Cladistic analysis allows inference of the structure of an ancestral steroid that acts as a ligand for the ancestral receptor.
Keywords: estrogens, steroid hormones, nuclear hormone receptors, ligands, Evolution, metabolic pathways, cladistics
Abstract
The origin of ancient ligand/receptor couples is often analyzed via reconstruction of ancient receptors and, when ligands are products of metabolic pathways, they are not supposed to evolve. However, because metabolic pathways are inherited by descent with modification, their structure can be compared using cladistic analysis. Using this approach, we studied the evolution of steroid hormones. We show that side-chain cleavage is common to most vertebrate steroids, whereas aromatization was co-opted for estrogen synthesis from a more ancient pathway. The ancestral products of aromatic activity were aromatized steroids with a side chain, which we named “paraestrols.” We synthesized paraestrol A and show that it effectively binds and activates the ancestral steroid receptor. Our study opens the way to comparative studies of biologically active small molecules.
INTRODUCTION
Reconstructing the past to better understand evolutionary processes is an important goal of evolutionary sciences. This approach relies greatly on actualism, the principle according to which laws and processes are uniform across time (1). Using this principle, paleontologists have reconstructed models of ancient birds, such as Archaeopteryx, and studied their flight abilities (2, 3). More recently, this approach has been used to reconstruct ancestral proteins to reveal historical mutations that have shaped their function through time (4). Unfortunately, we do not have yet the conceptual and technical framework to easily reconstruct small molecules produced by complex metabolic pathways. This is problematic for ligand-dependent proteins, such as the ancestral steroid receptor. That is why most reconstructions of hormone/receptor couples assume that only proteins evolve. This assumption is inconsistent with the distribution of biochemical features related to estrogen synthesis across animals (Fig. 1). This multistep pathway starts with side-chain cleavage of cholesterol to pregnenolone by the CYP11A enzyme and, after many additional reactions, ends with aromatization of testosterone into estradiol by the CYP19A enzyme (Fig. 1) (5). To date, side-chain cleavage of cholesterol to pregnenolone has been demonstrated only in vertebrates, consistent with the origin of CYP11A from a vertebrate-specific gene duplication (6). On the contrary, the aromatase CYP19A is specific to chordates, which is at odds with reports of aromatase activity in mollusks and cnidarians (7, 8). Mollusks can capture, store, and metabolize vertebrate sex steroids, which are abundant in sewage water. However, there is no conclusive evidence that they can endogenously synthesize vertebrate-type steroid hormones (9). We previously proposed that the observed aromatization reactions outside vertebrates occur on steroids that are different from vertebrate androgens (10), which is supported by the identification of these aromatized long-chained steroids in sponges and cnidarians (11, 12). However, steroidogenesis is more complicated than only those two key reactions. Unbiased hypothesis testing of the evolution of steroid metabolic activities has to take into account all the available knowledge on the well-characterized steroidogenic pathways in animals (5, 13–17). The standard and transparent procedure for this analysis is parsimony, which is widely used in the comparative anatomy field and has been exported into biochemistry for ordering biochemical events related to the universal metabolism (18, 19).
Fig. 1. Estradiol synthesis pathway in vertebrates and distribution across metazoans of related biochemical features.
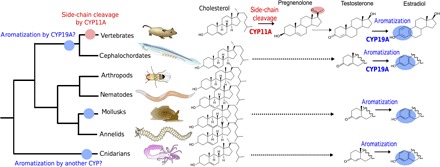
Vertebrate estradiol synthesis from cholesterol is a multistep pathway involving side-chain cleavage of cholesterol to pregnenolone by the CYP11A enzyme (red), a complex suite of reactions from pregnenolone to testosterone (dotted arrow), and aromatization by the CYP19A enzyme (blue). CYP19A is already present in chordates (blue dot on tree), whereas CYP11A is vertebrate-specific (red dot on tree). Aromatization has been described in cnidarians, mollusks, and cephalochordates (blue ovals). In cephalochordates, CYP19A may catalyze the aromatization reaction, whereas in mollusks and cnidarians, this definitely has to be a different enzyme, possibly a paralogous CYP (blue dots on mollusk and cnidarian branches of the tree). Because there is no consistent evidence for side-chain cleavage activities in those organisms, the exact endogenous substrate of those aromatizing enzymes remains elusive. It could be a steroid (hence the dotted line from cholesterol) but different from the vertebrate ones (hence the incomplete molecule structures).
RESULTS
Finding shared features of metabolic pathways using parsimony analysis
We used standard parsimony methods (cladistics)—in which biochemical pathways are considered as taxa, and the individual reactions (or types of reactions) catalyzed by identified enzymes shared by pathways are the compared character states (Fig. 2)—to investigate the evolutionary relationships between synthesis pathways of vertebrate sex and adrenal steroids, oxysterols and vitamin D, human and rodent bile acids, nematode dafachronic acids, and arthropod ecdysteroids. All these pathways start from cholesterol (except vitamin D synthesis, which starts from 7-dehydrocholesterol) and end with a final product, which is a ligand of a nuclear receptor (NR) (figs. S1 and S5). To elucidate the relative timing of the appearance of different metazoan steroidogenic pathways, we reconstructed their relationships by comparing the enzymatic reactions that are involved in each of them. In our data set of 72 taxa, each pathway from cholesterol to a steroid is a taxon—for example, cholesterol-to-estrone (C-to-E1; Fig. 2A) or cholesterol-to-estradiol (C-to-E2; Fig. 2A)—and enzymatic reactions along this pathway are the characters that are coded for each taxon (Fig. 2, A and B). We constructed a data matrix in which the pathways are systematically compared in terms of enzymatic reactions controlling the production of each ligand (Fig. 2, A and B, and fig. S1). The 151 characters were defined according to two criteria of homologies: type I and type II (Fig. 2, A and B, and fig. S1). Type I homologies are defined when two reactions in different metabolic pathways share the same enzyme with the same specificity for a substrate and the same product, for example, side-chain cleavage at carbon 20 (see numbering in Fig. 2C) in the synthesis of estrogen and estrone (character 31 in C-to-E1 and C-to-E2; Fig. 2, A and B) or dehydrogenation of cholesterol at carbon 7 in the synthesis of ecdysone or Δ7-dafachronic acid (character 58; Fig. 2, A and B). Type II homologies are cases in which pathways share enzymatic functions without sharing the specificity for a substrate in a more (character 134; Fig. 2, A and B) or less (character 147; Fig. 2, A and B) relaxed way (also see Materials and Methods), or when they share an enzyme adding the same residue at different positions on the carbon skeleton (character 151; fig. S2). There are many ways to go from cholesterol to some steroids; therefore, we used different numbers to refer to those various pathways (for example, C-to-CHENOAC_1 and C-to-CHENOAC_2; fig. S2).
Fig. 2. Parsimony analysis of metazoan steroidogenic pathways.
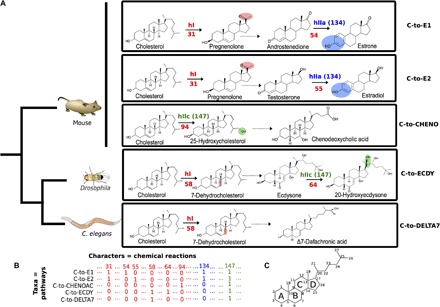
(A) Five examples of metazoan steroidogenic pathways. Some reactions are detailed to explain the character-coding procedure. Some pathways share identical reactions, defined here as type I homologies (hI, in red), such as side-chain cleavage of cholesterol to pregnenolone (reaction 31, in red) in the synthesis of estradiol and estrone (C-to-E1 and C-to-E2 pathways) or dehydrogenation of cholesterol to 7-dehydroxycholesterol (reaction 58, in red) in the C-to-ECDY and C-to-DELTA7 pathways. Some pathways also share similar reactions, defined as type II homologies (hII), with various degrees of similarity. Type IIa homologies (hIIa) describe the same chemical modification at the same place on two different substrates, such as aromatization (character 134, in blue) on androstenedione in the C-to-E1 pathway (reaction 54) or testosterone in the C-to-E2 pathway (reaction 55). Type IIc homologies (hIIc) describe the same chemical modification at different places on two different substrates, such as hydroxylation (character 147, in green) on carbon 25 from cholesterol (reaction 94) in the C-to-CHENO pathway or hydroxylation on carbon 20 from ecdysone in the C-to-ECDY pathway. C. elegans, Caenorhabditis elegans. (B) Extract of the data matrix corresponding to the pathways and steps described above. The presence or absence of characters is coded by 1 and 0, respectively (see complete matrix in fig. S1). (C) Nomenclature of carbon numbers and rings on the sterol skeleton.
