Abstract
Adhesion of cancer cells to endothelial cells is a key step in cancer metastasis; therefore, identifying the key molecules involved during this process promises to aid in efforts to block the metastatic cascade. We have previously shown that intercellular adhesion molecule-1 (ICAM-1) expressed by endothelial cells is involved in the interactions of bladder cancer cells (BCs) with the endothelium. However, the ICAM-1 ligands have never been investigated. In this study, we combined adhesion assays and atomic force microscopy (AFM) to identify the ligands involved and to quantify the forces relevant in such interactions. We report the expression of MUC1 and CD43 on BCs, and demonstrate that these ligands interact with ICAM-1 to mediate cancer cell-endothelial cell adhesion in the case of the more invasive BCs. This was achieved with the use of adhesion assays, which showed a strong decrease in the attachment of BCs to endothelial cells when MUC1 and CD43 were blocked by antibodies. In addition, AFM measurements showed a similar decrease, by up to 70%, in the number of rupture events that occurred when MUC1 and CD43 were blocked. When we applied a Gaussian mixture model to the AFM data, we observed a distinct force range for receptor-ligand bonds, which allowed us to precisely identify the interactions of ICAM-1 with MUC1 or CD43. Furthermore, a detailed analysis of the rupture events suggested that CD43 is strongly connected to the cytoskeleton and that its interaction with ICAM-1 mainly corresponds to force ramps followed by sudden jumps. In contrast, MUC1 seems to be weakly connected to the cytoskeleton, as its interactions with ICAM-1 are mainly associated with the formation of tethers. This analysis is quite promising and may also be applied to other types of cancer cells.
Introduction
Cancer metastasis is the primary cause of 90% of cancer-associated mortalities. The malignancy of cancer strongly depends upon the ability of primary tumors to metastasize to distant organs (1, 2). During metastasis, cancer cells manage to escape from primary tumors and penetrate into the blood flow (intravasation). Cancer cells that are carried in the blood flow can interact with the endothelium lining the walls of blood vessels, adhere, and migrate through the endothelium (extravasation) to form secondary tumors. Cancer cells and leukocytes follow similar mechanisms during the extravasation process: 1) rolling of cells on the endothelium, 2) adhesion of cells to the endothelium, and 3) spreading and transmigration of cells through the endothelium (3, 4, 5). The adhesion and migration of leukocytes through the endothelium have been studied in detail during inflammation (3, 6), but few results are available regarding the role of the key molecules involved in the adhesion and transmigration of cancer cells (6, 7, 8, 9, 10, 11).
The adhesion of cancer cells or leukocytes to endothelial cells (ECs) is an important step of the extravasation process and is mediated by several cell adhesion molecules (CAMs), including β1 integrins (12), vascular cell adhesion molecule-1 (VCAM-1) (13), L-selectin (14), and intercellular adhesion molecule-1 (ICAM-1) (6, 7, 15, 16). We previously showed that ICAM-1 expressed by ECs is involved in the adhesion of bladder cancer cells (BCs) (6). ICAM-1 (CD54) is a cell-surface glycoprotein consisting of five extracellular immunoglobulin-like domains (17). ICAM-1 is expressed on various cells, including ECs, leukocytes, B cells, T cells, fibroblasts, and some cancer cells, and its expression can be upregulated by cytokines such as TNF-α (6, 18). Leukocytes express LFA-1 and Mac-1 (β2 integrins), which are well-known ligands for ICAM-1. Cancer cells lack the expression of β2 integrins, but neutrophils can act as a bridge between cancer cells and ECs to help in the adhesion process (3). In addition, MUC1 (19) and CD43 (20) expressed by some cancer cells have been identified as ligands for ICAM-1. MUC1 (CD227) is a transmembrane glycoprotein that is usually expressed on the apical surface of secretory epithelia. It is overexpressed and underglycosylated in several common cancers, including breast, lung, colon, ovary, bladder cancers, and hence is used as a tumor marker (21). The extracellular domain of MUC1 mainly consists of 25–150 tandem repeats of the same 20 amino acid sequences (22) and these tandem repeats have been shown to recognize and bind to domain I of ICAM-1 (23). CD43 (leukosialin) is a transmembrane sialoglycoprotein that is normally expressed on the surface of T lymphocytes, monocytes, neutrophils, platelets, and some B lymphocytes. CD43 is also expressed by a variety of cancer cell lines and tumor tissues. It is undetected in normal tissues and for this reason is used as a marker for tumor detection (24, 25). The ligands involved in the adhesion of BCs to ECs are not yet as clearly identified, but CD43 and MUC1 are good candidates that could be involved in mediating the adhesion via ICAM-1. Furthermore, quantitative analysis of the forces involved in these interactions can help elucidate how cancer cells regulate their adhesion.
In recent years, single-cell force spectroscopy (SCFS) using atomic force microscopy (AFM) (6, 17, 18, 26, 27, 28) has been applied to study the adhesive interactions of cells with other cells, proteins, or functionalized surfaces. In particular, AFM is a powerful tool for identifying and quantifying receptor-ligand interactions (17, 28, 29, 30, 31, 32). Here, we used SCFS to unravel the key molecules involved in the adhesion of BCs to ECs. We combined adhesion assays and SCFS to characterize cancer cell-EC adhesion both qualitatively and quantitatively. We report the expression of MUC1 and CD43 in BCs with different levels of invasiveness (RT112, T24, and J82). When ICAM-1, CD43, and MUC1 were blocked with specific monoclonal antibodies (mAbs), adhesion of BCs to ECs was largely decreased for invasive cells such as T24 and J82, showing that these interactions are essential for invasive cells, while less invasive ones might interact through different receptors. Through SCFS measurements, we identified different force ranges for the interactions of MUC1 and CD43 with ICAM-1. To our knowledge, this is the first report on the force range corresponding to the interaction of MUC1 or CD43 with ICAM-1, with values of 43–53 pN, which compare well with those previously reported for the interaction of LFA-1 with ICAM-1 (17, 33). Finally, a detailed analysis of rupture events showed that MUC1 interacts with ICAM-1 mainly through tethers, whereas CD43 interacts with ICAM-1 mainly through jumps, this being related to a better attachment to the cytoskeleton in the latter case. Taken altogether, these results add to our understanding of the process of metastasis, when cancer cells bind to the endothelium through interactions between endothelial ICAM-1 and cancer cell ligands.
Materials and Methods
Cell lines and cell culture
Three human BCs (RT112/G2 grade, T24/G3 grade, and J82/G3 grade; ATCC, Rockville, MD) were cultured in RPMI 1640 medium (Gibco, Saint Aubin, France) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin mix (complete RPMI medium). The grade of BCs was defined based on the morphological and pathological characteristics of the original tumor, and represented progression from well to poorly differentiated phenotypes arising from superficial to invasive epithelial human bladder cancer (34, 35). The BCs were stably transfected with a plasmid expressing lifeact-GFP to stain F-actin, and these GFP-transfected cells were used for AFM experiments. Human vascular umbilical ECs (HUVECs) were purchased from Promocell (Heidelberg, Germany). The HUVECs were grown on culture dishes coated with 100 μg/mL collagen I (BD Biosciences, Le Pont de Claix, France) in complete endothelial growth medium (endothelial basal medium mixed with supplements) from Promocell. HUVEC subcultures from passages 2–6 were selected for our experiments. Cultures were grown at 37°C in a 5% CO2 humidified atmosphere.
Flow cytometry
BCs grown in a tissue culture flask were detached using a cell-dissociation solution (Sigma-Aldrich, Lyon, France). The cell suspension was incubated with primary antibodies (10 μg/mL) in ice for 45 min. Mouse anti-human MUC1 mAb clone E29 (DAKO, Glostrup, Denmark), mouse anti-human CD43 mAb clone L10 (Invitrogen, Saint Aubin, France), and mouse immunoglobulin (IgG; Vector Laboratories, CA) were used as the primary antibodies in this study. The cells were then incubated with goat anti-mouse IgG Alexa-488 (Life Technologies, Saint Aubin, France) secondary antibody in ice for 30 min. The expression levels of MUC1 and CD43 were measured using an Accuri C6 flow cytometer (BD Biosciences) and analyzed using the software provided with the flow cytometer. Both antibodies (E29 and L10) recognize an extracellular domain of MUC1 and CD43, respectively (23, 24). It has been reported that these antibodies can block MUC1 and CD43 and partially inhibit adhesion (23, 36).
