Beyond the potent stings of jellyfish, nematocysts have evolved independently in single-celled phytoplankton.
Keywords: Convergent evolution, cnidocyst, extrusome, secretory, secretion, minicollagen, mucocyst, trichocyst, red queen
Abstract
We examine the origin of harpoon-like secretory organelles (nematocysts) in dinoflagellate protists. These ballistic organelles have been hypothesized to be homologous to similarly complex structures in animals (cnidarians); but we show, using structural, functional, and phylogenomic data, that nematocysts evolved independently in both lineages. We also recorded the first high-resolution videos of nematocyst discharge in dinoflagellates. Unexpectedly, our data suggest that different types of dinoflagellate nematocysts use two fundamentally different types of ballistic mechanisms: one type relies on a single pressurized capsule for propulsion, whereas the other type launches 11 to 15 projectiles from an arrangement similar to a Gatling gun. Despite their radical structural differences, these nematocysts share a single origin within dinoflagellates and both potentially use a contraction-based mechanism to generate ballistic force. The diversity of traits in dinoflagellate nematocysts demonstrates a stepwise route by which simple secretory structures diversified to yield elaborate subcellular weaponry.
INTRODUCTION
Planktonic microbes are often viewed as passive food items for larger life-forms. In reality, eukaryotic microbes (for example, dinoflagellates and ciliates) have evolved a range of weapons, from armor and toxins to projectile organelles or “extrusomes” (1). The ballistic action of extrusomes has been known since early light microscopy, when predatory ciliates were seen to capture prey using syringe-like toxicysts. In turn, prey ciliates sometimes broke free of toxicysts by firing volleys of defensive extrusomes (mucocysts and trichocysts). These “arms races” are likely widespread in nature, given that ciliates and dinoflagellates consume up to 60% of primary production in aquatic environments (2) and considering the diversity of extrusomes found throughout the phytoplankton (in groups including cryptophytes, chrysophytes, raphidophytes, and euglenozoans) (1). However, extrusomes are perhaps the least studied of major organelle types.
Here, we used single-cell microscopic and genomic approaches to study ballistic organelles in cultivable (Polykrikos kofoidii; Figs. 1, C to E, and 2) and wild-caught dinoflagellates (Nematodinium sp.; Figs. 1, F to H, and 3). Dinoflagellates have a large arsenal of extrusomes, ranging from simple, ostensibly defensive organelles (for example, mucocysts and trichocysts) to elaborate organelles, for predation (for example, taeniocysts and nematocysts) (1). Cells of the genus Polykrikos capture other dinoflagellates using harpoon-like “nematocysts” (Fig. 1B and movie S1). Polykrikos can ensnare other dinoflagellates and tow them in for ingestion using a long filament (3, 4). The nematocyst capsule is sealed with a hatch-like operculum and contains a coiled tubule and a pointed stylet (5, 6). This constellation of features is shared by the nematocysts in the phylum Cnidaria (7, 8), which is among the earliest diverging predatory animal phyla. Dinoflagellates and cnidarians are very distantly related, so whether the ballistic similarities of these organelles reflect homology or convergent evolution has been a matter of some debate. In particular, it has been suggested that the origin of nematocysts in cnidarians was spurred by the acquisition of genes from dinoflagellates—specifically from groups, such as Symbiodinium, which live endosymbiotically within corals (9–11). To test this hypothesis, we used nematogenic genes identified from the Hydra nematocyst proteome (12) to search a collection of 30 eukaryotic genomes [including two genomes of Symbiodinium (13, 14)] and 120 dinoflagellate transcriptomes, plus a transcriptome that we previously obtained from the nematocyst-bearing dinoflagellate Polykrikos lebouriae (15).
Fig. 1. Diversity and independent origins of extrusomes.
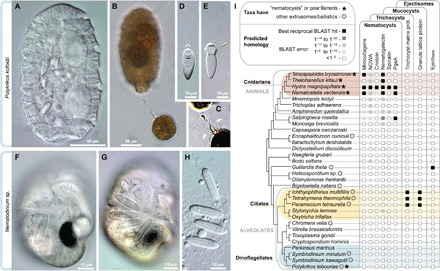
(A to E) Nematocysts in the dinoflagellate P. kofoidii, including a live whole cell (A) and a cell that was preserved in Lugol’s iodide solution while capturing a prey cell of A. tamarense (B). (C) Enhanced contrast image shows the defensive trichocysts deployed by A. tamarense (arrows) in response to attack by P. kofoidii. (D and E) Isolated nematocysts from P. kofoidii, seen as unfired (D) and discharged (E). (F to H) Nematocysts in the dinoflagellate Nematodinium sp., which have an eye-like ocelloid (F). A battery of nematocysts is visible in the live cell (G) and remains intact after cell lysis (H). (I) Genomic distribution of known extrusome proteins (vertical labels) across eukaryotes based on best reciprocal Basic Local Alignment Search Tool (BLAST) hits (black squares), which is a common predictor of protein homology. Taxa are listed within an established phylogenetic framework (38, 39).