Treating this matrix by parsimony analysis (70 of the 151 characters were parsimony-informative), we obtained a consensus of 216 equiparsimonious trees, each of 260 steps [consistency index (CI), 0.406; retention index (RI), 0.554]. To check the robustness of this consensus tree, we verified that searches under a varying number of iterations and different heuristics led to the same topology (see Materials and Methods). In Fig. 3, we present a 50% consensus tree, in which we highlight 12 strictly conserved nodes (A to L) and 2 suboptimal nodes (S1 and S2). The suboptimal node S1 is recovered when taxa C-to-15OHE2_1 and C-to-15OHE1_1 are excluded, and this also leads to the gain in the node grouping C-to-E1_1 and C-to-E2_1 and the branch leading to C-to-E1_5 and C-to-E2_5. The suboptimal node S2 and all downstream nodes are recovered when C-to-F2 and C-to-CRTSN2 are excluded.
Fig. 3. Relationships among animal steroidogenic pathways inferred from standard parsimony analysis.
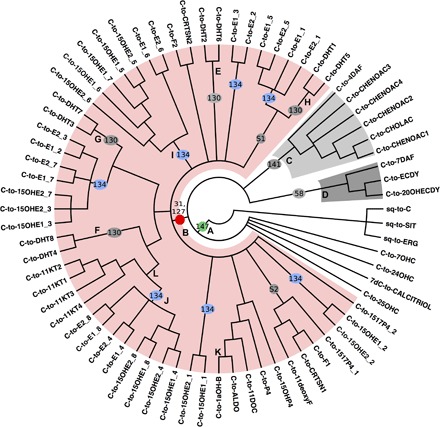
The 50% majority-rule consensus parsimony tree is rooted using eukaryotic sterol synthesis pathways. S1 and S2 indicate suboptimal nodes. Character numbers on branches refer to fig. S1B. All vertebrate sex and adrenal steroid synthesis pathways (highlighted in light red) are grouped together, sharing a side-chain cleavage reaction (31), whereas aromatized steroids are dispersed across the tree, indicating that aromatization of steroids without a side chain was recruited many times independently (blue ovals, 134). A shared trait of all analyzed animal steroid synthesis pathways is the occurrence of hydroxylation on some carbon residues (green dot, 147).
Side-chain cleavage, and not aromatization, as the common shared trait of vertebrate sex and adrenal steroid synthesis pathways
The unique synapomorphy uniting all bilaterian steroidogenic pathways is hydroxylation of the sterol backbone by a cytochrome P450 (CYP) enzyme (character 147 at node A, green dot; Fig. 3). Then, among bilaterian steroidogenic pathways, three major groups appear. A first large group gathers synthesis of vertebrate sex and adrenal steroids (highlighted in light red), which share side-chain cleavage and 3β-hydroxylation on a steroid without a lateral side chain (node B, red dot). The second group gathers synthesis pathways of bile acids and Δ4-dafachronic acid that share 3β-hydroxylation on a steroid with a lateral side chain (character 141 at node C, light gray). The third group gathers synthesis pathways of ecdysteroids and Δ7-dafachronic acid that share cholesterol 7,8-dehydrogenation (character 58 at node D, dark gray). The relationships between those three groups remain unresolved and form a seven-branch polytomy with the synthesis of calcitriol and three oxysterols. At node B, a large polytomy of 11 branches (with two suboptimal nodes) is found even under very permissive consensus conditions, which indicates either lack of data or that the shared common characters are not consistent enough to solve that region of the tree. 5α-Reduction of testosterone in the synthesis of dihydrotestosterone (C-to-DHT1-8) has been recruited four times independently (character 130 at nodes E to H), whereas steroid aromatization has been recruited seven times independently (character 134, blue dots). This suggests that enzymes capable of performing 5α-reduction and aromatization were already present before the appearance of enzymes performing side-chain cleavage and that they could later exploit the new possibilities offered by the cleaved steroids. It is therefore likely that these enzymes were originally acting on noncleaved steroids and were later recruited in the synthesis of steroids with a cleaved side chain. Aromatization occurs consistently in amphioxus ovary homogenates, in vitro; in contrast, side-chain cleavage was not biochemically detected under the same conditions, but was inferred from the gonadal expression of a putative CYP11 (20). This expressed gene was later shown not to be a CYP11A but a distant paralog, which was renamed CYP374 (6). There are some CYP11 genes in amphioxus (now named CYP11D and CYP11E) that branch at the basis of the vertebrate-specific CYP11A/CYP11B duplication (21); however, to date, we have no data about their expression pattern or the biochemical activity of their encoded products. Furthermore, the amphioxus genome contains an aromatase gene (22, 23) but lacks a one-to-one ortholog of the side-chain cleavage enzyme (6). Therefore, this analysis strongly suggests that steroids with a cleaved side chain are specific to vertebrates and absent in other animals. This implies that the steroid receptors present in other bilaterians (particularly the NR3Ds; Fig. 4B) bind steroids with a side chain. If this is true, then binding of 17β-estradiol to these receptors (24, 25) is probably a pharmacological effect.
Fig. 4. Paraestrol A synthesis and binding assays.
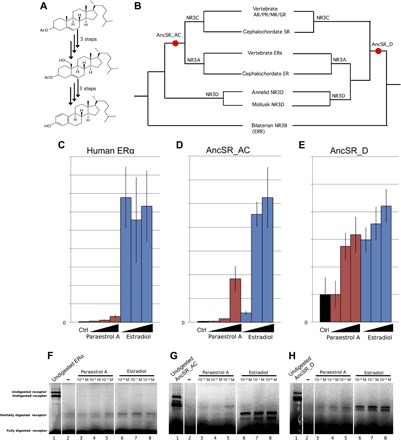
(A) Synthetic paraestrol A was produced in vitro using a six-step protocol, starting from cholesterol acetate (see the Supplementary Materials). (B) Two ancestral steroid receptors (AncSR_AC and AncSR_D) were reconstructed, taking into account phylogenetic uncertainty regarding the position of annelid and mollusk receptors (see sequence data set in fig. S3 and table S1). AR, androgen receptor; PR, progesterone; MR, mineralocorticoid receptor; GR, glucocorticoid receptor; SR, steroid receptor; ERR, estrogen-related receptor. (C to E) The ability of AncSR_AC and AncSR_D to activate the transcription of the luciferase reporter gene was tested in transfected human embryonic kidney (HEK) 293T cells with increasing concentrations of paraestrol A (10−6, 10−5, and 5 × 10−5 M) or estradiol (10−8 to 10−6 M). Human estrogen receptor α (ERα) was used as control (Ctrl). Activation folds are in arbitrary units, and their absolute values cannot be compared from one receptor to the other. (F to H) The binding ability of AncSR_AC and AncSR_D for paraestrol A and estradiol was tested by limited proteolytic assays. Lane 1, undigested protein; lanes 2 to 8, digested protein with ethanol as negative control (lane 2), paraestrol A at increasing concentrations (lanes 3 to 5 at 10−5 to 10−3 M, respectively), and estradiol (lanes 6 to 8 at 10−8 to 10−6 M, respectively).
Studies on steroid receptors from the NR3 subfamily have suggested that the estrogen-binding ability evolved before the ability to bind other side-chain cleaved steroids, such as androgens (24–26). As discussed above, in light of our new results, we infer that the ancestral ligand to the first steroid-binding receptor was not an 18-carbon estrogen but a molecule with a side chain in addition to the aromatic A-ring specific to vertebrate estrogens (Fig. 4A). Steroids with an aromatic A-ring and a lateral side chain have been identified in a tropical sponge (11), and the very molecule we propose as a proxy for this ancestral chordate steroid, 19-norcholesta-1,3,5(10)-trien-3-ol, was recently isolated from a soft coral (12). The synthesis of these molecules by nonbilaterian animals is probably related to defensive functions rather than to endocrine signaling (27). Because cholesterol hydroxylation already appeared at node A, the ancestral steroid may also have had an additional hydroxyl group at a position that we cannot exactly determine. The order of appearance of the biochemical activities (see below) indicates that 24- and 25-hydroxylation were among the first reactions to appear. 25-hydroxylase activity, which is reported in vertebrates, arthropods, annelids, and mollusks, is the most widespread in bilaterians (28). However, the side-chain cleavage mechanism involves the formation of 20- or 22-hydroxycholesterol as intermediates (29). Consequently, side-chain cleavage activity could derive from a 20- or 22-hydroxycholesterol, and the ancestral aromatized steroid might have been hydroxylated at one or many of those four positions. For the sake of simplicity, we tested the ligand-binding ability of an aromatized steroid with a side chain and with no hydroxylation. We define “paraestrols” as steroids comprising a cholesterol side chain and an aromatized A-ring to avoid confusion with other natural products bearing additional modifications on the side chain, such as ethyl or methyl groups on carbon 24 (11, 12). We further refer to 19-norcholesta-1,3,5(10)-trien-3-ol as paraestrol A, being the first member of this class to be synthesized de novo and experimentally characterized.