BC-EC adhesion assay
ECs (2 × 104) were seeded in 48-well plates (Nunc, Saint Aubin, France) coated with 100 μg/mL collagen I and grown to confluence for 3 days. BCs were labeled with calcein (Life Technologies) for 30 min at 37°C. Then, 2 × 105 labeled BCs were added per well and incubated for 30 min at 37°C. After different contact times were tested, this incubation time of 30 min was found to give an optimal number of adherent cells (data not shown). Nonadherent BCs were removed by gentle washing with complete RPMI medium. The remaining adherent cells were then lysed with detergent (10% SDS) and the fluorescence signal was quantified using a Victor3 multilabel counter (Perkin Elmer, Waltham, MD). For blocking experiments, ICAM-1 on ECs and MUC1 or CD43 on BCs were blocked with specific antibodies (20 μg/mL) for 15 min at 37°C before the addition of cancer cells. The percentage of adhesion was calculated as
Total fluorescence corresponds to the fluorescence of added cells (2 × 105), and fluorescence of adhered cells corresponds to the fluorescence value obtained after washing of the nonadherent cells (36).
AFM
AFM experiments were performed using a Nanowizard II atomic force microscope (JPK Instruments, Berlin, Germany) mounted on a Zeiss microscope (Carl Zeiss, Jena, Germany). This configuration allowed us to carry out AFM measurements and simultaneously observe the cells using phase-contrast or fluorescence modes. Long-range force measurements involving cell-cell interactions were performed using the CellHesion module (JPK Instruments), which enables vertical movement of the sample holder up to 100 μm thanks to the piezo-driven movement. In addition, the objective was mounted on a vertical piezo-translator (PIFOC, Physik Instrumente, Karlsruhe, Germany), which allowed us to move the objective concurrently with the microscope stage and focus on the cells during AFM measurements. All AFM measurements were carried out at 37°C ± 0.1°C using a petri dish heater (JPK Instruments). For AFM experiments, complete endothelial growth medium was supplemented with 20 mM Hepes at pH 7.4 (Sigma-Aldrich).
Preparation of the cantilever and substrates
V-shaped, 300-μm-long, tipless, gold-coated silicon nitride cantilevers with a nominal spring constant of ∼0.01 N/m (MLCT-O, Bruker, France) were used for force measurements. The AFM cantilever was soaked in acetone for 5 min, UV irradiated for 15 min, and incubated in 0.5 mg/mL biotin-BSA (Interchim, Montluçon, France) overnight at 37°C. The cantilever was then rinsed with phosphate-buffered saline (PBS) and incubated in 0.5 mg/mL streptavidin (Interchim) for 10 min at room temperature. Finally, the cantilever was rinsed with PBS and incubated in 0.5 mg/mL biotin-ConA (Interchim) for 10 min and then rinsed with PBS as previously described (6). This protocol mediates the binding of the cancer cell to the cantilever with a force greater than those measured for the interactions between cancer cells and HUVECs (33). The sensitivity of the cantilever was determined before each experiment by making contact with the glass coverslip, and the spring constant was obtained using the thermal noise method according to a previously described protocol (37). This was carried out on the same glass coverslip used for force measurements.
HUVEC substrate
For AFM experiments, HUVECs were grown on 35-mm-diameter petri dishes (TPP petri dishes, ThermoFisher Scientific, Villebon-sur-Yvette, France) modified as follows: a 30-mm-diameter glass coverslip was glued using UV curable adhesive (Norland #61) after a 26-mm hole was drilled at the bottom. The coverslip was cleaned with alcohol, dried with N2, and then treated with fibronectin (10 μg/mL). HUVECs were seeded on these dishes and cultured for 3 days at 37°C, 5% CO2 to achieve confluence. Before use, they were replaced with endothelial basal medium (without supplements) containing 20 mM Hepes (pH 7.4). The substrates (HUVECs, rICAM-1, or bovine serum albumin (BSA)) were used in basal medium without supplements during the attachment of cancer cells to the cantilever, as the proteins present in the supplements can bind to ConA and prevent attachment of the cancer cell to the cantilever.
Recombinant ICAM-1 substrate
To prepare recombinant ICAM-1 (rICAM-1) immobilized substrate, 20 μL of monomeric rICAM-1 (RD Systems, Lille, France; 25 μg/mL) in 0.1 M NaHCO3 was incubated overnight at 4°C in a petri dish. Unbound proteins were removed by washing with PBS and then the exposed surface was blocked using 100 μg/mL of BSA in PBS (Sigma-Aldrich). Finally, the BSA was replaced by RPMI 1640 medium without serum and used for further steps.
BSA substrate
For the BSA-immobilized substrate, 20 μL of BSA at 100 μg/mL in PBS was allowed to adsorb for 30 min at 37°C in a petri dish. Unbound proteins were removed by washing with PBS, and RPMI medium without serum was added to the BSA-coated dish. The different substrates are sketched in Fig. 1 A.
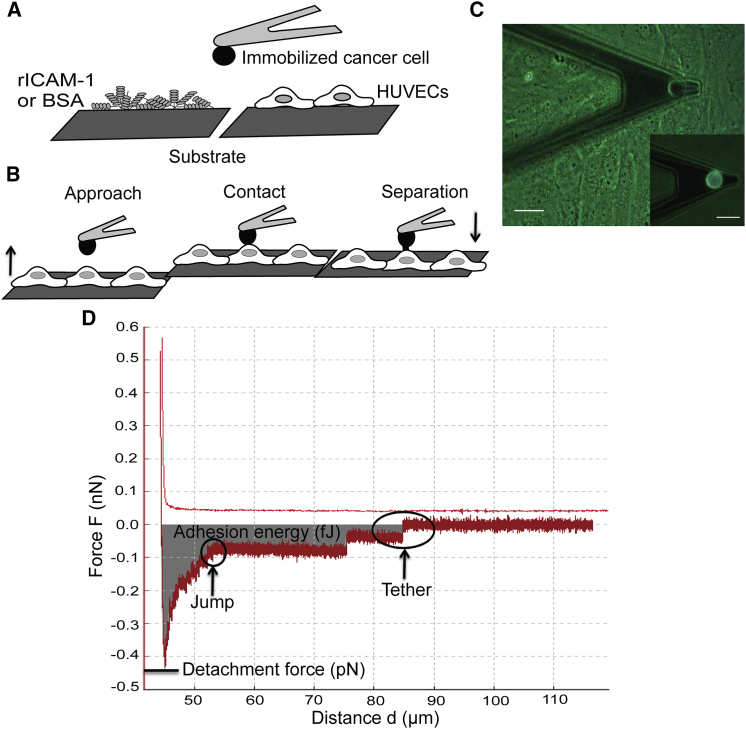
Figure 1.
Interactions between cancer cells and ECs, as measured by SCFS. (A) Sketch of the different substrates (rICAM-1, BSA, and HUVECs) used for SCFS experiments. (B and D) Sketch of the approach-retraction method and typical retraction force curve in terms of the piezo displacement. The HUVEC monolayer approaches the cancer cell at a constant velocity (5 μm/s). Then, the HUVECs come into contact with the cancer cell during 10 s (under 500 pN applied force) to create several bond complexes within the adhesion area. The HUVEC substrate is then retracted at a constant velocity to detach the adhesive bonds. The retraction curve shows force jumps and tethers corresponding to bond rupture forces. The adhesive energy (shaded area) represents the detachment work needed to completely unbind the cell from the substrate. The detachment force is the force necessary to stretch the cancer cell and the HUVEC until bonds start to detach. Note that some force jumps can follow a plateau corresponding to tether formation (see Discussion). (C) Picture of an AFM cantilever with an attached cancer cell above the HUVEC monolayer. (Inset) Fluorescence image of a fluorescent cancer cell attached to the cantilever. Scale bar, 20 μm. To see this figure in color, go online.
SCFS
SCFS measurements were performed by attaching a cancer cell to the cantilever, followed by an approach-retraction cycle. J82 cells were detached from the culture dish just before the AFM experiment using Trypsin/EDTA (Sigma-Aldrich), as EDTA alone was not able to detach the strongly adhering J82 cells efficiently. A previous study (38) showed that cells detached by EDTA alone recover their mechanical and adhesive properties faster than cells detached using trypsin alone (38), but that study also demonstrated that rupture force values (jumps and tethers) do not depend on the recovery time. The detached cells were suspended in complete RPMI medium and centrifuged at 1200 rpm for 5 min at 4°C. Finally, the cells were resuspended in RPMI medium without serum. J82 cells were injected ∼20–30 min after trypsinization into a petri dish containing a monolayer of HUVECs and allowed to settle. A ConA-functionalized cantilever tip was placed above a cancer cell (cancer cells are fluorescent and thus can be distinguished from HUVECs) and then the substrate was moved to touch the cell with a force of 1 nN for 10 s. The substrate was retracted slowly and the cantilever with the captured cell was allowed to establish firm adhesion for 10–15 min in the culture medium (Fig. 1 C, inset). Next, 1.5 mL of complete endothelial growth medium was added and the force measurements were performed as shown in Fig. 1 B.