Fig. 2. Reconstruction of the nematocysts in the dinoflagellate P. kofoidii.
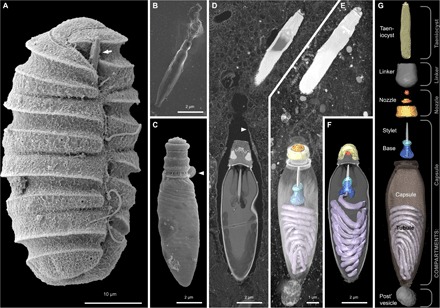
(A) SEM micrograph of a cell of P. kofoidii, including an armed taeniocyst (arrow) in the apical region that first contacts prey. (B) SEM micrograph of an isolated taeniocyst that has discharged its amorphous contents. (C) SEM micrograph of an isolated nematocyst that has become arrested very early in discharge; arrowhead, operculum. (D) FIB-SEM section of taeniocyst and nematocyst enclosed by a membranous chute (arrowhead) and (E) maximum intensity projection of the same region seen slightly from above and below (F). (G) Virtual dissection of the nematocyst-taeniocyst complex. Brackets indicate the membrane-bound compartments in which those components are grouped during early development. Later in development, compartments fuse to form the chute.
Fig. 3. Nematocyst discharge in Polykrikos.
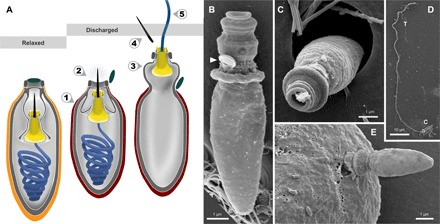
(A) The putative ballistic sequence of nematocyst discharge is as follows: (1) striated capsule contracts; (2) internal pressure forces stylet to pierce capsule and open operculum (dark green); (3) part of the capsule everts; (4) stylet exits through the nozzle and detaches; and (5) mucilaginous tubule (blue) uncoils and is projected through the stylet base (yellow). Orange, relaxed capsule wall; red, contracted capsule wall. (B to E) SEM micrographs of isolated discharged nematocysts (B to D) and a nematocyst piercing a prey cell (E). Arrowhead, operculum; C, capsule; T, tubule.
RESULTS
Contrary to the symbiogenic hypothesis, we found that core nematogenic proteins were restricted to cnidarians. Critically, the enzyme responsible for the osmotic propellant of cnidarian nematocysts [poly-γ-glutamate synthase, which is encoded by PgsAA (16)] was not found in any extrusome-bearing microbes, suggesting that dinoflagellates and other groups use different propellants to generate ballistic force. Animal PgsAA is derived from bacteria via lateral gene transfer (17). Other components of cnidarian nematocysts [for example, minicollagens (18), spinalin (19), and cnidoin (20)] appear to have arisen after multicellularity, with no evident gene donors or recipients among 130 transcriptomic and genomic data sets from dinoflagellates. This suggests that the similarities between the nematocysts of dinoflagellates and cnidarians reflect convergent evolution. Moreover, no homology was detected between dinoflagellate extrusome proteins and the known extrusome proteins from other microbial eukaryotes (for example, ciliates and cryptophytes). How then did dinoflagellates evolve these sophisticated ballistic organelles?
Because our current understanding of dinoflagellate nematocysts is limited to two-dimensional (2D) imagery, we assembled 3D reconstructions via focused ion beam scanning electron microscopy (FIB-SEM) on single cells of P. kofoidii. FIB-SEM allowed us to reconstruct nearly an entire cell of P. kofoidii in 3D, which contained four nematocysts, each connected to another organelle known as a taeniocyst (Fig. 2 and movie S2). Our renderings show that the similarity of dinoflagellate and cnidarian nematocysts has been overstated. While the capsule in cnidarians is topped simply by a hatch-like opening (the operculum) that becomes “uncorked” to allow firing (fig. S1), we found an additional—and far more elaborate—structure in Polykrikos, which lies beneath the operculum. This structure is composed of three concentric rings and appears to have a nozzle-like function.
By capturing the first high-resolution videos of nematocyst discharge (movies S2 and S3), as well as the SEM micrographs of nematocysts arrested at different stages of firing (fig. S2), we were able to make functional inferences about the ballistic mechanism of these complex organelles (Fig. 3). First, the coiled tubule must exit the capsule, which (unlike in cnidarians) has no openings (Figs. 2F and 3). Therefore, the role of the stylet is not only to pierce the prey but also to puncture first the capsule from within, thereby liberating the coiled tubule (Fig. 3). The tubule must pass through two concentric rings (in the novel “stylet base”), then through the center of the nozzle. As the tubule exits through this passage, it forces the operculum open and then uncoils. Once fired, the ballistic tubule gradually dissolves (fig. S2, M to Q). Interactions between Polykrikos and prey dinoflagellates reveal that the tubule does not function as the tow filament, as we initially suspected. Rather, the tubule discharges distally—toward the prey—and is perhaps intended to puncture it (Figs. 1B and 3E and movie S1). The tow filament, by contrast, is on the proximal end of the nematocyst and likely originates from the posterior vesicle.