Ancestral steroid receptors bound and activated by paraestrol A
We synthesized paraestrol A from cholesterol acetate in six steps (see Fig. 4A and the Supplementary Materials). Two different versions of the receptor were reconstructed (Fig. 4B), taking into account alternative possibilities concerning the relationships between steroid receptors from chordates and those from mollusks and annelids (6, 30). One possibility (Fig. 4B, left) is that the groups NR3A and NR3C are the result of a chordate-specific ancestral duplication. In this case, the ancestral steroid receptor would be the last common ancestor predating this chordate-specific gene duplication (AncSR_AC). Consequently, members of the NR3D group, classically defined as invertebrate ERs, would be orthologous to the last common ancestor of both estrogen and ketosteroid receptors. The alternative possibility (Fig. 4B, right) is that the last common ancestor from the chordate NR3A group was orthologous to the common ancestor of NR3D from annelids and mollusks. This implies that the gene orthologous to the chordate NR3C was secondarily lost in annelids and mollusks. In that case, the ancestral chordate steroid receptor would also be the ancestor for the annelid and mollusk NR3D (AncSR_D in Fig. 4B). Because this phylogeny is still being debated (6, 30), we predicted the sequence of the ligand-binding domains (LBDs) for each of the two possible ancestors (Fig. 5 and table S2) using an alignment of 142 proteins (table S1). When compared to other reconstructions at the same node (25, 26, 31), both ancestral proteins were reconstructed with the same confidence level, with a mean posterior probability (PP) of 0.76 on the whole LBD. The confidence is even higher concerning the 18 sites that are involved in ligand binding (PP = 0.94 for AncSR_D and PP = 0.93 for AncSR_AC). These values are similar to another reconstruction that used a larger data set including analysis of internal vertebrate branches (25). In that alternative reconstruction, corresponding to our AncSR_AC, the mean PP for the LBD is 0.70 and increases to 0.90 for the ligand-binding sites. Our reconstructions and that of Eick et al. (25) are identical with regard to the ligand-binding pocket (Fig. 5). There is an 82% identity among the three reconstructed sequences for the 230 aligned sites. Therefore, we assume that those small variations will have no significant effect on the biochemical activity of the receptor.
Fig. 5. Sequence alignments of the ligand-binding pockets of ancestral receptors compared to human ERα.
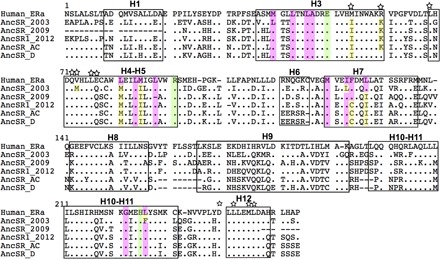
α-Helices that make up the LBD are boxed and numbered from 1 to 12 on the basis of the crystal structure from ERα (81). Amino acids from human ERα making direct hydrogen bonds with estradiol are highlighted in green. Amino acids making hydrophobic bonds with estradiol are highlighted in pink. Amino acids known to be involved in coactivator interaction are indicated with a star on top of each site. Differences with human ERα on ligand-binding or coactivator-binding sites are highlighted in yellow. Sequences of AncSR_AC and AncSR_D have been inferred in this study. The other ones come from previous analyses.
Using the synthesized AncSR_AC and AncSR_D sequences, we tested the ability of the resulting GAL4-LBD fusion protein to bind paraestrol A and to activate transcription in a ligand-dependent manner (Fig. 4, C to H). We performed transient transactivation assays using a UAS-tk-luciferase reporter system and constructs encoding fusions between the LBD of the receptors and the GAL4 DNA binding domain (GAL4-LBD). For comparison, we performed assays using human ERα and estradiol. With paraestrol A, both ancient receptors AncSR_AC and AncSR_D are able to activate transcription of the reporter gene in a dose-dependent manner (Fig. 4, D and E). Human ERα is much more sensitive to estradiol than to paraestrol A (Fig. 4C). This is also true for the AncSR_AC receptor, although the relative difference between both ligands is smaller when compared to ERα (Fig. 4, C and D). Thus, AncSR_AC behaves as a classic ligand-dependent transcription factor. By contrast, AncSR_D displays a constitutive transcriptional activity in the absence of ligand (Fig. 4E). The high constitutive activity observed with AncSR_D is reminiscent of the constitutive activity often observed with ERβ (32, 33), in contrast to ERα (34, 35). However, with paraestrol A at the micromolar range or with estradiol, AncSR_D activity can be further increased (Fig. 4E). Together, these results suggest that paraestrol A behaves as an activator of the ancestral steroid receptor.
The activity detected using transactivation assays could be due to the metabolic transformation of paraestrol A in mammalian cells. To check whether it really binds to ancestral receptors, we performed limited proteolytic assays (Fig. 4, F to H). This in vitro method tests whether the binding of a ligand can induce a conformational change in the receptor LBD, causing a partial protection to trypsin proteolysis. With estradiol, ERα and the two ancestral receptors are protected from proteolysis (Fig. 4, F to H). However, with paraestrol A, only AncSR_AC and AncSR_D are protected (Fig. 4, G and H). These results are consistent with the transactivation assays and confirm that paraestrol A is a bona fide ligand for the ancestral steroid receptor.
Paraestrol A side-chain wobbling in the ancestral receptor pocket
To obtain a deeper understanding of the ligand-receptor interaction and to check whether the conformation of paraestrol A is compatible with binding to the receptor, we performed ligand docking on a three-dimensional (3D) model of the reconstructed ancestral receptors (Fig. 6). Because these reconstructions are known to be sensitive to the initial template choice (35), we started with two different templates: human ERα [Protein Data Bank (PDB) code, 1ERE] and the ancestral corticoid receptor (AncCR; PDB code, 2Q3Y). Consistent with the principle of template influence, the inferred 3D structure was found to have a highly similar topology for both AncSR_AC and AncSR_D [root mean square deviation (RMSD) = 0.002 when modeled upon AncCR and RMSD = 0.006 upon ERα] modeled from the same template, whereas the distance was higher for both receptors comparing themselves upon modeling on the two different templates (RMSD = 1.124 between both AncSR_AC models and RMSD = 1.117). Despite these differences, the main features of the binding interaction are similar in both cases. Paraestrol A (shown in green) is docked inside the ligand-binding pocket of AncSR with a steroid core conformation similar to that adopted by 17β-estradiol (Fig. 6B, orange) in the ERα ligand-binding pocket. In particular, the polar residues E50 (in H3) and R91 (in H5) of AncSR_AC and Anc_D interact in an identical manner to the ERα LBD/17β-estradiol structure, where these residues anchor the ligand through interactions with the 3-OH hydroxyl group from the aromatized A-ring. In contrast, the 17β-OH group of estradiol is replaced by a long aliphatic chain in paraestrol A, whose conformation is shown here to be similar to that adopted by ponasterone or 20-hydroxyecdysone bound to the ecdysone receptor (1R1K and 2R40) (36, 37). The exact conformation of the side chain from paraestrol A cannot be predicted because it is flexible in nature and is likely to adapt to local structural constraints (Fig. 6, B and C). In particular, the comparison between the two types of homology models suggests that the region including helix H6 and loops H6-H7 and H11 (arrows) is very flexible and can easily accommodate the ligand tail located in this region. Other structural studies indicate that synthetic ligand side chains that are even bulkier than the side chain of paraestrol A can induce the formation of an extended subpocket in the LBD (38, 39). Furthermore, this gives a structural explanation for the smaller affinity of paraestrol A relative to 17β-estradiol: The absence of 17β-OH group in paraestrol A takes away the possibility of forming a hydrogen bond with H210 (homologous to H524) that stabilizes the interaction between 17β-estradiol and its receptor. Notably, interaction with H524 is not mandatory for ligand binding to E2. Some synthetic agonists, such as tamoxifen or other nonsteroidal compounds, also bind to ER without interacting with H524 (39, 40). However, those molecules bear hydroxylated carbons that form additional stabilizing interactions with other residues in the ligand-binding pocket. This is impossible with paraestrol A that bears no hydroxyl group on the side chain. Moreover, having a single additional hydroxyl group on the side chain of paraestrol A would not necessarily increase the binding affinity. The gain in binding energy is counteracted by higher desolvation cost in the case of the 25-OH group in ecdysteroids, whereas the presence of adjacent 20- and 22-OH moieties has a lower desolvation cost because of stabilizing intramolecular interactions between the adjacent hydroxyl groups (37). This phenomenon could also explain the observed micromolar affinity of 27-hydroxycholesterol to human ERs (41, 42). These docking studies demonstrate that paraestrol A is an excellent proxy for an ancestral steroid, supporting the hypothesis that a steroid from the paraestrol family was likely the starting point from which vertebrate-specific sex and adrenal steroids evolved. Furthermore, this model provides a structural explanation of how side-chain cleavage would increase the affinity during the transition from paraestrols to estrogens as ligands for steroid receptors.