After attachment of a cancer cell, the cantilever was set above the nucleus of a HUVEC (Fig. 1 C) and the substrate was moved at a speed of 5 μm/s. This position above the nucleus is a convenient location when using a cantilever with a cancer cell attached to it, and it is known that ICAM-1 is expressed all over the cells. During contact, a compression force of 500 pN was applied for 10 s in constant force mode and the substrate was then retracted with a speed of 5 μm/s. The influence of different compression forces (500 pN, 1 nN, and 2 nN) was studied while all other parameters were kept constant. The results (data not shown) indicated that the mean rupture force (i.e., the mean value of 10 events per force curve) did not change (whereas the number of rupture events increased) with compression force, as previously reported (33, 39). A contact time of 10 s was selected to obtain a sufficient number of rupture events (at least 10 in the control experiment). The force measurements were performed in open-loop mode with an acquisition frequency of 2048 Hz and a pulling distance of 70 μm. The open-loop mode works better in our conditions, as we do not need extremely high accuracy for the height. The drift over time was found to be 0.01 pN/μm, which is considered low enough to avoid any effects on the measurements. A force-distance (F-d) curve was obtained for each approach and retraction cycle (Fig. 1 B). Typically, with a single cancer cell attached to the cantilever, ∼30 F-d curves were acquired on 30 different HUVECs. The cancer cell was allowed to recover for 1 min between each force curve and 5 min between sets of six force curves. We used a recovery time greater than the contact time, which is commonly used by many researchers (26, 29). To block specific interactions involving cancer cells and HUVECs, cancer cells were incubated with antibodies specific for MUC1, CD43, or MUC1+CD43 at 25 μg/mL for 15 min before cell capture.
The retraction curve is characterized by the force required to separate the cancer cell from the HUVEC, referred to as the detachment force (Fig. 1 D). The detachment force is related to the elasticity of cells and the adhesion between them. The force jumps (or rupture events) in the force curve correspond to the breakup of bonds involved during a cell-cell interaction (Fig. 1 D). A typical force jump usually occurs at a constant loading rate (pN/s), ending with bond rupture. Sometimes the force jump follows a force plateau (loading rate close to zero), indicating the formation of membrane tethers. The retraction curve also provides information about the adhesion energy, which is the work required to detach the cancer cell. It is equal to the area under the retraction curve. This includes the work necessary to stretch the cells as well as the work required to break the molecular bonds (Fig. 1 D). All of these parameters (detachment force, rupture force, and adhesion energy) were obtained from the force curve using the built-in software (JPK Instruments), and the rupture events were identified using the protocol described in Kerssemakers et al. (40).
Latrunculin-A treatment
rICAM-1 substrates were prepared and J82 cells were attached to a functionalized cantilever as described above. Then, 1.5 mL of complete RPMI medium with Latrunculin-A (Lat-A; Sigma-Aldrich) was added into the petri dish to reach a final concentration of 0.2 μM. The cantilever with an attached J82 cell was rested for 15 min, allowing Lat-A to disturb the actin cytoskeleton. For these experiments, the parameters used for AFM were modified with a compression force of 2 nN instead of 500 pN, to obtain enough rupture events. Both the Lat-A experiments and control experiments were performed using these parameters. Constant height and closed-loop modes were used to compensate for cell deformation when applying Lat-A.
Data analysis
Considering that our data are comprised of rupture forces coming from different receptor-ligand interactions, a multicomponent mixture model is necessary to separate the contributions of the different receptors. To identify and separate the different subpopulations from the overall histograms, we used a Gaussian mixture model (GMM) analysis. In our study we used trimodal GMM for HUVECs/J82-control and bimodal GMM for data obtained while blocking MUC1 or CD43 to analyze the contributions of the different receptors. We used the built-in function fitgmdist from MATLAB (The MathWorks, Natick, MA) to identify the subpopulations from our data. This function uses maximum-likelihood estimations to find the parameters that best fit the experimental data, and provides the overall probability density function and subcomponents of each distribution.
All of the data for adhesion assays were generated by at least three independent experiments performed in triplicates. The data are reported as mean with standard error of the mean (mean ± SE) as the error bar. The significance of the data was calculated by one-way analysis of variance using GraphPad Prism (GraphPad Software, San Diego, CA). For each set of conditions, at least three independent AFM experiments were performed on three different days (overall, three cancer cells and 80–90 HUVECs). The data are reported as mean ± SE. Statistical analyses of the AFM data were performed using the generalized linear mixed model (GLMM) in the R software (2.14 release). With one cancer cell attached to the cantilever, ∼30 F-d curves were obtained on 30 different ECs. The data obtained from the F-d curves were independent in terms of ECs and dependent in terms of cancer cells. The AFM experiments performed were neither completely independent nor dependent, so to account for the heterogeneous data in the AFM experiments, we used a GLMM. Differences between the parameters calculated on untreated cells or while blocking MUC1, CD43, or both were tested using the mixed function of the apex package in the R software.
Results
Expression of MUC1 and CD43 by BCs
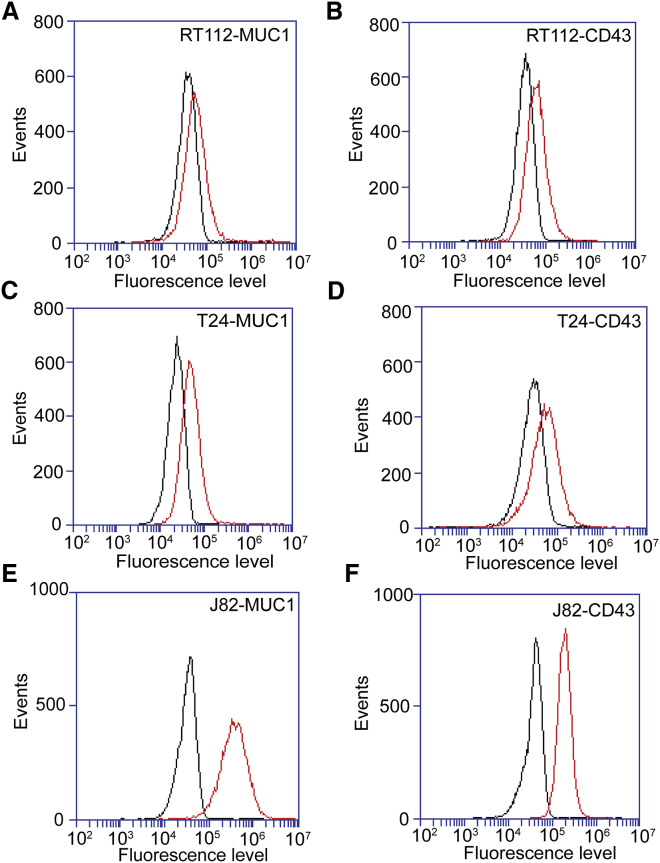
BCs (RT112, T24, and J82) were analyzed for the expression of MUC1 and CD43 by flow cytometry using MUC1 mAb E29 and CD43 mAb L10. The scatter plot obtained for MUC1 expression on J82 BCs as compared with the control is shown on Fig. S1. Control IgG was used to detect the background level (Fig. 2, black curves) in the measurements. As shown in Fig. 2, MUC1 and CD43 (red curves) were expressed by these cell lines at different levels. Weak expression of MUC1 and CD43 was observed on RT112 (Fig. 2, A and B) and T24 (Fig. 2, C and D), whereas J82 showed good expression of MUC1 and CD43 (Fig. 2, E and F) as compared with the other cell lines.
Figure 2.
Flow-cytometry analyses of MUC1 and CD43 expression in BCs. (A–F) Expression levels of MUC1 and CD43 (red curve) as determined by flow-cytometry analysis in comparison with an irrelevant antibody (black curve): RT112 cells (A and B), T24 cells (C and D), and J82 cells (E and F). To see this figure in color, go online.
MUC1 and CD43 mediate BC-EC adhesion
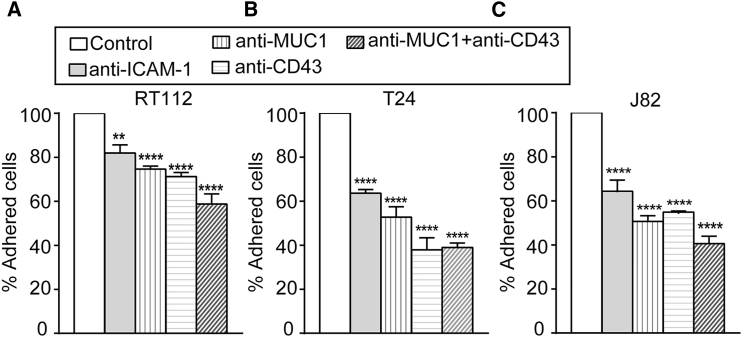
As demonstrated above, BCs express MUC1 and CD43, which are ligands for ICAM-1 (19, 20). To ascertain whether this expression mediates the adhesion of BCs to ECs via interaction with ICAM-1, adhesion assays were performed. mAbs were used to inhibit ICAM-1 expressed on ECs and MUC1 or CD43 expressed on BCs (RT112, T24, and J82) before the adhesion assay. The percentage of adhesion after blocking ICAM-1 on ECs, and MUC1, CD43, or MUC1+CD43 on BCs was calculated and compared with that obtained using a control antibody (mouse IgG). The percentage of cells that adhered in control versus the total number of cells added was 75%, 55%, and 66%, respectively, for RT112, T24, and J82. We normalized the control to 100% and calculated the inhibition when receptors were blocked on different cell lines. Blocking ICAM-1 on ECs showed an ∼36% decrease in adhesion for T24 and J82 (Fig. 3, B and C), and an ∼18% decrease in adhesion for RT112 (Fig. 3 A). Likewise, blocking MUC1 or CD43 on T24 and J82 showed an ∼50% decrease in adhesion (Fig. 3, B and C), and RT112 showed an ∼25% decrease in adhesion (Fig. 3 A). Blocking MUC1+CD43 induced an additional decrease in adhesion compared with blocking them separately in all 3 BCs (Fig. 3, A–C). The decrease in adhesion when blocking ICAM-1, MUC1, or CD43 was 50% smaller in RT112 as compared with T24 and J82. These results indicate that BC-EC adhesion is mainly mediated by the interactions of MUC1 and CD43 with endothelial ICAM-1 for invasive cells (T24 and J82). In contrast, RT112 adhesion on ECs is less dependent on ICAM-1.