Positioned distally to each nematocyst is a previously described organelle—the taeniocyst—that emerges from a finger-like projection near the top of each Polykrikos cell (Fig. 2A). Although the taeniocyst makes first contact with the prey, its functions are unclear. We provide evidence that taeniocysts are also ballistic structures. On five occasions, we observed that taeniocysts violently discharge when isolated from the cell (movie S5) by launching their contents through an apical channel (Fig. 2B). The taeniocyst and nematocyst seem to work in tandem, with (i) the taeniocyst intially adhering to the prey, followed by (ii) discharge of the nematocyst, which punctures the prey, and last (iii), the prey is retrieved using a tow filament. According to observations by Westfall and Bradbury (5), each mature nematocyst-taeniocyst complex resides within a single membrane-bound compartment (for example, the “chute”) (5). Our observations were largely in agreement with the previous study, although we did not observe any vesicular structures in the chute between the nematocyst and taeniocysts and instead found a novel “linker” organelle. This cylindrical structure connected the taeniocyst and the nematocyst (Fig. 2), further indicating that these two extrusomes types are deployed in succession, in a coordinated ballistic response. No comparable arrangement is found in cnidarians.
In summary, the structure and function of nematocysts in Polykrikos appear fundamentally different from—and more complex than—those in cnidarians. Therefore, to understand how these ballistics evolved, we examined the diversity of nematocysts in other dinoflagellates. We performed FIB-SEM on a second dinoflagellate, Nematodinium sp., a rare genus known from other ultrastructural studies to have a different nematocyst arrangement than in polykrikoids (21, 22). Extrusome features in Nematodinium are so divergent as to question whether dinoflagellate nematocysts are monophyletic. Nematodinium lacks taeniocysts and coiled ballistic tubules—instead, each nematocyst consists of a ring of parallel subcapsules reminiscent of a Gatling gun (21, 22).
We performed FIB-SEM on a single high-pressure frozen Nematodinium cell containing eight nematocysts, of which we fully reconstructed three (Fig. 4C). Our 3D reconstructions confirmed the intricacy of nematocysts in Nematodinium sp. and revealed several novel features (Fig. 4). Although nematocysts of Nematodinium were thought to be capped by an “operculum” made of homogeneous material (21–23), we showed that this was not the case. Instead, the nematocyst is topped by an elegant rosette-like structure, within a membrane-bound apical compartment (Fig. 4L). Beneath this, we found structures previously imaged only by Greuet (which he did not name) (22) and noticed that they contained concentric rings similar to the nozzle in the nematocysts of Polykrikos. Although these putative nozzles were too small for us to internally reconstruct using FIB-SEM, their concentric rings were evident in our transmission electron microscope (TEM) sections (fig. S4C). As in Polykrikos, each nozzle inserts into a capsule that is positioned directly below it and is held in place by gasket-like rings (Fig. 4, K and L).
Fig. 4. Reconstruction of the nematocysts in Nematodinium sp.
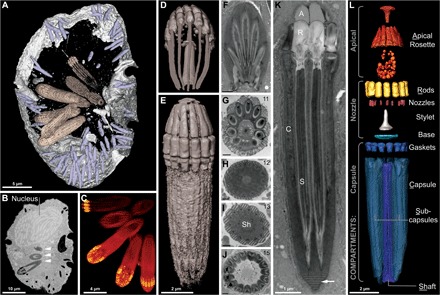
(A to E) Images derived from single-cell FIB-SEM. (A) Partial reconstruction of a cell with nematocysts shown in beige and mucocysts shown in purple. (B) Longitudinal FIB-SEM section through a cell showing the nucleus and nematocysts (arrowheads). (C) 3D maximum intensity projection showing a battery of nematocysts. (D) 3D tilted reconstruction of a nematocyst. (E) Longitudinal reconstruction of a nematocyst. (F to K) Single-cell TEM micrographs of a nematocyst in oblique section (F), cross section (G to J), and longitudinal section (K) showing the striated material (arrow) and variable symmetries across different nematocysts, with the number of subcapsules printed in the upper right corner. (L) A virtual dissection of the nematocyst components. Brackets indicate the membrane-bound compartments bounding the components indicated. (F) to (J) scale bar, 500 nm.
The most compelling case for homology between the nematocysts of Polykrikos and Nematodinium is the identical pattern of striated material within their capsules (Figs. 4K and 5, D to F, and fig. S2, B and C). This shared trait and their development in tiered compartments (figs. S2 to S4), plus the placement of nematocyst-bearing dinoflagellates within a single Gymnodiniales (9) clade (fig. S6), suggest that these nematocysts had a single origin within unarmored dinoflagellates. Therefore, nozzles and stylet bases represent shared derived characters in at least the most recent ancestor of Polykrikos and Nematodinium.
