activated CaMKIIα holoenzyme with either 2 or 10 subunits are indistinguishable in terms of CaM KD, Kmax, basal activity, and cooperativity. Thus, we conclude that the fundamental functional units of CaMKII holoenzyme are its paired catalytic-domains.
Author Contributions
P.S., K.A.D., H.L.P., T.A.N., and S.S.V. designed the research; P.S., K.A.D., and T.A.N. performed the research; H.L.P. and J.V.V. contributed analytic tools; P.S., K.A.D., T.A.N., and S.S.V. analyzed the data; and P.S., T.A.N., and S.S.V wrote the article.
Acknowledgments
We are grateful for the constructive criticism and advice from the other members of the Laboratory of Molecular Physiology, National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism (NIAAA).
The intramural program of the National Institute on Alcohol Abuse and Alcoholism, the National Institutes of Health, Bethesda, Maryland, funded this work.
Editor: David Piston.
References
- 1.Shonesy B.C., Jalan-Sakrikar N., Colbran R.J. CaMKII: a molecular substrate for synaptic plasticity and memory. Prog. Mol. Biol. Transl. Sci. 2014;122:61–87. doi: 10.1016/B978-0-12-420170-5.00003-9. [DOI] [PubMed] [Google Scholar]
- 2.Lu H.E., MacGillavry H.D., Blanpied T.A. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 2014;34:7600–7610. doi: 10.1523/JNEUROSCI.4364-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemieux M., Labrecque S., De Koninck P. Translocation of CaMKII to dendritic microtubules supports the plasticity of local synapses. J. Cell Biol. 2012;198:1055–1073. doi: 10.1083/jcb.201202058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer K.U., De Koninck P., Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- 5.Strack S., McNeill R.B., Colbran R.J. Mechanism and regulation of calcium/calmodulin-dependent protein kinase II targeting to the NR2B subunit of the N-methyl-D-aspartate receptor. J. Biol. Chem. 2000;275:23798–23806. doi: 10.1074/jbc.M001471200. [DOI] [PubMed] [Google Scholar]
- 6.Strack S., Choi S., Colbran R.J. Translocation of autophosphorylated calcium/calmodulin-dependent protein kinase II to the postsynaptic density. J. Biol. Chem. 1997;272:13467–13470. doi: 10.1074/jbc.272.21.13467. [DOI] [PubMed] [Google Scholar]
- 7.Strack S., Colbran R.J. Autophosphorylation-dependent targeting of calcium/calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl-D-aspartate receptor. J. Biol. Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- 8.Tsui J., Inagaki M., Schulman H. Calcium/calmodulin-dependent protein kinase II (CaMKII) localization acts in concert with substrate targeting to create spatial restriction for phosphorylation. J. Biol. Chem. 2005;280:9210–9216. doi: 10.1074/jbc.M407653200. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen T.A., Sarkar P., Vogel S.S. Covert changes in CaMKII holoenzyme structure identified for activation and subsequent interactions. Biophys. J. 2015;108:2158–2170. doi: 10.1016/j.bpj.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen T.A., Sarkar P., Vogel S.S. Fluorescence polarization and fluctuation analysis monitors subunit proximity, stoichiometry, and protein complex hydrodynamics. PLoS One. 2012;7:e38209. doi: 10.1371/journal.pone.0038209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rellos P., Pike A.C., Knapp S. Structure of the CaMKIIδ/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8:e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg O.S., Deindl S., Kuriyan J. Oligomerization states of the association domain and the holoenzyme of Ca2+/CaM kinase II. FEBS J. 2006;273:682–694. doi: 10.1111/j.1742-4658.2005.05088.x. [DOI] [PubMed] [Google Scholar]
- 13.Hudmon A., Schulman H. Neuronal Ca2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 14.Hanson P.I., Meyer T., Schulman H. Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron. 1994;12:943–956. doi: 10.1016/0896-6273(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 15.Thaler C., Koushik S.V., Vogel S.S. Structural rearrangement of CaMKIIα catalytic domains encodes activation. Proc. Natl. Acad. Sci. USA. 2009;106:6369–6374. doi: 10.1073/pnas.0901913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoelz A., Nairn A.C., Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol. Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 17.Kanaseki T., Ikeuchi Y., Yamauchi T. Structural features of Ca2+/calmodulin-dependent protein kinase II revealed by electron microscopy. J. Cell Biol. 1991;115:1049–1060. doi: 10.1083/jcb.115.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris E.P., Török K. Oligomeric structure of α-calmodulin-dependent protein kinase II. J. Mol. Biol. 2001;308:1–8. doi: 10.1006/jmbi.2001.4584. [DOI] [PubMed] [Google Scholar]