Figure 3.
Quantification of BC-EC adhesion. (A–C) In adhesion assays, we quantified the percentage (mean ± SE) of 3 BCs (RT112 (A), T24 (B), and J82 (C)) adhering to ECs while blocking ICAM-1 on ECs and blocking MUC1, CD43, and MUC1+CD43 on cancer cells. One-way analysis of variance was performed to determine significance with respect to the control; ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01.
Effect of blocking MUC1 and CD43 as measured by SCFS
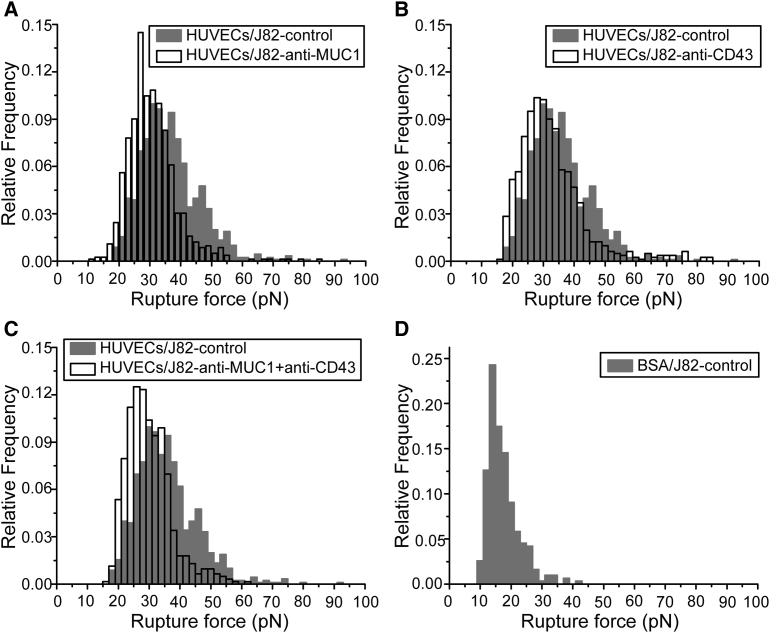
To measure the adhesion forces involved in BC-EC adhesion, we performed SCFS. We used J82 cells for our AFM experiments because they express a higher level of MUC1 and CD43 as compared with two other cell lines (see Fig. 2). Initially, we quantified the interactions that were not mediated through ICAM-1 (nonspecific interactions or mediated by other receptor-ligand interactions, hereafter called nonspecific interactions) using BSA. A single J82 cell was attached to the functionalized tipless cantilever, put in contact with BSA on the substrate, and then retracted. Force curves were analyzed to identify and measure the forces corresponding to the rupture events. The rupture forces obtained from the force curves are represented on a histogram with a bin size of 2 pN. We selected the best bin size to fit our data using the Freedom-Diaconis rule (41, 42) with the R software. The rupture force histogram (Fig. 4 D) revealed that the nonspecific interactions were mediated by a range of rupture forces <30 pN. AFM experiments were then performed using a HUVEC monolayer as the substrate and a J82 cell attached to the cantilever. Force curves were analyzed and a rupture force histogram for nontreated cells (control) was obtained. Similarly, AFM measurements were performed by attaching a J82 cell blocked with antibodies specific for MUC1, CD43, or MUC1+CD43, and rupture force histograms for anti-MUC1, anti-CD43, and anti-MUC1+anti-CD43 were obtained.
Figure 4.
SCFS analysis of BC-EC adhesion. Force histograms showing the distribution of rupture events for adhesion of the HUVEC monolayer with J82 BCs in different conditions were obtained from force curves (applied force 500 pN, time of contact 10 s, velocity 5 μm/s). (A–C) Histograms obtained while blocking MUC1 (A), CD43 (B), and MUC1+CD43 (C) on J82 cells (white histogram) were compared with the control without antibody (gray histogram). (D) The rupture force histogram for nonspecific interactions was obtained with the use of a BSA-coated substrate.
The effect of blocking the receptors involved in the interaction (MUC1, CD43, or both) was quantified by comparing their force distribution with that of the control. While blocking MUC1, the force histogram showed a significant decrease in the number of larger rupture force events as compared with the control (Fig. 4 A). This decrease in rupture events was quantified by considering the rupture events above (>36 pN), since the events <36 pN were found to be unresponsive when MUC1 or CD43 was blocked (Table S1). The number of force curves obtained under each condition is represented by N, and the number of rupture events (>36 pN) obtained from the force curves is represented by n (Table 1). Analysis of the total number of rupture events showed that the J82 control (4.8 events per curve) had almost 2.7 times more events compared with the case after MUC1 was blocked (1.8 events per curve). The inhibition of adhesion due to blocking of MUC1 was quantified as ∼64% (Table 1). Likewise, blocking CD43 (2.8 events per curve) showed 1.7 times fewer events compared with the control (Fig. 4 B), with ∼42% normalized inhibition (Table 1). Blocking MUC1+CD43 (1.3 events per curve) showed 3.6 times fewer events compared with the control (Fig. 4 C), and inhibition increased to ∼72% (Table 1).
Table 1.
Analysis of Rupture Force Distribution
| Substrate | J82 Cell Condition | % Inhibition Compared with Control | N | n | Mean Number of Rupture Events per Curve (M = n/N) |
|---|---|---|---|---|---|
| HUVECs | control | 88 | 426 | 4.8 ± 0.9 | |
| anti-MUC1 | 63.9 | 91 | 159 | 1.8 ± 0.7 | |
| anti-CD43 | 42.2 | 89 | 249 | 2.8 ± 1.0 | |
| anti-MUC1+anti-CD43 | 72.2 | 90 | 121 | 1.3 ± 0.3 | |
| rICAM-1 | Control | 79 | 217 | 2.8 ± 0.9 | |
| anti-MUC1 | 49.1 | 70 | 98 | 1.4 ± 0.1 | |
| anti-CD43 | 44. 4 | 86 | 132 | 1.5 ± 0.3 | |
| anti-MUC1+anti-CD43 | 68.4 | 85 | 74 | 0.9 ± 0.1 |
The % inhibition in number of rupture events while blocking different receptors involved in the interaction was quantified by comparison with the control for both HUVECs and rICAM-1 as the substrate. N represents the number of force curves, n is the total number of rupture events >36 pN when using HUVECs as the substrate or >30 pN when using rICAM-1 as the substrate, and M represents the mean rupture events per curve. The % inhibition obtained by blocking a specific receptor was quantified using the formula [1 − (MAb/Mcont)] × 100. MAb represents the mean number of rupture events obtained while blocking MUC1, CD43, and MUC1+CD43 using specific antibodies, and Mcont represents the mean number of rupture events for the control.
ICAM-1 mediates the interaction of J82 cells with HUVECs
To study the interactions of ICAM-1 alone with BC ligands (MUC1 and CD43), we used an rICAM-1 protein-coated substrate instead of a HUVEC monolayer. SCFS experiments were performed with a J82 cell attached to the cantilever and rICAM-1 adsorbed on the substrate. Force histograms obtained while blocking MUC1, CD43, or MUC1+CD43 were compared with that obtained for the control (Fig. S2), and inhibition was quantified (Table 1) by considering rupture events >30 pN, since events of <30 pN were found to be unresponsive when MUC1 and CD43 were blocked (Table S1). Blocking MUC1 (1.4 events per curve) or CD43 (1.5 events per curve) as compared with the control (2.8 events per curve) showed an inhibition of ∼49% and ∼44%, respectively (Table 1). Blocking both MUC1 and CD43 (0.9 events per curve) showed an increase in normalized inhibition by up to ∼68% (Table 1). The inhibition percentages obtained by blocking MUC1, CD43, or MUC1+CD43 were found similar to those obtained using rICAM-1 and a HUVEC monolayer as the substrate. These results suggest that under these conditions, the interaction of J82 cancer cells with ECs is mainly mediated by ICAM-1 expressed on ECs, in agreement with the results obtained from adhesion assays and our previous work (6).