What drives the ballistics in dinoflagellate nematocysts? In cnidarians, an osmotic propellant is synthesized by PgsAA, which was absent in our 130 dinoflagellate data sets. It is possible that dinoflagellates use an as-yet uncharacterized osmotic propellant. An alternate possibility is that they generate force by contracting fibers in the capsule wall to create pressure within the capsule. Our ultrastructural interpretations provide some support for this, because the striated material was found with two different periodicities, which potentially represent a contracted and an uncontracted state (fig. S5, D to F). These are unlikely to be fixation artifacts because they were consistent across specimens prepared by standard chemical fixation and freeze-substitution.
Previously, it was also unclear which part of the nematocyst, if any, in Nematodinium is the actual projectile, because these nematocysts were resistant to induced firing, unlike in Polykrikos. However, several minutes after cell lysis, these nematocysts spontaneously ruptured and expanded, providing insight into their ballistic mechanism. Whereas other nematocyst components remained static, we observed rapid elongation of the subcapsules; within 1 s, the subcapsules extended up to twice their initial lengths (fig. S5, K to N). In TEM, subcapsules appear to release an accordion-like membrane with a diffuse lattice (fig. S5I). This is similar to mucocysts—defensive organelles that share the same size, shape, and lattice-like ultrastructure (fig. S5J)—but any homology between nematocysts and mucocysts remains to be demonstrated.
DISCUSSION
These findings illustrate a new level of hierarchical complexity in organelles, given that each nematocyst is essentially a bouquet of smaller ballistic organelles. The shared similarities between single projectile nematocysts and the multibarreled ones (which are variable, with 11 to 15 barrels; Fig. 4, G to J) suggest how complex ballistics could have arisen from simpler preexisting secretory structures (fig. S7). We suspect that derived features, such as stylets [capable of piercing armor (Fig. 3E)], injectable tubules (potentially to deliver toxins), and pressurized ballistics (seemingly to increase speed), may be counteradaptations to prey armor and defensive ballistics (for example, mucocysts and trichocysts), resulting from an evolutionary arms race. We previously showed that wild-caught cells of Nematodinium sp. had eaten other dinoflagellates—including their discharged trichocysts—having evidently overcome these defenses (1). Illuminating the interplay between dinoflagellate predators and prey will be ecologically relevant, because Polykrikos modulates the populations of important planktonic organisms (24–26), including the armored dinoflagellate Alexandrium tamarense, which is among the most prevalent agents of toxic algal blooms (27, 28). We successfully cultivated P. kofoidii on A. tamarense, as well as a second armored dinoflagellate, Lingulodinium polyedra. In the process, we observed at least one P. kofoidii cell ensnare its prey (Fig. 1B) despite a counterassault from the prey’s trichocysts (Fig. 1C).
Although our hypothesis that a cellular arms race drove the elaboration of extrusomes has yet to be tested, it is clear that obligate predation has become a successful strategy for these dinoflagellates [that is, polykrikoids have lost photosynthesis multiple times (15)]. Despite the misconception that phytoplankton are passive cells, eukaryotic algae have given rise to (and arose from) multiple predatory lineages and, in the process, have independently evolved sophisticated ballistic organelles that exceed those of animals in complexity.
MATERIALS AND METHODS
Proteome mining for nematogenic proteins
Our database consisted of published protein predictions from across a representative group of 30 eukaryotes (Fig. 1I). This included a novel proteome that we predicted from the dinoflagellate P. lebouriae (15), which bears nematocysts. Among the other representatives were public genomes sequenced from extrusome-bearing eukaryotes [Symbiodinium (13, 14), Paramecium, Tetrahymena, Cryptomonas, Hydra, Nematostella, and Thelohanellus], as well as two genomes from taxa with complex secretory structures (Toxoplasma and Cryptosporidium), and two genomes from parasites that invade hosts via ballistic spore cells (Helicosporidium and Encephalitozoon). To compensate for the low coverage of the only available myxozoan genome (Thelohanellus kitauei), we also used a published proteome predicted from a transcriptome of the myxozoan Tetracapsuloides bryosalmonae.
Only 4 of these 31 data sets were from dinoflagellates (few dinoflagellates have been sequenced owing to their massive genome sizes), so to ensure that dinoflagellate proteins were thoroughly searched, we also queried against public predicted proteomes from the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP)—to which we contributed three dinoflagellate transcriptomes (Togula jolla, Protoceratium reticulatum, and Polarella glacialis). In total, the MMETSP contains 120 dinoflagellate transcriptomes from across 44 species and 27 ciliate transcriptomes from across 19 species, many of which have extrusomes (29).
Collection
Cells of Nematodinium sp. and Actiniscus pentasterias were collected off a seaplane station dock in Sidney, British Columbia, Canada (48.652545°N, 123.447200°W) in June 2014, and cells of P. kofoidii and Gymnodinium fasciculatum were collected off a pier in Vancouver, British Columbia, Canada (49.272704°N, 123.187827°W), in July 2015, and off the dock of Friday Harbor Marine Labs, Washington, USA (48.545755, −123.012741) in June 2016 by towing a 20-μm-mesh plankton net through the surface water. Contents were immediately passed through a 150-μm-mesh plankton net to exclude larger organisms, leaving in a fraction that consisted mostly of predatory dinoflagellates. Within 4 hours of collection, cells were visually identified using an inverted light microscope and individually picked and transferred by pulled glass micropipettes into dishes of filtered seawater. Cells of Nematodinium sp. were identified by the presence of both nematocysts and an eye-like ocelloid (Fig. 1F). Cells of P. kofoidii were discerned as binucleate “pseudocolonies” with four girdle flagella, which lacked plastids and have nematocysts (Fig. 1A). Cells of A. pentasterias were identified by the pair of five-pointed silica stars around their nuclei and their apical “docidosome” extrusomes (30). Cells likely belonging to G. fasciculatum were identified by the presence of docidosomes, lack of plastids, and gymnodinoid shape (round, unarmored cells with equatorial girdle).