Fig. 6. Paraestrol A docking into the pocket of the ancestral steroid receptor.
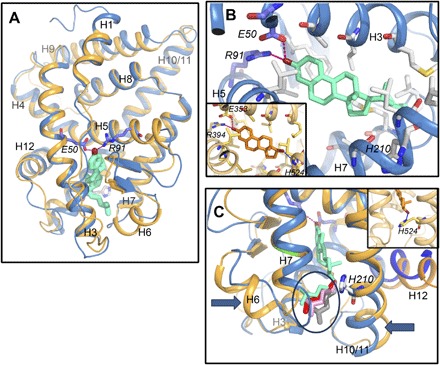
(A) Overall view of the homology model of AncSR LBD based on the crystal structure of ERα LBD in complex with 17β-estradiol (PDB code, 1ERE). Paraestrol A (green) is docked inside and binds to the receptor through the residues E50 and R91. Similar results are obtained using the crystal structure of the ancestral corticoid receptor in complex with desoxycorticosterone (PDB code, 2Q3Y) as a template (light orange trace). Both traces are from AncSR_D, with the traces from AncSR_AC being identical. (B) Detailed view on the binding pocket. Compared to the binding of 17β-estradiol (orange, inset), paraestrol A lacks the possibility of binding H524 from the receptor through its 17β-hydroxylated carbon. (C) Because of lack of binding to H210, the residue homologous to H524 in ERα (orange, inset), the aliphatic chain of paraestrol A wobbles inside the pocket. A few among many possible conformations are represented by different colors (encircled).
Dating the origins of steroid synthesis pathways
As previously developed for the universal metabolism (18, 19), the cladistic method tracing metabolic pathway evolution also allows us to date the appearance of biochemical steps relative to each other. We used synapomorphies related to families of reactions to assign different time periods to the tree branches (fig. S4). Our approach assumes that enzyme specificity has evolved from low-specificity proteins catalyzing a whole range of activities at low levels to enzyme subfamilies with potent and highly specialized activities (43–46). If this interpretation is correct, then the putative common ancestry of pathways can be postulated not only on the basis of shared enzymes with high specificities (such as reaction 31 at node B in Fig. 3) but also on the basis of very similar reactions (type II homologies). Thus, if downstream branches that do not bear changes in type II homologies follow a branch of the tree, then those downstream branches are of the same period as the main branch. When a new type II homology occurs on a branch, it defines the next period (for example, node B; fig. S4; see also Materials and Methods). According to this method, type II homologies distinguish between specific time periods during which innovations—in terms of enzymatic involvement in steroidogenesis—have occurred. On the basis of this, we defined seven relative time periods (Fig. 7B and fig. S5).
Fig. 7. Sequential ordering of animal steroid metabolic pathways.
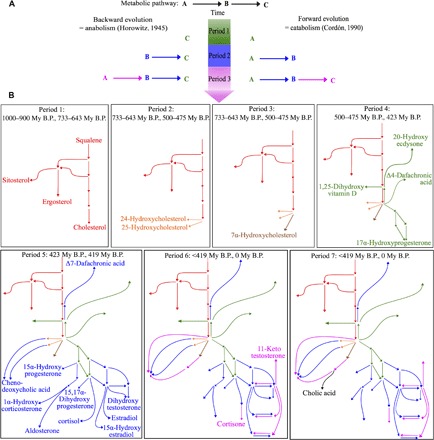
(A) Two theoretical models about metabolic pathway evolution: anabolic pathways evolving backward, with the more downstream reactions appearing first (47), and catabolic pathways evolving forward, with the more upstream reactions appearing first (50). (B) Sequential view of pathway appearance during successive periods in animal steroidogenesis. Colors indicate successive periods corresponding to the appearance of new types of enzymatic activities. Final compounds appearing at each period are indicated. All compounds appear in fig. S5. My B.P., million years before the present.
Theories on biochemical pathway evolution (Fig. 7A) predict that anabolic pathways evolve backward (47), which has been verified for glycolysis (48) or for the mandelate pathway (49), whereas catabolic pathways evolve forward (50), which has been proven, for example, for the urea cycle (51). The definition of periods allowed us to decipher the order of appearance of different steroidogenic pathways (Fig. 7B and fig. S5). Starting from the cholesterol synthesis backbone (Fig. 7B, period 1, red), the first pathway to appear was the synthesis of 24- and 25-hydroxycholesterol (period 2, orange), followed by the synthesis of 7-hydroxycholesterol (period 3, brown), and later by the synthesis of vitamin D3, Δ4-dafachronic acid, ecdysone, progesterone, and 11-deoxycortisol (period 4, green). Chenodeoxycholic acid and the bulk of vertebrate sex and adrenal steroids also appeared during the same period (period 5, blue). A later phase of diversification led to the synthesis of chenodeoxycholic acid through two additional pathways: cortisone and 11-ketotestosterone (period 6, purple). The last pathway to appear was the synthesis of cholic acid (period 7, black). Because the more upstream reactions in the pathways were the first to appear, steroidogenesis thus evolved in a forward direction, through a successive appearance of steps in successive generations of animals. This result is consistent with a detailed analysis of the distribution of bile acids across vertebrates, suggesting that the bile acid synthesis pathway evolved from C27 bile alcohols, structurally equivalent to oxysterols, to C24 bile acids that necessitate further catalytic steps to be made (52). Thus, from an evolutionary perspective, steroid synthesis should be viewed as a cholesterol degradation pathway rather than as a synthesis pathway, as suggested for bile acids (53). This is also in line with the nested position of steroidogenic enzymes within detoxification enzymes in phylogenetic trees based on genomic data (6). According to this view, steroidogenesis derived from catabolic pathways that are implicated in xenobiotic detoxification and steroid hormones could be viewed as co-opted cholesterol metabolites (54).
Differences in steroid metabolism among metazoans enable one to put temporal limits to some of the defined periods (fig. S4). On the basis of the assumption that a pathway specific for a given clade cannot be older than the oldest member of this clade, we identified five chronological anchor points that allow rooting of the relative periods in an absolute time frame. After initial divergence from fungi, around 1100 to 900 My B.P., the first bilaterian-specific steroidogenic pathway cannot be older than the divergence between protostomes and deuterostomes, estimated to 733 to 643 My B.P. (fig. S4, node A) (55). The presence of a clade of vertebrate side-chain steroidogenic pathways (fig. S4, node B) indicates that the last common ancestor of gnathostomes and lampreys was already able to synthesize at least one sex or adrenal steroid. This defines the second chronological anchor point between 500 and 475 My B.P. (56). Then, the elasmobranch-specific ability to make 1α-hydroxysteroid (fig. S4, node K) appeared after the divergence between chondrichtyians and osteichtyians 423 My B.P. (57). Finally, synthesis of the teleostean-specific 11-ketotestosterone (fig. S4, node L) could not be older than the split between teleosts and tetrapods 419 My B.P. (57). Dating the appearance of new enzymatic activities and new synthesis pathways also enables one to integrate the sets of receptors and enzymes that were present at the same time point on the basis of genomic analyses (6, 58, 59).