Force ranges corresponding to MUC1 and CD43 during their interaction with ICAM-1
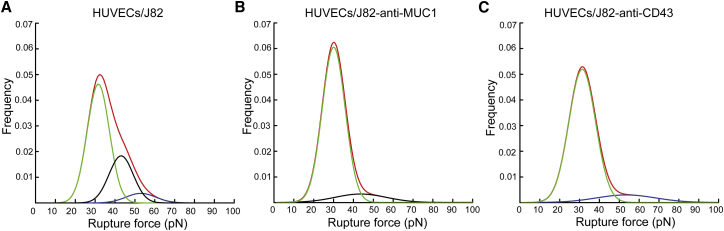
Given the strong inhibition obtained by blocking both MUC1 and CD43, we can consider that the rupture forces obtained are mainly due to the interaction between ICAM-1 (on ECs) and MUC1 and CD43 (on BCs); hence, we can expect three main peaks corresponding to 1) a nonspecific interaction and other receptor-ligand interactions, 2) interaction of ICAM-1 with CD43, and 3) interaction of ICAM-1 with MUC1. To investigate the force ranges corresponding to MUC1 and CD43 during their interaction with ICAM-1, we analyzed our data using a GMM function. We used a common approach based on qualitative assessment and fit the force histogram by changing the number of Gaussians to observe which model best fit our data (43, 44). Fig. S3 shows that the fit obtained using three Gaussians (adjusted R-square 0.981) represents our data better than two Gaussians (adjusted R-square 0.974) on the HUVECs/J82 interaction force histogram. These findings from the qualitative analysis also go along with our initial assumptions that the force histogram obtained in the control would likely have three subpopulations. First, we used a trimodal GMM function in our control (HUVECs/J82 cell) to identify three Gaussian peaks. The GMM analysis showed the overall probability distribution (red) and three distinctive peaks (Fig. 5 A) with mean rupture forces of ∼31, ∼41, and ∼50 pN (Table 2). Specific force ranges corresponding to MUC1 and CD43 interactions were obtained by analyzing the force histogram obtained while blocking MUC1 or CD43. When blocking MUC1, we considered that most of the specific interactions involving MUC1 were suppressed, so we analyzed the data using bimodal GMM to identify two Gaussian peaks (Fig. 5 B). The results showed a first peak (green) with a mean rupture force of ∼29 pN corresponding to a nonspecific interaction, and a second peak (black) with a mean rupture force of ∼43 pN corresponding to the interaction of ICAM-1 with CD43 (Table 2). Similarly, considering that most of the specific interactions involving CD43 were suppressed while CD43 was blocked, we analyzed the data to identify two Gaussian peaks (Fig. 5 C). This result also showed a first peak (green) due to a nonspecific interaction at ∼31 pN and a second peak (blue) with a mean rupture force of ∼53 pN corresponding to the interaction of ICAM-1 with MUC1 (Table 2). This GMM analysis demonstrates that ICAM-1 interacts with MUC1 and CD43 with distinct force ranges.
Figure 5.
MUC1 and CD43 expressed on BCs interact with ECs with different force ranges. (A) GMM function analysis on the control, showing three different subpopulations: nonspecific interactions (green) and interactions of MUC1 (blue) and CD43 (black). (B) After blocking of MUC1, showing two subpopulations: nonspecific interactions (green) and interaction of CD43 (black). (C) After blocking of CD43, showing two subpopulations: nonspecific interactions (green) and interaction of MUC1 (blue). To see this figure in color, go online.
Table 2.
Force Range Obtained from the GMM Analysis of SCFS Data for the Interaction of BCs with ECs
| Substrate | J82 Cell Condition | Peak 1 (Green) |
Peak 2 (Black) |
Peak 3 (Blue) |
|---|---|---|---|---|
| Mean ± SE | Mean ± SE | Mean ± SE | ||
| HUVECs | Control | 31.0 ± 0.25 | 40.9 ± 0.34 | 50.4 ± 0.84 |
| anti-MUC1 | 29.3 ± 0.21 | 42.9 ± 1.42 | ||
| anti-CD43 | 31.1 ± 0.25 | 53.3 ± 1.6 |
GMM analysis revealed that the interaction of CD43 is mediated by a mean rupture force of ∼43 pN and the interaction of MUC1 is mediated by a mean rupture force of ∼53 pN. Colors indicated for peaks 1, 2, and 3 refer to the colors used in Fig. 5.
MUC1 and CD43 attach differently to the cytoskeleton
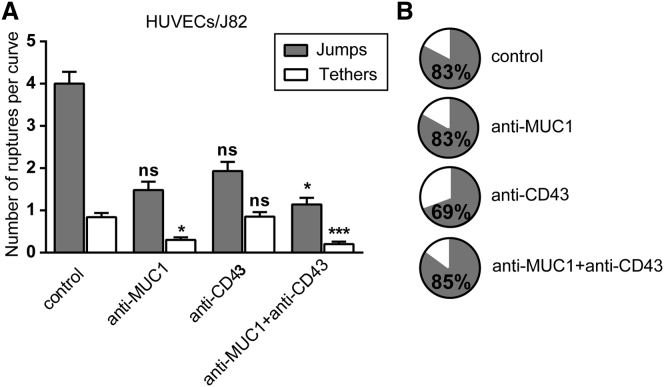
Jumps and tethers observed in the force curves can provide information about whether or not the receptor is connected to the cytoskeleton. Therefore, we determined the number of jumps and tethers within the force curve under the different conditions. We considered rupture events followed by the force plateau with a distance higher than 2 μm as tethers in our analysis (45, 46). To obtain comparable numbers for different conditions, we obtained the number of jumps or tethers per curve by dividing the total number of jumps or tethers (>36 pN) by the number of curves for each condition.
The data obtained from HUVECs/J82 are shown in Fig. 6. The analysis of the number of tethers for different conditions revealed some interesting information (Fig. 6 A). When blocking MUC1 and MUC1+CD43, we observed a significant decrease (∼64% and ∼76%) in the number of tethers. This result suggests that most MUC1-mediated interactions may occur through tethers. Furthermore, blocking CD43 did not change the number of tethers as compared with the control (Fig. 6 A), in agreement with our early inference. These results strongly suggest that MUC1 on cancer cells is weakly connected to the cytoskeleton. The pie charts in Fig. 6 B represent the relative presence of jumps and tethers for the different conditions. We observed the same proportion of jumps when MUC1 was blocked (∼83%) and in the control (∼83%) (Fig. 6 B). Conversely, when CD43 was blocked, the amount of jumps decreased (∼70%), indicating that the interaction of CD43 with ICAM-1 may correspond to jumps, and that CD43 is closely connected to the cytoskeleton.
Figure 6.
MUC1 and CD43 interact with ICAM-1 through tethers and jumps. (A) Histogram showing the (mean ± SE) number and type of ruptures (jumps and tethers >36 pN) while blocking the receptors involved in the interaction, using HUVECs as the substrate. GLMM-R software was used to determine significance with respect to the control; ∗∗∗p < 0.001, ∗p < 0.05, and n.s. p > 0.5. (B) Pie charts showing the percentage of jumps and tethers for each condition. The percentage of jumps is indicated.
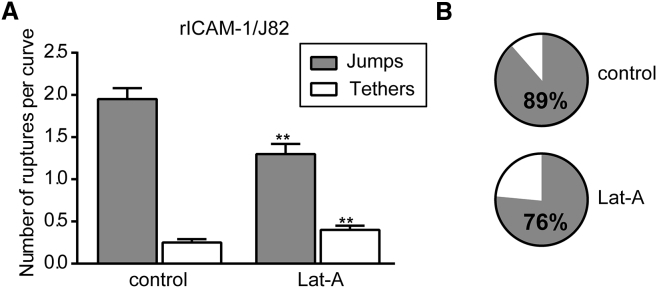
Effect of Lat-A treatment on jumps and tethers
Tethers were separated considering a force plateau of >2 μm (as described above), and the mean number of jumps and tethers for the control and after Lat-A treatment was obtained by analyzing rupture events >30 pN (Fig. 7). Fig. 7 A shows an ∼63% increase in tethers after Lat-A treatment as compared with the control. The effect on jumps was smaller, with an ∼34% decrease after Lat-A treatment. This demonstrates that disruption of the actin cytoskeleton by Lat-A effectively increases the occurrence of tethers while decreasing, although to a lesser extent, the number of jumps. The pie chart in Fig. 7 B also clearly shows the increased proportion of tethers after Lat-A treatment.
Figure 7.
Effect of Lat-A on the number of tethers. (A) Histogram showing the number (mean ± SE) and types of ruptures (jumps and tethers >30 pN) for control or after Lat-A treatment. GLMM was performed to check the significance with respect to control. ∗∗p ≤ 0.01. (B) Pie charts showing the proportion of jumps and tethers for both conditions. The percentage of jumps is indicated.