Cultures
Behavioral observations of P. kofoidii and its prey—L. polyedra and a nonlytic strain of A. tamarense—were made between 2009 and 2010 on the polyclonal culture previously established by Tillmann and Hoppenrath (31). Interactions between predators and prey in well culture plates or culture flasks were viewed through differential interference contrast optics using an Axiovert 200 M inverted light microscope (Zeiss). For higher-resolution imagery, individual cells were transferred using a micropipette onto glass slides and imaged with Zeiss Axioskop 2. Under these conditions, discharge of nematocysts was video recorded using a Sony DSP 3-CCD camera (Sony Deutschland).
Standard chemical fixation of single cells for TEM
Each isolated cell of Nematodinium sp. and P. kofoidii was micropipetted onto a poly-l-lysine–coated slide. Cells were fixed with 2% glutaraldehyde in filtered seawater for 30 min on ice. After two washes in filtered seawater, cells were postfixed in 1% OsO4 for 30 min. Cells were dehydrated through a graded series of ethanol (50, 70, 85, 90, 95, 100, and 100%), infiltrated with acetone-resin mixtures (acetone, 2:1, 1:1, and 1:2; Epon 812 resin), and embedded in Epon 812 resin. Polymerization at 60°C produced a resin-embedded cell affixed to the glass slide. Using a power drill, resin was shaved to a 1-mm block (3), which was removed from the glass slide with a fine razor. The block, containing a single cell, was super glued to a resin stub in the desired orientation for sectioning. Thin sections were produced with a diamond knife, poststained with uranyl acetate and lead citrate, and viewed under a Hitachi H7600 TEM.
High-pressure freezing and freeze substitution of single cells for TEM and FIB-SEM
Using a micropipette, cells of Nematodinium sp. and P. kofoidii were individually transferred into a droplet of filtered seawater. Cells were frozen immediately to minimize fixation artifacts, using a Leica EM HPM100 high-pressure freezer (Leica). Subsequently, freeze substitution was used to remove the aqueous content of the cells and replace it with an acetone solution containing 5% water, 1% osmium tetroxide, and 0.1% uranyl acetate, at −80°C for 48 hours, −20°C for 6 hours, then graded back to 4°C for 13 hours. The prepared samples were washed twice in 100% acetone. Two cells were recovered by micropipette. Each cell was placed on a separate ThermoNox coverslip, where it adhered to a patch of poly-l-lysine. In preparation for FIB-SEM, cells were infiltrated with a 1:1 mix of acetone and Embed 812 resin for 2 hours, then 100% resin overnight. A second ThermoNox coverslip was applied, sandwiching each cell in a thin layer of resin between the coverslips. Resin was polymerized at 65°C for 24 hours. Afterward, the top coverslip was removed with a razorblade to expose the resin face overlying the cell.
Focused ion beam scanning electron microscopy
One cell each of Nematodinium sp. and P. kofoidii was imaged by an FEI Helios NanoLab 650 DualBeam FIB-SEM. The ion beam milled through the cell in 250-nm increments, yielding 169 image slices for Nematodinium and 946 for Polykrikos. Images were aligned as a z stack in Amira 5.5. Features of interest, including the nozzle, stylet, and tubule, were semiautomatically segmented, that is, manually traced in approximately one of every three slices, before automatic interpolation filled in the volumes between the slices, following the manufacter’s instructions. Surfaces of these structures were generated, then smoothed and colorized to produce 3D models of nematocysts and their components. We also produced 3D models without segmentation, as maximum intensity projections of the image stack. Replicates of each organelle were imaged, with six nematocysts present in Nematodinium and four in Polykrikos, across various stages of development.
Confocal microscopy
Cells of P. kofoidii were fixed in 4% paraformaldehyde in filtered seawater for 10 min, then rinsed three times in 0.1 M phosphate-buffered saline (PBS) solution before storage in PBS with 0.05% NaN3 (sodium azide, as a preservative) at 4°C. Fixed cells of P. kofoidii were washed from PBS: NaN3 solution with 3 × 15 min exchanges of 0.1 M PBS, followed by permeabilization in PBT (0.1 M PBS + 0.1% Triton X-100) for 30 min at 4°C. For antibody staining, the cells were incubated in blocking solution (PBT + 1% bovine serum albumin) at 4°C for 30 min and posteriorly incubated at 4°C for 12 hours with a primary mouse anti-tubulin acetylated antibody (Sigma-Aldrich) at a 1:100 concentration in blocking solution. Primary antibody solution was then removed with multiple exchanges of PBT. Specimens were then incubated with a secondary anti-mouse Alexa Fluor 647 antibody (Molecular Probes) at a concentration of 1:100 in blocking solution, at 4°C for 12 hours. Secondary antibodies were removed with multiple exchanges of PBT. Filamentous actin fibers were posteriorly labeled by incubating the cells in a 1:100 dilution of Alexa Fluor 488–conjugated phalloidin (Molecular Probes) in PBT for 1 hour followed by 3 × 15 min exchanges of PBS before imaging by confocal laser-scanning microscopy. Incubations were always performed in the dark while rocking at 4°C in glass well plates.