DISCUSSION
Beyond molecular actualism: Ancient metabolic pathways as a helping hand for enzyme recruitment
Although the forward and backward models of metabolic evolution provide consistent explanations for the origin of multiple very ancient pathways related to the universal metabolism, most of the late metabolic pathway evolution is thought to have occurred through patchwork evolution due to multiple recruitments of enzymes from preexisting pathways (60). There is an apparent contradiction between the strictly forward pattern that we observe for the evolution of current steroid synthesis pathways and the genomic and biochemical data. If some enzymatic activities (for example, aromatization) were recruited, which is consistent with our parsimony analysis, then it would fit a patchwork evolution model. This contradiction can be resolved by integrating receptors and paraestrols into an evolutionary model (Fig. 8). At the time when oxysterol synthesis appeared in basal chordates (corresponding to periods 2 and 3), oxysterols could probably bind to already existing ancestral receptors from the NR1H/I group. We propose that, during this period, the ligands for the AncSR were paraestrols synthesized by the ancestral CYP19 aromatase and other enzymes, such as an ancestral HSD3B (Fig. 8A). Later, during period 4, the appearance of the CYP11A side-chain cleavage enzyme (Fig. 8B) was concomitant with the appearance of progesterone synthesis in basal vertebrates (Fig. 8B), which is also consistent with the binding of progestagens to the unduplicated member of the NR3C family in lampreys (61). Finally, during period 5, opportunistic branching of the CYP19A aromatase from C27 sterols to side-chain cleaved steroid substrates led to the synthesis of modern estrogens and subsequent extinction of the paraestrol pathway (Fig. 7C). Explaining this complete replacement, instead of maintaining both pathways in parallel, requires an understanding of which molecular properties allowed estrogens to outcompete paraestrols (62). Our binding assays and structural modeling data suggest that side-chain cleavage helped to increase the binding affinity to ERs. This gain in affinity may have facilitated the transition from a promiscuous food sensing system to a highly specific coupling between nutritional status and reproduction (63). This model provides an alternative to the “ligand exploitation model,” which was built on the assumption that estradiol was the first estrogen, and tried to explain how intermediates in its synthesis-like progesterone or testosterone existed long before their receptor appeared (64). Paraestrols can be synthesized without progesterone, testosterone, or any side-chain intermediate as precursors, and this gives the possibility for coexistence of progestagens and paraestrols in basal vertebrates before the emergence of modern estrogens (Fig. 8B). Notably, if 17β-estradiol and similar aromatized steroids are canonical estrogens that act as high-affinity ligands for ERs, then it becomes increasingly clear that other nonaromatized steroids could also be potential physiological ER ligands (65, 66). For example, 5α-androstane-3β,17β-diol (also called 3β-Adiol) and Δ5-androstenediol bind ERα and ERβ with nanomolar affinity (67, 68). Synthesis of both compounds does not require aromatase, and synthesis of Δ5-androstenediol does not even require 3β-HSD activity. More recently, 27-hydroxycholesterol has also been found to be an endogenous ligand for the ER, binding it with micromolar affinity that is physiologically relevant (41, 42). Synthesis of 27-hydroxycholesterol represents the simplest possible synthesis pathways, with just one hydroxylation step from cholesterol. Therefore, it would be extremely interesting to delineate the contribution of these alternative ligands better, which may correspond to more ancient molecules that are active in the estrogenic pathway.
Fig. 8. Origin of vertebrate estrogens through opportunistic connections from ancient paraestrol synthesis pathways.
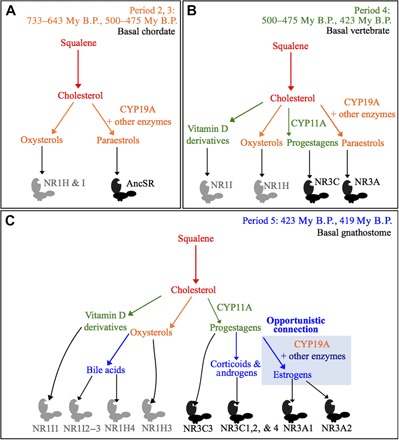
Coevolution between metabolic pathways, in colors corresponding to different periods (A to C) and steroid receptors of the NR1 (gray) and NR3 (black) subfamilies. We infer that, in addition to the sequential appearance of actual metabolites at different time steps (periods 2 and 3, oxysterols; period 4, progestagens; period 5, androgens), the history of NR ligand synthesis pathways involved metabolites combining features that are separated in actual molecules, such as paraestrols in periods 2 to 4. Those intermediary metabolites may have facilitated a gradual shift in substrate specificity for ancient enzymes, such as the CYP19A aromatase, and hence the building of new pathways through opportunistic connections of old enzymes with new substrates during period 5.
It was previously suggested that the most abundant metabolites can act as helping hands for pathway evolution, providing readily available substrates for newly arising catalytic activities (69). Here, we expand this view with a detailed case study showing that newly appearing metabolites can become substrates for older enzymes, leading to both expansion and extinction of pathways. Thus, just as fossils share a combination of characters that are absent in modern organisms, we argue that the actual set of observable metabolites represents only a subset of the full metabolite space. Our method provides the possibility of defining new classes of molecules that can be helpful to orient screens in analytical chemistry and study coevolution between proteins and their ligands.
MATERIALS AND METHODS
Taxonomic sampling
This work focused on metabolic pathways, leading to the synthesis of steroids that are all end product ligands of nuclear hormone receptors: vertebrate sex and adrenal steroids, bile acids, oxysterols and vitamin D, nematode dafachronic acids, and arthropod ecdysteroids. All these molecules were synthesized from a common precursor, cholesterol, except vitamin D, in which synthesis starts from 7-deoxycholesterol, and the three outgroups that are the synthesis pathways from squalene to ergosterol, cholesterol, and sitosterol. Some pathway examples are given in Fig. 2A. A complete list of the 72 pathways is shown in fig. S1A.
Characters and homologies
A complete list of characters is shown in fig. S1B. Compared character states are individual reactions (or types of reactions) catalyzed by identified enzymes shared by pathways. They were defined according to four homology criteria, as illustrated in Fig. 2 and fig. S2. The criteria for formulating a primary homology are as follows: shared specific enzymatic activity (I), shared enzymatic function without shared specificity for a substrate (IIa), shared functional family (IIc), and a new kind of homology that takes into account the sharing of the same enzyme, which performs two slightly different reactions (IIe). Type “hIIb” and “hIId” homologies defined previously (18) are not relevant here.
Type hI homologies
Primary homologies of “type hI” were defined as sharing the same reaction, with absolute specificity for the substrate, for two or more different pathways. For example, cholesterol side-chain cleavage by CYP11A1 into pregnenolone (character 31) occurred in the pathway transforming cholesterol into estrone (C-to-E1, red oval; Fig. 2) and in the pathway transforming cholesterol into estradiol (C-to-E2, red oval; Fig. 2). These two pathways shared the CYP11A1 enzyme, without difference in specificity for its substrate. In other words, the specificity is taken into account when defining type I homologies.
Type hII homologies
Type hII homologies corresponded to a more relaxed homology criterion, where two reactions did not share the same molecule as a substrate. Three subtypes (a, c, and e) were used here.
Type hIIa homologies
Primary homologies of “type hIIa” occurred when two pathways shared similar enzymatic function without considering their respective specificity for a substrate. This was the case for aromatization of androstenedione to estrone (character 54 in C-to-E1; Fig. 2) or aromatization of testosterone to estradiol (character 55 in C-to-E2; Fig. 2). Both reactions can be grouped together as character 134 (Fig. 2, blue).
Type hIIc homologies
Primary homologies of “type hIIc” are for shared functional family of enzymatic reactions, such as hydroxylations on either carbon 25 of the sterol skeleton in the C-to-CHENO pathway (Fig. 2) or carbon 20 in the C-to-ECDY pathway (Fig. 2). Both reactions can be grouped together as character 147 (Fig. 2, green).
Type hIIe homologies
We defined here a fifth type of secondary homology (type “hIIe”) for sharing of the same enzyme that performs two slightly different reactions. Character 151 provides an example, where the CYP27A1 enzyme hydroxylated either carbon 25 or carbon 26 on the cholesterol side chain during the synthesis of bile acids and calcitriol (fig. S2). There is a risk associated with this type of homology: The enzyme could have been recruited by one of the two pathways. However, the above reaction was situated very early (first step of cholesterol hydroxylation), favoring the hypothesis of an initial versatility of the enzyme.