We performed a GMM analysis on the data obtained after separating the jumps and tethers and considering rupture events >30 pN (thereby removing peak 1 corresponding to the nonspecific interactions). The data from the control and Lat-A treatment were then fitted with two Gaussians (corresponding to peak 2 and peak 3 in Fig. 5), and the relative area obtained for the two peaks is shown in Table 3. As demonstrated above, peak 2 and peak 3 should mostly correspond to the interactions of CD43 with ICAM-1, and MUC1 with ICAM-1, respectively. When we looked at the area of the peaks obtained when considering jumps alone, we observed a decrease in the area of peak 2 and in the ratio after treatment with Lat-A as compared with the control (Table 3). This result indicates that the link between CD43 molecules and the actin cytoskeleton was disturbed after Lat-A treatment, leading to a decrease in the number of jumps. By contrast, when we analyzed the area of tethers, we did not observe any change in the area of peak 2 or in the ratio after Lat-A treatment as compared with the control (Table 3), confirming the fact that the events we are considering as tethers are not connected to the cytoskeleton.
Table 3.
Area Obtained from the GMM Analysis of SCFS Data
| Substrate | J82 Cell Condition | Jumps |
Tethers |
||||
|---|---|---|---|---|---|---|---|
| Peak 2 (Area) | Peak 3 (Area) | Ratio |
Peak 2 (Area) | Peak 3 (Area) | Ratio |
||
| Peak 2/Peak 3 | Peak 2/Peak 3 | ||||||
| rICAM-1 | control | 0.61 | 0.39 | 1.6 | 0.41 | 0.59 | 0.7 |
| Lat-A | 0.48 | 0.52 | 0.9 | 0.42 | 0.58 | 0.7 | |
GMM analysis showed that the number of jumps related to the interaction of CD43 with ICAM-1 decreased after Lat-A treatment as compared with the control.
Discussion
The mechanisms and key molecules involved in the adhesion of leukocytes to the endothelium have been investigated in great detail (18, 33, 47, 48), but only a few works are available regarding the adhesion forces and proteins involved in cancer cell-EC adhesion (11, 45, 49). Previously, we identified ICAM-1 as a key molecule for BC adhesion to the endothelium (16), and investigated the role of ICAM-1 in BC adhesion using SCFS (6). These results led us to hypothesize that MUC1 and CD43 are ligands expressed by BCs to mediate their adhesion with endothelial ICAM-1. In this work, we characterized the influence of MUC1 and CD43 on BC-EC adhesion in detail using adhesion assays and SCFS.
As a model for ECs, we used HUVECs (derived from the veins of human umbilical cords), even though it is known that metastasis occurs mainly in the microcirculation. We think that this should not be a problem, as we are interested in the adhesion of cells mediated through ICAM-1, which is expressed similarly on all cell types. In this study, we used three bladder cell lines with different degrees of invasiveness, expressing MUC1 and CD43 at various levels. RT112 and T24 have a weak expression of MUC1. In this work, we were able to detect a weak expression of MUC1 on RT112 and T24. This is in contrast to our previous study (6), in which we found no detectable expression of these two proteins on both cell lines. This might be due to the fact that in the study presented here, instead of C595 mAb we used E29 mAb, which has been shown to bind to several locations in the multiple-repeats domain of MUC1 (50), thus increasing its sensitivity. This hypothesis is supported by the higher MUC1 expression obtained for J82 in this study as compared with the previous one (6). The expression of MUC1 is controversial in the literature, as one study found weak expression in T24 at the mRNA level (51), whereas another study reported no detectable level of MUC1 mRNA in T24 (52). In contrast, J82 cells showed a good expression of MUC1, as observed in previous work (51). MUC1 expression was also reported in other BCs, such as KU7, UMUC2, SCaBER, TCCSUP, and HT-1367 (51, 52). The expression of CD43 has not been studied extensively, but some studies have shown a de novo expression of CD43 in cancer tissues, particularly of the bladder (53, 54).
It has already been shown that MUC1 mediates the adhesion of breast cancer cells to ECs and increases transendothelial migration (55), and that CD43 plays a role in tumor-mesothelial cell adhesion via its interaction with ICAM-1 (36). Our results show that the adhesion of BCs (T24 and J82) on ECs was greatly reduced when blocking ICAM-1, MUC1, or CD43, whereas the adhesion of RT112 to ECs was not much affected by blocking these receptors. Our results indicate that BC-EC adhesion is mainly mediated through ICAM-1 for invasive cells (T24 and J82), as previously reported using AFM (6). The inhibition obtained using antibodies against MUC1 and CD43 was not complete (i.e., additive), and this can be explained by the fact that 1) these antibodies are not able to completely inhibit the interactions (as previously reported (23, 36), and 2) other interactions not involving ICAM-1-CD43 or ICAM-1-MUC1 interactions might be at play. For example, E-selectin can interact with both MUC1 and CD43 (22, 56), and integrins have also been shown to play a role in this process (12). This indicates that the interactions that were not inhibited by these antibodies could be either background or interactions not involving MUC1 or CD43.
SCFS has been very useful for quantifying the adhesive forces involved between cell-cell and cell-surface interactions (29, 57). The adhesion forces involved in the interactions of a leukocyte ligand (LFA-1) with ICAM-1 were studied in detail using this method (17, 58, 59). Recent work by our team showed that ICAM-1 expressed on ECs is a key molecule in mediating adhesion to BCs (6). In this work, we applied SCFS to unravel the differences in force range originating from the interactions of ICAM-1 with either MUC1 or CD43.
SCFS experiments performed by blocking MUC1 or CD43 on BCs showed a significant decrease in the number of rupture events (Table 1), in agreement with our adhesion assays. When we replaced the HUVEC monolayer with rICAM-1 to specifically study the interactions mediated by ICAM-1 with its cancer cell ligands, we found that the decrease in rupture events after inhibition with the antibodies was similar to that obtained with ECs, clearly indicating that the interactions of J82 cells with ECs were mainly mediated by ICAM-1. In contrast, the mean adhesion energy and mean detachment force showed no significant difference when MUC1, CD43, or both were blocked as compared with the control (Fig. S4). This might be due to the fact that these two parameters also depend on cell rheology in a complex manner.
By applying a GMM analysis to data from experiments using blocking antibodies against MUC1 or CD43, we were able to discriminate three different force ranges when BCs interacted with ECs. Nonspecific interactions or those that did not involve interactions between ICAM-1 and MUC1 or CD43 could be assigned to a mean rupture force of ∼30 pN, a value in agreement with the data obtained using a BSA-coated substrate (which we found to be the best control), and the interactions of CD43 and MUC1 with ICAM-1 were found to have mean rupture forces of ∼43 and ∼53 pN, respectively. In our previous work (6), the histogram of rupture forces resulting from the interaction of HUVECs and J82 revealed a double Gaussian distribution with mean values of ∼42 and ∼70 pN. When ICAM-1 was blocked on ECs, only one major peak centered at ∼29 pN remained. This value is in close agreement with the 30 pN peak found for nonspecific interactions in the study presented here (i.e., with BSA). On the basis of these results, we propose that CD43 and MUC1 are the two main ligands that interact with ICAM-1 when BCs interact with ECs. The slight difference in peak values can be explained by the use of a higher sampling rate (204.8 Hz in our previous study versus 2048 Hz in this work), leading to an increase in resolution and a better discrimination of very close rupture events such as double bonds (60, 61). In addition, in this work we analyzed a much greater amount of data. The parameters that might affect the mean rupture force, such as the local loading rate and cooperativity, were also investigated. We found that there was no effect of the local loading rate (measured on the F-d curve before rupture) on the mean rupture forces (Fig. S5), and that the cooperativity effects could be neglected due to the short contact time used in our experiments, as already suggested in previous work (29).
The force curves obtained by SCFS also revealed two different types of rupture events. The first type, called jump, is characterized by a linear increase of force before rupture (Fig. 1 D), whereas for the second type, or tether, no force increase appears before the rupture event (i.e., it is a force plateau) (Fig. 1 D). The cytoplasmic domains of MUC1 and CD43 were reported to interact with the actin cytoskeleton through ERM (erzin, radixin, and moesin) protein linkers (62, 63). In accordance with previous studies that considered only the jumps in the force curves to represent the unbinding of adhesive units (29, 61), we analyzed the relative frequency of these events in our different experiments. The rupture forces obtained by considering only jumps showed a force distribution similar to that obtained by considering both jumps and tethers (Fig. S6). The same was true when different receptors (MUC1, CD43, or MUC1+CD43) were blocked (data not shown). The GMM analysis of the data obtained from jumps also showed a similar force range for nonspecific interactions and interactions of MUC1 and CD43 with ICAM-1 (Table S2). It has been shown that interactions that give rise to a force increase followed by jumps correspond to receptors anchored to the cytoskeleton, whereas force plateaus or tethers that are followed by rupture originate from receptors that are weakly or not attached to the actin cell cortex (28, 61). This means that the relative presence of jumps and tethers in the rupture events can provide information regarding how tightly receptors are connected to the cytoskeleton (46, 64, 65, 66). In our case, a detailed analysis of the number of jumps and tethers showed that MUC1 on J82 BCs seems to be weakly connected to the cytoskeleton and that its interactions are mainly mediated through tethers. On the other hand, CD43 could be more closely linked to the cytoskeleton and associated with the early jumps obtained on the force curve.