Molecular phylogenetic analyses
A single cell each of A. pentasterias and G. fasciculatum was individually lysed in a polymerase chain reaction tube and amplified with primers described in Gomez et al. (32) (Nematodinium sp. and polykrikoids were not sequenced because populations from this area have already been barcoded). The 18S and 28S ribosomal DNA (rDNA) sequences were short fragments, but they were included for lack of any other sequence data from these species.
A dinoflagellate phylogeny was estimated using 18S and 28S rDNA sequences, concatenated as 2389 nucleotide alignment, across 50 representative dinoflagellate taxa, including all nematocyst-bearing dinoflagellates. Nucleotides were aligned with MUSCLE (33), and fast-evolving and ambiguously aligned regions were removed using Gblocks 0.91b (34). The nucleotide substitution model (GTRGAMMA) was estimated using the Models package in Mega 6.0.5 (35). A maximum likelihood phylogeny was run with 500 bootstraps in RaxML (36). A second, Bayesian analysis was run for 10,000 generations in MrBayes 3.2 (37), using the high-heating setting of (Nchains = 3).
Supplementary Material
Acknowledgments
This work was made possible by the University of British Columbia technicians, G. Martens, D. Horne, and B. Ross, who carried out high-pressure freezing of the specimens and G. Owen who operated the FIB-SEM imaging of Nematodinium. G.S.G would like to thank E. Gavelis for carrying out plankton tows as well as the faculty and staff at Friday Harbor Laboratories for accommodating us while we collected cells from the plankton. Funding: S.Ö. was funded by the DFG (Oe 416/4-1). Author contributions: G.S.G., P.J.K., and B.S.L. conceived the project and wrote the paper. G.S.G. performed phylogenomic analysis of nematogenic proteins, collected uncultivated dinoflagellates, chemically fixed and imaged cells under TEM, reconstructed FIB-SEM data in 3D, produced illustrations for the paper, and induced nematocyst firing in Polykrikos and Nematodinium, which was imaged through SEM and light microscopy. U.T. cultured Polykrikos along with prey, which he recorded through light microscopic images, film, and SEM. K.C.W. and C.R. imaged the Polykrikos through FIB-SEM, which was funded by S.M. T.H. and S.Ö. provided a proteomic data set, which their laboratory groups obtained from Hydra. All authors participated in the drafting process. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Proteomic data from Hydra is available upon request from T.H. and S.U., and transcriptomic data from Polykrikos lebouriae is available upon request from G.S.G. Other data sets used in phylogenomic analysis were from public genomes, and transcriptomes were from the Marine Microbial Eukaryote Transcriptome Sequencing project, which can be freely downloaded from iMicrobe interactive data commons. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1602552/DC1
fig. S1. A synthesis of fundamental differences between the nematocysts in cnidarians and dinoflagellates.
fig. S2. Ultrastructure and discharge of nematocysts in P. kofoidii.
fig. S3. Nematocyst development in P. kofoidii.
fig. S4. Nematocyst development in Nematodinium sp.
fig. S5. Contractile and projectile traits in the nematocysts of Nematodinium sp.
fig. S6. Molecular phylogeny of dinoflagellates with complex extrusomes.
fig. S7. Model of nematocyst homology and evolution in dinoflagellates.
fig. S8. Cytoskeletal associations with the nematocyst-taeniocyst complex in P. kofoidii.
movie S1. FIB-SEM reconstruction of the nematocyst-taeniocyst complex in P. kofoidii.
movie S2. Discharge of nematocyst isolated from P. kofoidii.
movie S3. Discharge of nematocyst isolated from P. kofoidii.
movie S4. P. kofoidii hunting L. polyedra.
movie S5. Discharge of a taeniocyst isolated from P. kofoidii.