Most of the characters corresponded to a single enzymatic reaction and a single enzyme. Five characters corresponded to multiple enzymes (88, 93, 100, 110, and 116 in figs. S1 and S5) involved in bile acid β-oxidation, which is a highly conserved process in which all the steps occurred in the same order for all bile acid precursors. Therefore, coding these steps separately would overweigh this part of the matrix.
Some characters referring to type II homologies contain question marks in the matrix. These are included when the pathway does not exhibit the appropriate substrate for the reaction, or when coding the reaction is meaningless. For example, coding the possibility of a reduction from a ketone in β on carbon 17 (reaction 129) has a meaning only for pathways where there is, at one step or another, a ketone on this carbon, and is meaningless for pathways where carbon 17 is the junction point of the side chain (such as in ecdysone synthesis).
Phylogenetic reconstruction
Tree search
The matrix contains 72 taxa (fig. S1A) and 151 characters (fig. S1C). Seventy characters are parsimony-informative. The matrix was analyzed using TNT version 1.1 (70), made freely available by the Willi Hennig Society. Characters were treated as unordered and unweighted in the search for the most parsimonious tree. Tree searches were carried out using the tree bisection-reconnection algorithm and the parsimony ratchet for a more extensive exploration of the possible tree space (71). Memory was set to hold up to 1,000,000 trees, with a gradual increase from 10,000 to 150,000 iterations, to ensure that the number of most parsimonious trees was decreasing or null. For the parsimony ratchet, the probability of upweighting was set to vary from the default 4% up to 50%, under a search of 10,000 iterations, to check that changes in weighting do not influence the length of the output tree. CI and RI were calculated as indicated by Farris (72). Check for rogue taxa was carried out using the prunnelsen option, which counts the number of nodes gained in either the strict or semistrict consensus and reports those prunings, which improve the results (70).
Rooting
Synthesis pathways from squalene to cholesterol, ergosterol, or sitosterol (73) were used as outgroups, assuming that the synthesis of sterol precursors by various eukaryotes is more ancient than that of steroid hormones from those precursors in animals.
Defining time span criteria
From the root to the tip of branches, phylogenetic trees provide a relative order of transformations (referred to as enzymatic innovations here) through time. We call a deep, more inclusive internal branch or clade an “upstream node” and a more terminal, less inclusive internal branch or clade a “downstream node.” Time spans in metabolism are defined as the time along the tree separating two type II character changes. This criterion is based on the assumption that the apparition of type II homologies correlates with the apparition of new kinds of enzymatic reactions catalyzed by the same enzyme. To define periods, the following criteria must be taken into account:
The order of nodes. In the absence of other criteria, two sister nodes are of the same relative period.
The nature of enzymatic changes. If downstream branches that do not bear changes in the type II homologies follow a branch, then the downstream branches are of the same period. When a new type II homology occurs on a branch, it defines the next period, except when it is already present in an earlier branch (homoplasy). For example, the node I is blue (fig. S4, period 5) because of character 134. This character also appears at node J, where there is no period change in comparison with the upstream node L because the period of node L (period 6) is already posterior to the period when character 134 appeared. Losses of type II characters were not used to define periods because they could be the result of very different processes (mutations, changes in cellular localization, changes in expression regulation, etc.).
Highly specific type I homologies. In two cases (characters 31 and 58), a single enzyme with high specificity innovates new enzymatic mechanisms. It is recorded as a type I homology, but because it also corresponds to enzymatic changes used to define type II homologies, it is taken into account for defining periods.
Biochemical characterization of paraestrol A
Paraestrol A synthesis
Paraestrol A [19-norcholesta-1,3,5(10)-trien-3-ol; CAS no. 5918-28-5; CHEBI:1177495] was first synthesized in a pharmacological context (74) and later isolated as a natural product from a coral (12). Kocovsky and Baines (75) first described the practical chemical removal of the 10β-methyl group from (affordable) cholesterol-like precursors. Our synthetic protocol (see the Supplementary Materials) had been slightly adapted and yielded paraestrol A (336 mg) in 5.3% over six steps from cholesterol acetate (5.0 g). Lipinski parameters and related properties for paraestrol A were measured using Advanced Chemistry Development (ACD/Labs) Software version 11.02. 17β-Estradiol was purchased from Sigma-Aldrich. Stock solutions of paraestrol A and estradiol were prepared in ethanol at 10−2 M.
Ancestral receptor reconstruction
LBDs of ancestral receptors from the NR3 subfamily were inferred according to maximum likelihood criteria using the codeml module of Phylogenetic Analysis by Maximum Likelihood 4 (PAML 4) (76). Because of the uncertainty about the exact relationships between the three families (10), two different receptors were inferred using two different tree input files (fig. S3). Sequence accession numbers are listed in table S1. Inferred sequences are given and compared in Fig. 5 and table S2, with information on ligand-binding residues (31) and interactions with cofactors (40). The receptor LBDs were synthesized de novo by Eurofins MWG Operon and cloned into the pG4MpolyII vector, which contains the GAL4 amino acids 1 to 147, to create chimeric GAL4-LBD proteins. Because the region in the N terminus of helix H1 is highly variable and unpredictable, we used the corresponding eight amino acids from the human ERα as a linker region between GAL4 and the LBD in all constructions.
Transactivation assays
HEK 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (Invitrogen by Life Technologies). The cells were transfected at 70% confluence in 96-well plates using ExGen 500 (Euromedex) according to the manufacturer’s instructions, with 60 ng of total DNA, including the reporter plasmid 17M-Glob-Luc and the CMV-βGal plasmid as an internal control to normalize for variations of transfection efficiency. After 6 hours, cells were treated with ligands. Culture medium was replaced by DMEM supplemented with 10% charcoal-treated fetal calf serum and containing paraestrol A or estradiol diluted to different final concentrations (10−6 to 10−4 M and 10−6 to 10−8 M, respectively). After 24 hours of treatment, cells were lysed and assayed for luciferase activity.
Limited proteolytic digestion
These experiments were performed following the study of Vivat et al. (77). Receptors were translated and 35S-labeled in vitro using the TNT-coupled reticulocyte lysate system (Promega). The ability of ligands to protect receptors from trypsin digestion was assessed at 10−3 to 10−5 M for paraestrol A and 10−6 to 10−8 M for estradiol.
Homology modeling and ligand docking
Homology modeling was carried out using SWISS-MODEL (78). Both receptors AncSR_AC and AncSR_D were modeled on the basis of (i) the crystal structure of ERα LBD in complex with 17β-estradiol (PDB code, 1ERE) and (ii) the crystal structure of the ancestral corticoid receptor in complex with desoxycorticosterone (PDB code, 2Q3Y). Overlap between the structures was quantified using RMSD, estimated using the PyMOL Molecular Graphics System version 1.4.1 (Schrödinger LLC). Model quality was good on the basis of the QMEAN Z-scores (79): −1.47 and −0.87 for AncSR_AC and −1.525 and −0.62 for AncSR_D based on 1ERE and 2Q3Y, respectively. Paraestrol A was generated using the online version of PRODRG (80).
Ligand docking inside the AncSR was carried out manually by placing the steroid core of paraestrol A in a location similar to that of 17β-estradiol. The aliphatic chain conformation of paraestrol A was initially made similar to that of 20-hydroxyecdysone or ponasterone bound to the ecdysone receptor (36, 37) and further allowed to move around the C20 atom. The side chains of a few residues in the proximity of the aliphatic tail were selected in a conformation different from that seen in the original homology model to better accommodate the paraestrol A aliphatic chain (D34, L39, M40, L43, F102, I214, V115, M118, H210, and K219).