The association of these ligands with the cytoskeleton was further studied after disruption of the actin cytoskeleton using Lat-A treatment on cancer cells (66, 67). An analysis of the number of jumps and tethers showed a decrease in the number of jumps and an increase in the number of tethers after Lat-A treatment compared with the control (Fig. 7). These results indicate that disruption of the actin cytoskeleton transforms some jumps (bonds connected to the cytoskeleton) to tethers, but probably not all. A GMM analysis was performed to study the influence of Lat-A on the links between the cytoskeleton and the molecules involved (CD43 and MUC1). Our results show that Lat-A treatment decreased the area and ratio of jumps for peak 2, and that some of the CD43 molecules that were connected to the cytoskeleton (and giving rise to jumps) were disrupted after the treatment.
Conclusions
To conclude, we have shown that MUC1 and CD43 are two adhesive proteins that are expressed on BCs at different levels. Through adhesion assays and SCFS, we demonstrated that MUC1 and CD43 interact with endothelial ICAM-1 to mediate the adhesion of BCs to ECs. Combining SCFS measurements and GMM analysis, we showed that MUC1 interactions involved larger force levels than CD43 interactions, and that most of the MUC1 interactions were mediated through tethers. By analyzing the ratio of tethers and jumps that could be attributed to ICAM-1 interactions with either MUC1 or CD43, we concluded that most of the MUC1 molecules present on J82 BCs were weakly connected to the cytoskeleton. The interactions with the cytoskeleton appeared to be stronger for CD43, leading to jumps in the force curves. Experiments using the actin-depolymerizing drug Lat-A confirmed this differential ligand association with the cytoskeleton.
Given the fact that the expression of MUC1 and CD43 has also been reported in many other cancer cells, we believe that our results will help elucidate the mechanisms that occur during early adhesion of cancer cells to the endothelium. It will be interesting to further study the involvement of these interactions in other processes, particularly tumor cell transmigration.
Author Contributions
V.S.R. performed all experiments and the AFM analysis. Statistical analysis on R was performed by A.D. All of the authors designed the experiments. V.S.R., A.D., and C.V. wrote the manuscript.
Acknowledgments
We thank Alexei Grichine for help with the GMM analysis, Yara Abidine for help with the AFM, and our team members for fruitful discussions.
This work was supported ANR grant No. 12-BS09-020-01 (TRANSMIG). The Nanoscience Foundation provided ssupport for the AFM platform. The DYFCOM team at LIPhy is a member of LabeX Tec21 (Investissements d’Avenir, grant agreement No. ANR-11-LABX-0030).
Editor: Christopher Yip.
Footnotes
Six figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30164-9.
Supporting Material
References
- 1.Reymond N., d’Água B.B., Ridley A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 2.Jeon J.S., Zervantonakis I.K., Charest J.L. In vitro model of tumor cell extravasation. PLoS One. 2013;8:e56910. doi: 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strell C., Entschladen F. Extravasation of leukocytes in comparison to tumor cells. Cell Commun. Signal. 2008;6:10–22. doi: 10.1186/1478-811X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bersini S., Jeon J.S., Kamm R.D. In vitro models of the metastatic cascade: from local invasion to extravasation. Drug Discov. Today. 2014;19:735–742. doi: 10.1016/j.drudis.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mierke C.T. Physical break-down of the classical view on cancer cell invasion and metastasis. Eur. J. Cell Biol. 2013;92:89–104. doi: 10.1016/j.ejcb.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Laurent V.M., Duperray A., Verdier C. Atomic force microscopy reveals a role for endothelial cell ICAM-1 expression in bladder cancer cell adherence. PLoS One. 2014;9:e98034. doi: 10.1371/journal.pone.0098034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chotard-Ghodsnia R., Haddad O., Duperray A. Morphological analysis of tumor cell/endothelial cell interactions under shear flow. J. Biomech. 2007;40:335–344. doi: 10.1016/j.jbiomech.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miles F.L., Pruitt F.L., Cooper C.R. Stepping out of the flow: capillary extravasation in cancer metastasis. Clin. Exp. Metastasis. 2008;25:305–324. doi: 10.1007/s10585-007-9098-2. [DOI] [PubMed] [Google Scholar]
- 9.Shenoy A.K., Lu J. Cancer cells remodel themselves and vasculature to overcome the endothelial barrier. Cancer Lett. 2016;380:534–544. doi: 10.1016/j.canlet.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mierke C.T. Role of the endothelium during tumor cell metastasis: is the endothelium a barrier or a promoter for cell invasion and metastasis? J. Biophys. 2008;2008:183516. doi: 10.1155/2008/183516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y., Hoeppner L.H., Mukhopadhyay D. Neuropilin-2 promotes extravasation and metastasis by interacting with endothelial α5 integrin. Cancer Res. 2013;73:4579–4590. doi: 10.1158/0008-5472.CAN-13-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyder C., Gloria-Maercker E., Dittmar T. Role of the β1-integrin subunit in the adhesion, extravasation and migration of T24 human bladder carcinoma cells. Clin. Exp. Metastasis. 2005;22:99–106. doi: 10.1007/s10585-005-4335-z. [DOI] [PubMed] [Google Scholar]
- 13.Klemke M., Weschenfelder T., Samstag Y. High affinity interaction of integrin alpha4beta1 (VLA-4) and vascular cell adhesion molecule 1 (VCAM-1) enhances migration of human melanoma cells across activated endothelial cell layers. J. Cell. Physiol. 2007;212:368–374. doi: 10.1002/jcp.21029. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M., Yanaba K., Sato S. Regulation of local and metastatic host-mediated anti-tumour mechanisms by L-selectin and intercellular adhesion molecule-1. Clin. Exp. Immunol. 2006;143:216–227. doi: 10.1111/j.1365-2249.2005.02989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche Y., Pasquier D., Duperray A. Fibrinogen mediates bladder cancer cell migration in an ICAM-1-dependent pathway. Thromb. Haemost. 2003;89:1089–1097. [PubMed] [Google Scholar]
- 16.Haddad O., Chotard-Ghodsnia R., Duperray A. Tumor cell/endothelial cell tight contact upregulates endothelial adhesion molecule expression mediated by NFkappaB: differential role of the shear stress. Exp. Cell Res. 2010;316:615–626. doi: 10.1016/j.yexcr.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Wojcikiewicz E., Moy V.T. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Chen A., Goligorsky M.S. Atomic force microscopy measurement of leukocyte-endothelial interaction. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H359–H367. doi: 10.1152/ajpheart.00491.2003. [DOI] [PubMed] [Google Scholar]
- 19.Regimbald L.H., Pilarski L.M., Hugh J.C. The breast mucin MUCI as a novel adhesion ligand for endothelial intercellular adhesion molecule 1 in breast cancer. Cancer Res. 1996;56:4244–4249. [PubMed] [Google Scholar]
- 20.Rosenstein Y., Park J.K., Burakoff S.J. CD43, a molecule defective in Wiskott-Aldrich syndrome, binds ICAM-1. Nature. 1991;354:233–235. doi: 10.1038/354233a0. [DOI] [PubMed] [Google Scholar]
- 21.Simms M.S., Hughes O.D., Bishop M.C. MUC1 mucin as a tumour marker in bladder cancer. BJU Int. 1999;84:350–352. doi: 10.1046/j.1464-410x.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 22.Geng Y., Yeh K., King M.R. Three to tango: MUC1 as a ligand for both E-selectin and ICAM-1 in the breast cancer metastatic cascade. Front. Oncol. 2012;2:76. doi: 10.3389/fonc.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi T., Takahashi T., Imai K. MUC1 mucin core protein binds to the domain 1 of ICAM-1. Digestion. 2001;63(Suppl. 1):87–92. doi: 10.1159/000051917. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Rodriguez J., Andersson C.X., Hansson G.C. The leukocyte antigen CD43 is expressed in different cell lines of nonhematopoietic origin. Tumour Biol. 2002;23:193–201. doi: 10.1159/000067252. [DOI] [PubMed] [Google Scholar]
- 25.Tuccillo F.M., Palmieri C., Scala G. Cancer-associated CD43 glycoforms as target of immunotherapy. Mol. Cancer Ther. 2014;13:752–762. doi: 10.1158/1535-7163.MCT-13-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrichs J., Helenius J., Müller D.J. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nat. Protoc. 2010;5:1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 27.Franz C.M., Taubenberger A., Müller D.J. Studying integrin-mediated cell adhesion at the single-molecule level using AFM force spectroscopy. Sci. STKE. 2007;2007:pl5. doi: 10.1126/stke.4062007pl5. [DOI] [PubMed] [Google Scholar]