REFERENCES AND NOTES
- 1.Hausmann K., Extrusive organelles in protists. Int. Rev. Cytol. 52, 197–276 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Schmoker C., Hernández-León S., Calbet A., Microzooplankton grazing in the oceans: Impacts, data variability, knowledge gaps and future directions. J. Plankton Res. 35, 691–706 (2013). [Google Scholar]
- 3.Matsuoka K., Cho H.-J., Jacobson D. M., Observations of the feeding behavior and growth rates of the heterotrophic dinoflagellate Polykrikos kofoidii (Polykrikaceae, Dinophyceae). Phys. Chem. Chem. Phys. 39, 82–86 (2000). [Google Scholar]
- 4.Lee M. J., Jeong H. J., Lee K. H., Jang S. H., Kim J. H., Kim K. Y., Mixotrophy in the nematocyst–taeniocyst complex-bearing phototrophic dinoflagellate Polykrikos hartmannii. Harmful Algae 49, 124–134 (2015). [Google Scholar]
- 5.Westfall J. A., Bradbury P. C., The fine structure of the nematocyst taeniocyst complex in polykrikos-kofoidi. J. Protozool. 29, 474–475 (1982). [Google Scholar]
- 6.Hoppenrath M., Yubuki N., Bachvaroff T. R., Leander B. S., Re-classification of Pheopolykrikos hartmannii as Polykrikos (Dinophyceae) based partly on the ultrastructure of complex extrusomes. Eur. J. Protistol. 46, 29–37 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Öezbek S., The cnidarian nematocyst: A miniature extracellular matrix within a secretory vesicle. Protoplasma 248, 635–640 (2011). [DOI] [PubMed] [Google Scholar]
- 8.David C. N., Özbek S., Adamczyk P., Meier S., Pauly B., Chapman J., Hwang J. S., Gojobori T., Holstein T. W., Evolution of complex structures: Minicollagens shape the cnidarian nematocyst. Trends Genet. 24, 431–438 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hoppenrath M., Bachvaroff T. R., Handy S. M., Delwiche C. F., Leander B. S., Molecular phylogeny of ocelloid-bearing dinoflagellates (Warnowiaceae) as inferred from SSU and LSU rDNA sequences. BMC Evol. Biol. 9, 116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. S. Hwang, S. Nagai, S. Hayakawa, Y. Takaku, T. Gojobori, The search for the origin of cnidarian nematocysts in dinoflagellates, in Evolutionary Biology from Concept to Application, P. Pontarotti, Ed. (Springer, 2008), pp. 135–152. [Google Scholar]
- 11.Shostak S., Kolluri V., Symbiogenetic origins of cnidarian cnidocysts. Symbiosis 19, 1–29 (1995). [Google Scholar]
- 12.Balasubramanian P. G., Beckmann A., Warnken U., Schnölzer M., Schüler A., Bornberg-Bauer E., Holstein T. W., Özbek S., Proteome of Hydra nematocyst. J. Biol. Chem. 287, 9672–9681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S., Cheng S., Song B., Zhong X., Lin X., Li W., Li L., Zhang Y., Zhang H., Ji Z., Cai M., Zhuang Y., Shi X., Lin L., Wang L., Wang Z., Liu X., Yu S., Zeng P., Hao H., Zou Q., Chen C., Li Y., Wang Y., Xu C., Meng S., Xu X., Wang J., Yang H., Campbell D. A., Sturm N. R., Dagenais-Bellefeuille S., Morse D., The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., Takeuchi T., Hisata K., Tanaka M., Fujiwara M., Hamada M., Seidi A., Fujie M., Usami T., Goto H., Yamasaki S., Arakaki N., Suzuki Y., Sugano S., Toyoda A., Kuroki Y., Fujiyama A., Medina M., Coffroth M. A., Bhattacharya D., Satoh N., Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Gavelis G. S., White R. A. III, Suttle C. A., Keeling P. J., Leander B. S., Single-cell transcriptomics using spliced leader PCR: Evidence for multiple losses of photosynthesis in polykrikoid dinoflagellates. BMC Genomics 16, 528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber J., Nematocysts (stinging capsules of Cnidaria) as Donnan-potential-dominated osmotic systems. Eur. J. Biochem. 184, 465–476 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N., Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 18, R858–R859 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Adamczyk P., Meier S., Gross T., Hobmayer B., Grzesiek S., Bächinger H. P., Holstein T. W., Özbek S., Minicollagen-15, a novel minicollagen isolated from Hydra, forms tubule structures in nematocysts. J. Mol. Biol. 376, 1008–1020 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Koch A. W., Holstein T. W., Mala C., Kurz E., Engel J., David C. N., Spinalin, a new glycine- and histidine-rich protein in spines of Hydra nematocysts. J. Cell Sci. 111, 1545–1554 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Beckmann A., Xiao S., Müller J. P., Mercadante D., Nüchter T., Kröger N., Langhojer F., Petrich W., Holstein T. W., Benoit M., Gräter F., Özbek S., A fast recoiling silk-like elastomer facilitates nanosecond nematocyst discharge. BMC Biol. 13, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mornin L., Francis D., Fine structure of Nematodinium armatum a naked dinoflagellate. J. Microsc. 6, 759 (1967). [Google Scholar]
- 22.Greuet C., Étude ultrastructurale et évolution des cnidocystes de Nematodinium, Péridinien Warnowiidae Lindemann. Proc. Natl. Acad. Sci. U.S.A. 7, 345–355 (1971). [Google Scholar]