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of C. Cunchillos, who laid the foundation of comparative anatomy of metabolic pathways. We thank J. Burden, C. Dauphin-Villemant, R. Lafont, E. Ragsdale, and the reviewers for comments on the manuscript; D. Russell for checking the bile acid pathways; and B. Demeneix for the initial impulse to this project. Funding: This work was funded by the French Ministry of Research, Agence Nationale de la Recherche (ANR-10-BLAN-1234), European Union FP6 (CASCADE, CRESCENDO), École Normale Supérieure de Lyon, CNRS, and Fondation pour la Recherche Médicale (FDT20100920225). Author contributions: G.V.M., F.B., D.M., J.H., G.L., and V.L. conceived and designed the experiments. G.V.M. performed the cladistic analysis and ancestral receptor sequence reconstruction. D.P. performed the chemical synthesis. J.G.-M. performed the biochemical experiments. I.M.L.B. performed the molecular modeling. All authors discussed the results and contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1601778/DC1
Supplementary Materials and Methods
fig. S1. Data matrix containing 72 taxa (rows) and 151 characters (columns).
fig. S2. Complements regarding the coding procedure.
fig. S3. Phylogenetic relationships among the sequences that were used to reconstruct AncSR_AC and AncSR_D.
fig. S4. Temporal relationships among steroid synthesis pathways, inferred from a 50% majority-rule consensus parsimony tree.
fig. S5. Detailed view of animal steroid metabolic pathways.
table S1. Accession numbers for the 142 sequences used for the predictions of the ancestral receptors.
table S2. Sequences of the LBDs for the two reconstructed receptors.
REFERENCES AND NOTES
- 1.Romano M., Reviewing the term uniformitarianism in modern Earth sciences. Earth Sci. Rev. 148, 65–76 (2015). [Google Scholar]
- 2.Chatterjee S., Templin R. J., The flight of Archaeopteryx. Naturwissenschaften 90, 27–32 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Nudds R. L., Dyke G. J., Narrow primary feather rachises in Confuciusornis and Archaeopteryx suggest poor flight ability. Science 328, 887–889 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Harms M. J., Thornton J. W., Evolutionary biochemistry: Revealing the historical and physical causes of protein properties. Nat. Rev. Genet. 14, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne A. H., Hales D. B., Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 25, 947–970 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Markov G. V., Tavares R., Dauphin-Villemant C., Demeneix B. A., Baker M. E., Laudet V., Independent elaboration of steroid hormone signaling pathways in metazoans. Proc. Natl. Acad. Sci. U.S.A. 106, 11913–11918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twan W.-H., Hwang J.-S., Chang C.-F., Sex steroids in scleractinian coral, Euphyllia ancora: Implication in mass spawning. Biol. Reprod. 68, 2255–2260 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Osada M., Tawarayama H., Mori K., Estrogen synthesis in relation to gonadal development of Japanese scallop, Patinopecten yessoensis: Gonadal profile and immunolocalization of P450 aromatase and estrogen. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139, 123–128 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Scott A. P., Do mollusks use vertebrate sex steroids as reproductive hormones? Part I: Critical appraisal of the evidence for the presence, biosynthesis and uptake of steroids. Steroids 77, 1450–1468 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Markov G. V., Laudet V., Origin and evolution of the ligand-binding ability of nuclear receptors. Mol. Cell. Endocrinol. 334, 21–30 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Wang G.-Y.-S., Crews P., Geodisterol, a novel polyoxygenated sterol with an aromatic A ring from the tropical marine sponge Geodia sp. Tetrahedron Lett. 37, 8145–8146 (1996). [Google Scholar]
- 12.Yan X.-H., Liu H.-L., Huang H., Li X.-B., Guo Y.-W., Steroids with aromatic A-rings from the Hainan soft coral Dendronephthya studeri Ridley. J. Nat. Prod. 74, 175–180 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Bury N. R., Sturm A., Evolution of the corticosteroid receptor signalling pathway in fish. Gen. Comp. Endocrinol. 153, 47–56 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lowartz S., Petkam R., Renaud R., Beamish F. W. H., Kime D. E., Raeside J., Leatherland J. F., Blood steroid profile and in vitro steroidogenesis by ovarian follicles and testis fragments of adult sea lamprey, Petromyzon marinus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 134, 365–376 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Huang X., Warren J. T., Gilbert L. I., New players in the regulation of ecdysone biosynthesis. J. Genet. Genomics 35, 1–10 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T., Li Y., Suino-Powell K., Xu H. E., Auchus R. J., Antebi A., Mangelsdorf D. J., Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124, 1209–1223 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Wollam J., Magner D. B., Magomedova L., Rass E., Shen Y., Rottiers V., Habermann B., Cummins C. L., Antebi A., A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLOS Biol. 10, e1001305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunchillos C., Lecointre G., Integrating the universal metabolism into a phylogenetic analysis. Mol. Biol. Evol. 22, 1–11 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Cunchillos C., Lecointre G., Ordering events of biochemical evolution. Biochimie 89, 555–573 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Mizuta T., Asahina K., Suzuki M., Kubokawa K., In vitro conversion of sex steroids and expression of sex steroidogenic enzyme genes in amphioxus ovary. J. Exp. Zool. 309A, 83–93 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Baker M. E., Nelson D. R., Studer R. A., Origin of the response to adrenal and sex steroids: Roles of promiscuity and co-evolution of enzymes and steroid receptors. J. Steroid Biochem. Mol. Biol. 151, 12–24 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Castro L. F. C., Santos M. M., Reis-Henriques M. A., The genomic environment around the aromatase gene: Evolutionary insights. BMC Evol. Biol. 5, 43 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callard G. V., Tarrant A. M., Novillo A., Yacci P., Ciaccia L., Vajda S., Chuang G.-Y., Kozakov D., Greytak S. R., Sawyer S., Hoover C., Cotter K. A., Evolutionary origins of the estrogen signaling system: Insights from amphioxus. J. Steroid Biochem. Mol. Biol. 127, 176–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bridgham J. T., Brown J. E., Rodríguez-Marí A., Catchen J. M., Thornton J. W., Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLOS Genet. 4, e1000191 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eick G. N., Colucci J. K., Harms H. J., Ortlund E. A., Thornton J. W., Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLOS Genet. 8, e1003072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton J. W., Need E., Crews D., Resurrecting the ancestral steroid receptor: Ancient origin of estrogen signaling. Science 301, 1714–1717 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Lafont R., Mathieu M., Steroids in aquatic invertebrates. Ecotoxicology 16, 109–130 (2007). [DOI] [PubMed] [Google Scholar]
- 28.R. Lafont, J. Koolman, Diversity of ecdysteroids in animal species, in Ecdysone: Structures and Functions, G. Smagghe, Ed. (Springer, 2009), pp. 47–71. [Google Scholar]
- 29.Strushkevich N., MacKenzie F., Cherkesova T., Grabovec I., Usanov S., Park H.-W., Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl. Acad. Sci. U.S.A. 108, 10139–10143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eick G. N., Thornton J. W., Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol. Cell. Endocrinol. 334, 31–38 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Keay J., Thornton J. W., Hormone-activated estrogen receptors in annelid invertebrates: Implications for evolution and endocrine disruption. Endocrinology 150, 1731–1738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremblay G. B., Tremblay A., Labrie F., Giguère V., Ligand-independent activation of the estrogen receptors α and β by mutations of a conserved tyrosine can be abolished by antiestrogens. Cancer Res. 58, 877–881 (1998). [PubMed] [Google Scholar]
- 33.Koehler K. F., Helguero L. A., Haldosén L.-A., Warner M., Gustafsson J.-Å., Reflections on the discovery and significance of estrogen receptor β. Endocr. Rev. 26, 465–478 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Maggi A., Liganded and unliganded activation of estrogen receptor and hormone replacement therapies. Biochim. Biophys. Acta 1812, 1054–1060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginalski K., Comparative modeling for protein structure prediction. Curr. Opin. Struct. Biol. 16, 172–177 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Billas I. M. L., Iwema T., Garnier J.-M., Mitschler A., Rochel N., Moras D., Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature 426, 91–96 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Browning C., Martin E., Loch C., Wurtz J.-M., Moras D., Stote R. H., Dejaegere A. P., Billas I. M. L., Critical role of desolvation in the binding of 20-hydroxyecdysone to the ecdysone receptor. J. Biol. Chem. 282, 32924–32934 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Nettles K. W., Bruning J. B., Gil G., O’Neill E. E., Nowak J., Hughs A., Kim Y., DeSombre E. R., Dilis R., Hanson R. N., Joachimiak A., Greene G. L., Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 8, 563–568 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nwachukwu J. C., Srinivasan S., Zheng Y., Wang S., Min J., Dong C., Liao Z., Nowak J., Wright N. J., Houtman R., Carlson K. E., Josan J. S., Elemento O., Katzenellenbogen J. A., Zhou H.-B., Nettles K. W., Predictive features of ligand-specific signaling through the estrogen receptor. Mol. Syst. Biol. 12, 864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L., The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Nelson E. R., Wardell S. E., Jasper J. S., Park S., Suchindran S., Howe M. K., Carver N. J., Pillai R. V., Sullivan P. M., Sondhi V., Umetani M., Geradts J., McDonnell D. P., 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 342, 1094–1098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Q., Ishikawa T., Sirianni R., Tang H., McDonald J. G., Yuhanna I. S., Thompson B., Girard L., Mineo C., Brekken R. A., Umetani M., Euhus D. M., Xie Y., Shaul P. W., 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 5, 637–645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen R. A., Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425 (1976). [DOI] [PubMed] [Google Scholar]