- 28.Puech P.H., Taubenberger A., Heisenberg C.P. Measuring cell adhesion forces of primary gastrulating cells from zebrafish using atomic force microscopy. J. Cell Sci. 2005;118:4199–4206. doi: 10.1242/jcs.02547. [DOI] [PubMed] [Google Scholar]
- 29.Taubenberger A., Cisneros D.A., Franz C.M. Revealing early steps of α2β1 integrin-mediated adhesion to collagen type I by using single-cell force spectroscopy. Mol. Biol. Cell. 2007;18:1634–1644. doi: 10.1091/mbc.E06-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulchek T.A., Friddle R.W., Noy A. Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc. Natl. Acad. Sci. USA. 2005;102:16638–16643. doi: 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alsteens D., Garcia M.C., Dufrêne Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfreundschuh M., Alsteens D., Müller D.J. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM. Nat. Commun. 2015;6:8857–8863. doi: 10.1038/ncomms9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Wojcikiewicz E.P., Moy V.T. Dynamic adhesion of T lymphocytes to endothelial cells revealed by atomic force microscopy. Exp. Biol. Med. (Maywood) 2006;231:1306–1312. doi: 10.1177/153537020623100804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champelovier P., Simon A., Seigneurin D. Is interferon gamma one key of metastatic potential increase in human bladder carcinoma? Clin. Cancer Res. 2003;9:4562–4569. [PubMed] [Google Scholar]
- 35.Masters J.R.W., Hepburn P.J., Franks L.M. Tissue culture model of transitional cell carcinoma: characterization of twenty-two human urothelial cell lines. Cancer Res. 1986;46:3630–3636. [PubMed] [Google Scholar]
- 36.Ziprin P., Alkhamesi N.A., Darzi A.W. Tumour-expressed CD43 (sialophorin) mediates tumourmesothelial cell adhesion. Biol. Chem. 2004;385:755–761. doi: 10.1515/BC.2004.092. [DOI] [PubMed] [Google Scholar]
- 37.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 38.Schubert R., Strohmeyer N., Müller D.J. Assay for characterizing the recovery of vertebrate cells for adhesion measurements by single-cell force spectroscopy. FEBS Lett. 2014;588:3639–3648. doi: 10.1016/j.febslet.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Benoit M., Gabriel D., Gaub H.E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 40.Kerssemakers J.W.J., Munteanu E.L., Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 41.Bura E., Zhmurov A., Barsegov V. Nonparametric density estimation and optimal bandwidth selection for protein unfolding and unbinding data. J. Chem. Phys. 2009;130:015102. doi: 10.1063/1.3050095. [DOI] [PubMed] [Google Scholar]
- 42.Bizzari A.R., Cannistraro S. Dynamic Force Spectroscopy and Biomolecular Recognition. CRC Press; Boca Raton, FL: 2012. Biological applications of dynamic force spectroscopy; pp. 163–192. [Google Scholar]
- 43.Imoukhuede P.I., Popel A.S. Quantitative fluorescent profiling of VEGFRs reveals tumor cell and endothelial cell heterogeneity in breast cancer xenografts. Cancer Med. 2014;3:225–244. doi: 10.1002/cam4.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weddell J.C., Imoukhuede P.I. Quantitative characterization of cellular membrane-receptor heterogeneity through statistical and computational modeling. PLoS One. 2014;9:e97271. doi: 10.1371/journal.pone.0097271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puech P.H., Poole K., Müller D.J. A new technical approach to quantify cell-cell adhesion forces by AFM. Ultramicroscopy. 2006;106:637–644. doi: 10.1016/j.ultramic.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Tulla M., Helenius J., Heino J. TPA primes alpha2beta1 integrins for cell adhesion. FEBS Lett. 2008;582:3520–3524. doi: 10.1016/j.febslet.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 47.Barreiro O., Yáñez-Mó M., Sanchez-Madrid F. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol. 2002;157:1233–1245. doi: 10.1083/jcb.200112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., Craig S.E., Moy V.T. Molecular basis for the dynamic strength of the integrin alpha4beta1/VCAM-1 interaction. Biophys. J. 2004;87:3470–3478. doi: 10.1529/biophysj.104.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Héroult M., Schaffner F., Augustin H.G. EphB4 promotes site-specific metastatic tumor cell dissemination by interacting with endothelial cell-expressed ephrinB2. Mol. Cancer Res. 2010;8:1297–1309. doi: 10.1158/1541-7786.MCR-09-0453. [DOI] [PubMed] [Google Scholar]
- 50.Karsten U., Serttas N., Goletz S. Binding patterns of DTR-specific antibodies reveal a glycosylation-conditioned tumor-specific epitope of the epithelial mucin (MUC1) Glycobiology. 2004;14:681–692. doi: 10.1093/glycob/cwh090. [DOI] [PubMed] [Google Scholar]
- 51.Fujii T., Shimada K., Konishi N. ALKBH2, a novel AlkB homologue, contributes to human bladder cancer progression by regulating MUC1 expression. Cancer Sci. 2013;104:321–327. doi: 10.1111/cas.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaur S., Momi N., Batra S.K. Altered expression of transmembrane mucins, MUC1 and MUC4, in bladder cancer: pathological implications in diagnosis. PLoS One. 2014;9:e92742. doi: 10.1371/journal.pone.0092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santamaría M., López-beltrán A., Molina I.J. Specific monoclonal antibodies against leukocyte-restricted tumor cells react with nonhematopoietic tumor. Cancer Res. 1996;56:3526–3529. [PubMed] [Google Scholar]
- 54.Hurford M.T., Gujral S., Schwarting R. Extramedullary myeloid cell tumor of the urinary bladder in a patient with myelodysplastic syndrome. Pathol. Res. Pract. 1999;195:699–703. doi: 10.1016/S0344-0338(99)80061-2. discussion 705–706. [DOI] [PubMed] [Google Scholar]
- 55.Rahn J.J., Chow J.W., Hugh J.C. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin. Exp. Metastasis. 2005;22:475–483. doi: 10.1007/s10585-005-3098-x. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto M., Atarashi K., Hirata T. CD43 functions as a ligand for E-Selectin on activated T cells. J. Immunol. 2005;175:8042–8050. doi: 10.4049/jimmunol.175.12.8042. [DOI] [PubMed] [Google Scholar]
- 57.Fierro F.A., Taubenberger A., Illmer T. BCR/ABL expression of myeloid progenitors increases β1-integrin mediated adhesion to stromal cells. J. Mol. Biol. 2008;377:1082–1093. doi: 10.1016/j.jmb.2008.01.085. [DOI] [PubMed] [Google Scholar]
- 58.Wojcikiewicz E.P., Zhang X., Moy V.T. Contributions of molecular binding events and cellular compliance to the modulation of leukocyte adhesion. J. Cell Sci. 2003;116:2531–2539. doi: 10.1242/jcs.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wojcikiewicz E.P., Abdulreda M.H., Moy V.T. Force spectroscopy of LFA-1 and its ligands, ICAM-1 and ICAM-2. Biomacromolecules. 2006;7:3188–3195. doi: 10.1021/bm060559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taninaka A., Hirano Y., Shigekawa H. Force measurement enabling precise analysis by dynamic force spectroscopy. Int. J. Mol. Sci. 2012;13:453–465. doi: 10.3390/ijms13010453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helenius J., Heisenberg C.P., Müller D.J. Single-cell force spectroscopy. J. Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 62.Bennett R., Jr., Järvelä T., Vaheri A. Mucin MUC1 is seen in cell surface protrusions together with ezrin in immunoelectron tomography and is concentrated at tips of filopodial protrusions in MCF-7 breast carcinoma cells. J. Histochem. Cytochem. 2001;49:67–77. doi: 10.1177/002215540104900107. [DOI] [PubMed] [Google Scholar]
- 63.Serrador J.M., Nieto M., Sánchez-Madrid F. CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood. 1998;91:4632–4644. [PubMed] [Google Scholar]
- 64.Friedrichs J., Legate K.R., Benoit M. A practical guide to quantify cell adhesion using single-cell force spectroscopy. Methods. 2013;60:169–178. doi: 10.1016/j.ymeth.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Celik E., Faridi M.H., Gupta V. Agonist leukadherin-1 increases CD11b/CD18-dependent adhesion via membrane tethers. Biophys. J. 2013;105:2517–2527. doi: 10.1016/j.bpj.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sariisik E., Popov C., Benoit M. Decoding cytoskeleton-anchored and non-anchored receptors from single-cell adhesion force data. Biophys. J. 2015;109:1330–1333. doi: 10.1016/j.bpj.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun M., Graham J.S., Grandbois M. Multiple membrane tethers probed by atomic force microscopy. Biophys. J. 2005;89:4320–4329. doi: 10.1529/biophysj.104.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.