- 23.Greuet C., Hovasse R., About genesis of nematocysts of Polykrikos-schwartzi-Butschli. Proc. Natl. Acad. Sci. U.S.A. 13, 145–149 (1977). [Google Scholar]
- 24.Tillmann U., Interactions between planktonic microalgae and protozoan grazers. J. Eukaryot. Microbiol. 51, 156–168 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Matsuyama Y., Miyamoto M., Kotani Y., Grazing impacts of the heterotrophic dinoflagellate Polykrikos kofoidii on a bloom of Gymnodinium catenatum. Aquat. Microb. Ecol. 17, 91–98 (1999). [Google Scholar]
- 26.Jeong H. J., Park K. H., Kim J. S., Kang H., Kim C. H., Choi H.-J., Kim Y. S., Park J. Y., Park M. G., Reduction in the toxicity of the dinoflagellate Gymnodinium catenatum when fed on by the heterotrophic dinoflagellate Polykrikos kofoidii. Aquat. Microb. Ecol. 31, 307–312 (2003). [Google Scholar]
- 27.John U., Tillmann U., Hülskötter J., Alpermann T. J., Wohlrab S., Van de Waal D. B., Intraspecific facilitation by allelochemical mediated grazing protection within a toxigenic dinoflagellate population. Proc. Biol. Sci. 282, 20141268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wohlrab S., Tillmann U., Cembella A., John U., Trait changes induced by species interactions in two phenotypically distinct strains of a marine dinoflagellate. ISME J. 10, 2658–2668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Zhao X., Liu X., Warren A., Zhao F., Miao M., Phylogenomics of non-model ciliates based on transcriptomic analyses. Protein Cell 6, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen G., Light and electron microscopical observations of the dinoflagellate Actiniscus pentasterias (Dinophyceae). J. Phycol. 29, 486–499 (1993). [Google Scholar]
- 31.Tillmann U., Hoppenrath M., Life cycle of the pseudocolonial dinoflagellate Polykrikos kofoidii (Gymnodiniales, Dinoflagellata). J. Phycol. 49, 298–317 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Gómez F., Moreira D., López-García P., Molecular phylogeny of noctilucoid dinoflagellates (Noctilucales, Dinophyceae). Protist 161, 466–478 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Ronquist F., Huelsenbeck J. P., MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Burki F., Inagaki Y., Bråte J., Archibald J. M., Keeling P. J., Cavalier-Smith T., Sakaguchi M., Hashimoto T., Horak A., Kumar S., Klaveness D., Jakobsen K. S., Pawlowski J., Shalchian-Tabrizi K., Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol. Evol. 1, 231–238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janouskovec J., Tikhonenkov D. V., Burki F., Howe A. T., Kolísko M., Mylnikov A. P., Keeling P. J., Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. U.S.A. 112, 10200–10207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holstein T., The morphogenesis of nematocytes in Hydra and Forskalia: An ultrastructural-study. J. Ultrastruct. Res. 75, 276–290 (1981). [DOI] [PubMed] [Google Scholar]
- 41.Lom J., Depuytor P., Observations sur l’ ultrastructure des trophozoites de myxosporidies. C. R. Hebd. Seances Acad. Sci. 260, 2588–2598 (1965). [Google Scholar]
- 42.Vesk M., Lucas I. A. N., The rhabdosome: A new type of organelle in the dinoflagellate Dinophysis. Protoplasma 134, 62–64 (1986). [Google Scholar]
- 43.Kang N. S., Jeong H. J., Moestrup Ø., SHIN W., NAM S. W., Park J. Y., De Salas M. F., Kim K. W., Noh J. H., Description of a new planktonic mixotrophic dinoflagellate Paragymnodinium shiwhaense n. gen., n. sp from the coastal waters off western Korea: Morphology, pigments, and ribosomal DNA gene sequence. J. Eukaryotic Microbiol. 57, 121–144 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Kang N. S., Jeong H. J., Moestrup Ø., Park T. G., Gyrodiniellum shiwhaense n. gen., n. sp., a new planktonic heterotrophic dinoflagellate from the coastal waters of western Korea: Morphology and ribosomal DNA gene sequence. J. Eukaryotic Microbiol. 58, 284–309 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1602552/DC1
fig. S1. A synthesis of fundamental differences between the nematocysts in cnidarians and dinoflagellates.
fig. S2. Ultrastructure and discharge of nematocysts in P. kofoidii.
fig. S3. Nematocyst development in P. kofoidii.
fig. S4. Nematocyst development in Nematodinium sp.
fig. S5. Contractile and projectile traits in the nematocysts of Nematodinium sp.
fig. S6. Molecular phylogeny of dinoflagellates with complex extrusomes.
fig. S7. Model of nematocyst homology and evolution in dinoflagellates.
fig. S8. Cytoskeletal associations with the nematocyst-taeniocyst complex in P. kofoidii.
movie S1. FIB-SEM reconstruction of the nematocyst-taeniocyst complex in P. kofoidii.
movie S2. Discharge of nematocyst isolated from P. kofoidii.
movie S3. Discharge of nematocyst isolated from P. kofoidii.
movie S4. P. kofoidii hunting L. polyedra.
movie S5. Discharge of a taeniocyst isolated from P. kofoidii.