- 19.Lantsman K., Tombes R.M. CaMK-II oligomerization potential determined using CFP/YFP FRET. Biochim. Biophys. Acta. 2005;1746:45–54. doi: 10.1016/j.bbamcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Vogel S.S., Nguyen T.A., Wieb van der Meer B. Springer Series in Chemical Physics. Springer; Heidelberg, Germany: 2015. An introduction to interpreting time resolved fluorescence anisotropy curves; p. 624. [Google Scholar]
- 21.Vogel S.S., Thaler C., Koushik S.V. Time resolved fluorescence anisotropy. In: Periasamy A., Clegg R.M., editors. FLIM Microscopy in Biology and Medicine. Taylor & Francis; Boca Raton, FL: 2010. pp. 245–290. [Google Scholar]
- 22.Koushik S.V., Vogel S.S. Energy migration alters the fluorescence lifetime of Cerulean: implications for fluorescence lifetime imaging Forster resonance energy transfer measurements. J. Biomed. Opt. 2008;13:031204. doi: 10.1117/1.2940367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Müller J.D., Gratton E. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys. J. 2002;82:133–144. doi: 10.1016/S0006-3495(02)75380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Wei L.N., Müller J.D. Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15492–15497. doi: 10.1073/pnas.2533045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Müller J.D. Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:3147–3152. doi: 10.1073/pnas.0606557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B., Chen Y., Müller J.D. Fluorescence fluctuation spectroscopy of mCherry in living cells. Biophys. J. 2009;96:2391–2404. doi: 10.1016/j.bpj.2008.12.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slaughter B.D., Huff J.M., Li R. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS One. 2008;3:e1931. doi: 10.1371/journal.pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wouters F.S., Bastiaens P.I. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells. Curr. Biol. 1999;9:1127–1130. doi: 10.1016/s0960-9822(99)80484-9. [DOI] [PubMed] [Google Scholar]
- 29.Harpur A.G., Wouters F.S., Bastiaens P.I. Imaging FRET between spectrally similar GFP molecules in single cells. Nat. Biotechnol. 2001;19:167–169. doi: 10.1038/84443. [DOI] [PubMed] [Google Scholar]
- 30.Rizzo M.A., Springer G.H., Piston D.W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 31.Förster T. Energy migration and fluorescence. 1946. J. Biomed. Opt. 2012;17:011002. doi: 10.1117/1.JBO.17.1.011002. [DOI] [PubMed] [Google Scholar]
- 32.Kneen M., Farinas J., Verkman A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suhling K., Siegel J., Davis D.M. Imaging the environment of green fluorescent protein. Biophys. J. 2002;83:3589–3595. doi: 10.1016/S0006-3495(02)75359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 35.Nagai T., Ibata K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 36.Eggeling C., Kask P., Jäger S. Rapid analysis of Forster resonance energy transfer by two-color global fluorescence correlation spectroscopy: trypsin proteinase reaction. Biophys. J. 2005;89:605–618. doi: 10.1529/biophysj.104.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y., Johnson J., Mueller J.D. Observing protein interactions and their stoichiometry in living cells by brightness analysis of fluorescence fluctuation experiments. Methods Enzymol. 2010;472:345–363. doi: 10.1016/S0076-6879(10)72026-7. [DOI] [PubMed] [Google Scholar]
- 38.Vámosi G., Mücke N., Tóth K. EGFP oligomers as natural fluorescence and hydrodynamic standards. Sci. Rep. 2016;6:33022. doi: 10.1038/srep33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohl T., Schwille P. Fluorescence correlation spectroscopy with autofluorescent proteins. Adv. Biochem. Eng. Biotechnol. 2005;95:107–142. doi: 10.1007/b102212. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias D.A., Violin J.D., Tsien R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar P., Koushik S.V., Gryczynski Z. Photophysical properties of Cerulean and Venus fluorescent proteins. J. Biomed. Opt. 2009;14:034047. doi: 10.1117/1.3156842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clegg R.M. Forster resonance energy transfer—FRET what it is, why do it, and how it’s done. In: T. W. J. Gadella., editor. FRET and FLIM Techniques. Elsevier; Amsterdam, the Netherlands: 2009. pp. 1–57. [Google Scholar]
- 43.Vogel S.S., van der Meer B.W., Blank P.S. Estimating the distance separating fluorescent protein FRET pairs. Methods. 2014;66:131–138. doi: 10.1016/j.ymeth.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Meer B.W. Kappa-squared: from nuisance to new sense. J. Biotechnol. 2002;82:181–196. doi: 10.1016/s1389-0352(01)00037-x. [DOI] [PubMed] [Google Scholar]
- 45.van der Meer B.W., van der Meer D.M., Vogel S.S. FRET—Förster Resonance Energy Transfer. Wiley; Weinheim, Germany: 2013. Optimizing the orientation factor κ2 for more accurate FRET measurements; pp. 63–104. [Google Scholar]
- 46.Vogel S.S., Thaler C., Koushik S.V. Fanciful FRET. Sci. STKE. 2006;2006:re2. doi: 10.1126/stke.3312006re2. [DOI] [PubMed] [Google Scholar]