- 44.Khersonsky O., Roodveldt C., Tawfik D. S., Enzyme promiscuity: Evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Carbonell P., Lecointre G., Faulon J.-L., Origins of specificity and promiscuity in metabolic networks. J. Biol. Chem. 286, 43994–44004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voordeckers K., Brown C. A., Vanneste K., van der Zande E., Voet A., Maere S., Verstrepen K. J., Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLOS Biol. 10, e1001446 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horowitz N. H., On the evolution of biochemical syntheses. Proc. Natl. Acad. Sci. U.S.A. 31, 153–157 (1945). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fothergill-Gilmore L. A., Michels P. A. M., Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59, 105–235 (1993). [DOI] [PubMed] [Google Scholar]
- 49.Petsko G. A., Kenyon G. L., Gerlt J. A., Ringe D., Kozarich J. W., On the origin of enzymatic species. Trends Biochem. Sci. 18, 372–376 (1993). [DOI] [PubMed] [Google Scholar]
- 50.F. Cordón, Tratado Evolucionista De Biologia (Aguilar, 1990). [Google Scholar]
- 51.Takiguchi M., Matsubasa T., Amaya Y., Mori M., Evolutionary aspects of urea cycle enzyme genes. BioEssays 10, 163–166 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Hagey L. R., Møller P. R., Hofmann A. F., Krasowski M. D., Diversity of bile salts in fish and amphibians: Evolution of a complex biochemical pathway. Physiol. Biochem. Zool. 83, 308–321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell D. W., Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50, S120–S125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker M. E., Xenobiotics and the evolution of multicellular animals: Emergence and diversification of ligand-activated transcription factors. Integr. Comp. Biol. 45, 172–178 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Peterson K. J., Cotton J. A., Gehling J. G., Pisani D., The Ediacaran emergence of bilaterians: Congruence between the genetic and the geological fossil records. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1435–1443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.P. Janvier, Living primitive fishes and fishes from deep time, in Primitive Fishes, D. J. McKenzie, A. P. Farrell, C. J. Brauner, Eds. (Academic Press, 2007), pp. 1–51. [Google Scholar]
- 57.Zhu M., Zhao W., Jia L., Lu J., Qiao T., Qu Q., The oldest articulated osteichthyan reveals mosaic gnathostome characters. Nature 458, 469–474 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Bertrand S., Brunet F. G., Escriva H., Parmentier G., Laudet V., Robinson-Rechavi M., Evolutionary genomics of nuclear receptors: From twenty-five ancestral genes to derived endocrine systems. Mol. Biol. Evol. 21, 1923–1937 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Bridgham J. T., Eick G. N., Larroux C., Deshpande K., Harms M. J., Gauthier M. E. A., Ortlund E. A., Degnan B. M., Thornton J. W., Protein evolution by molecular tinkering: Diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLOS Biol. 8, e1000497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caetano-Anollés G., Yafremava L. S., Gee H., Caetano-Anollés D., Kim H. S., Mittenthal J. E., The origin and evolution of modern metabolism. Int. J. Biochem. Cell Biol. 41, 285–297 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Baker M. E., Asnaashari P., Chang D. J., McDonnell S., 3D models of lamprey progesterone receptor complexed with progesterone, 7α-hydroxy-progesterone and 15α-hydroxy-progesterone. Steroids 76, 169–176 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Firn R. D., Jones C. G., A Darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot. 60, 719–726 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Della Torre S., Benedusi V., Fontana R., Maggi A., Energy metabolism and fertility—A balance preserved for female health. Nat. Rev. Endocrinol. 10, 13–23 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Thornton J. W., Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. U.S.A. 98, 5671–5676 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker M. E., Insights from the structure of estrogen receptor into the evolution of estrogens: Implications for endocrine disruption. Biochem. Pharmacol. 82, 1–8 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Baker M. E., What are the physiological estrogens? Steroids 78, 337–340 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Kuiper G. G. J. M., Carlsson B., Grandien K., Enmark E., Häggblad J., Nilsson S., Gustafsson J.-Å., Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138, 863–870 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Weihua Z., Lathe R., Warner M., Gustafsson J.-Å., An endocrine pathway in the prostate, ERβ, AR, 5α-androstane-3β,17β-diol, and CYP7B1, regulates prostate growth. Proc. Natl. Acad. Sci. U.S.A. 99, 13589–13594 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt S., Sunyaev S., Bork P., Dandekar T., Metabolites: A helping hand for pathway evolution? Trends Biochem. Sci. 28, 336–341 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Goloboff P. A., Farris J. S., Nixon K. C., TNT: A free program for phylogenetic analysis. Cladistics 24, 774–786 (2008). [Google Scholar]
- 71.Nixon K. C., The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15, 407–414 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Farris J. S., The retention index and the rescaled consistency index. Cladistics 5, 417–419 (1989). [DOI] [PubMed] [Google Scholar]
- 73.Summons R. E., Bradley A. S., Jahnke L. L., Waldbauer J. R., Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 951–968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romo J., Rosenkranz G., Djerassi C., Steroids. X.1 Aromatization experiments in the cholesterol series. J. Org. Chem. 15, 1289–1292 (1950). [Google Scholar]
- 75.Kocovsky P., Baines R. S., Stereoelectronically controlled, thallium (III)-mediated C-19 degradation of 19-hydroxy steroids. An expedient route to estrone and its congeners via 19-nor-10β-hydroxy intermediates. J. Org. Chem. 59, 5439–5444 (1994). [Google Scholar]
- 76.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Vivat V., Zechel C., Wurtz J.-M., Bourguet W., Kagechika H., Umemiya H., Shudo K., Moras D., Gronemeyer H., Chambon P., A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 16, 5697–5709 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arnold K., Bordoli L., Kopp J., Schwede T., The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Benkert P., Biasini M., Schwede T., Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schüttelkopf A. W., van Aalten D. M. F., PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol Crystallogr. 60, 1355–1363 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B., Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5998–6003 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leng T., Zhang J., Xie J., Zhou S., Huang Y., Zhou Y., Zhu W., Yan G., Synthesis and anti-glioma activity of 25(R)-spirostan-3β,5α,6β,19-tetrol. Steroids 75, 224–229 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Kalvoda J., Heusler K., Ueberwasser H., Anner G., Wettestein A., 19-Norsteroide IV. Über die reduktive Ätherspaltung bei 5α-Halogen-6β,19-oxido-steroiden. Über Steroide, 198. Mitteilung. Helv. Chim. Acta 46, 1361–1369 (1963). [Google Scholar]
- 84.Kalvoda J., Heusler K., Die Hypojodit-Reaktion (Verfahren zur intramolekularen Substitution an nicht-aktivierten C-Atomen). Synthesis 1971, 501–526 (1971). [Google Scholar]
- 85.Kočovský P., Stieborová I., Synthesis of strophanthidin. Tetrahedron Lett. 30, 4295–4298 (1989). [Google Scholar]
- 86.Kocovsky P., Pour M., Stereo- and regiocontrol of electrophilic additions to cyclohexene systems by neighboring groups. Competition of electronic and stereoelectronic effects and comparison of the reactivity of selected electrophiles. J. Org. Chem. 55, 5580–5589 (1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1601778/DC1
Supplementary Materials and Methods
fig. S1. Data matrix containing 72 taxa (rows) and 151 characters (columns).
fig. S2. Complements regarding the coding procedure.
fig. S3. Phylogenetic relationships among the sequences that were used to reconstruct AncSR_AC and AncSR_D.
fig. S4. Temporal relationships among steroid synthesis pathways, inferred from a 50% majority-rule consensus parsimony tree.
fig. S5. Detailed view of animal steroid metabolic pathways.
table S1. Accession numbers for the 142 sequences used for the predictions of the ancestral receptors.
table S2. Sequences of the LBDs for the two reconstructed receptors.


