ABSTRACT
Severe acute respiratory syndrome (SARS) is a respiratory disease, caused by a coronavirus (SARS-CoV), that is characterized by atypical pneumonia. The nucleocapsid protein (N protein) of SARS-CoV plays an important role in inhibition of type I interferon (IFN) production via an unknown mechanism. In this study, the SARS-CoV N protein was found to bind to the SPRY domain of the tripartite motif protein 25 (TRIM25) E3 ubiquitin ligase, thereby interfering with the association between TRIM25 and retinoic acid-inducible gene I (RIG-I) and inhibiting TRIM25-mediated RIG-I ubiquitination and activation. Type I IFN production induced by poly I·C or Sendai virus (SeV) was suppressed by the SARS-CoV N protein. SARS-CoV replication was increased by overexpression of the full-length N protein but not N amino acids 1 to 361, which could not interact with TRIM25. These findings provide an insightful interpretation of the SARS-CoV-mediated host innate immune suppression caused by the N protein.
IMPORTANCE The SARS-CoV N protein is essential for the viral life cycle and plays a key role in the virus-host interaction. We demonstrated that the interaction between the C terminus of the N protein and the SPRY domain of TRIM25 inhibited TRIM25-mediated RIG-I ubiquitination, which resulted in the inhibition of IFN production. We also found that the Middle East respiratory syndrome CoV (MERS-CoV) N protein interacted with TRIM25 and inhibited RIG-I signaling. The outcomes of these findings indicate the function of the coronavirus N protein in modulating the host's initial innate immune response.
KEYWORDS: SARS coronavirus, nucleocapsid, interferon, TRIM25, RIG-I
INTRODUCTION
As the first line of defense against viruses, type I interferon (IFN) plays a critical role in initiating host antiviral responses. Following virus infection, the host innate immune system is triggered by the recognition of virus-specific components (pattern recognition receptors [PRRs]), such as DNA, single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), or glycoproteins (1). The Toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are the most common host PRRs that respond to RNA viruses (2). The cytoplasmic virus receptor RIG-I directly recognizes and binds to viral 5′-PPP RNA and short dsRNA, which are found in cells infected with a variety of RNA viruses, through its helicase and repressor domain (RD) (1, 3). After recognition, the N-terminal caspase recruitment domains (CARDs) of RIG-I are modified by ubiquitin, mediated by the E3 ligase tripartite motif protein 25 (TRIM25) (4). The domains then initiate an antiviral signaling cascade by interacting with the downstream partner MAVS/VISA/IPS-1/Cardif (4, 5), leading to the phosphorylation and activation of the transcription factors IRF3 and NF-κB, eventually leading to the production of type I IFN and many other cytokines. IFN-β secretion induces IFN-stimulated genes (ISGs), which exert antiviral effector functions (6).
Viruses have evolved the capacity to evade host immune recognition and to suppress the host IFN system (7). Viruses encode viral proteins that interfere with PRR signaling pathways to gain an early advantage against host defense. For example, the influenza A virus NS1 protein, Ebola virus VP35 protein, and vaccine virus E3L protein bind viral dsRNA to evade host immune recognition (8). Virus-encoded proteins also target IFN genes and IFN-induced genes to prevent host antiviral effector responses (2, 9).
Severe acute respiratory syndrome coronavirus (SARS-CoV), which emerged in 2003, causes high mortality in humans. Similar to other known coronaviruses, SARS-CoV has a 29.7-kb genome that encodes four structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) (10, 11) proteins. The N protein enters host cells together with viral RNA. Both the N and C termini of the N protein bind viral RNA to form the helical ribonucleocapsid (RNP), which plays a fundamental role in viral assembly (12, 13). The multimerization of N proteins occurs primarily through the C terminus (14). The N-terminal portion of the N protein (amino acids [aa] 168 to 208) is important for the interaction with the viral M protein, indicating that this region may be crucial for viral packaging (15, 16). The N protein of SARS-CoV has been demonstrated to upregulate the expression of the proinflammatory factor COX2 and to interact with the proteasome subunit p42, which affects a variety of basic cellular processes and inflammatory responses (17, 18). Notably, N also inhibits the synthesis of IFN-β, which plays a vital role in innate immunity against RNA virus infections by a mechanism that is not fully understood (19–21).
In this study, we found that the SARS-CoV N protein interacted with TRIM25 and interfered with the association between TRIM25 and RIG-I. Thus, TRIM25-mediated RIG-I ubiquitination was inhibited by the SARS-CoV N protein and contributed to the suppression of type I IFN production.
RESULTS
The SARS-CoV N protein interacts with TRIM25.
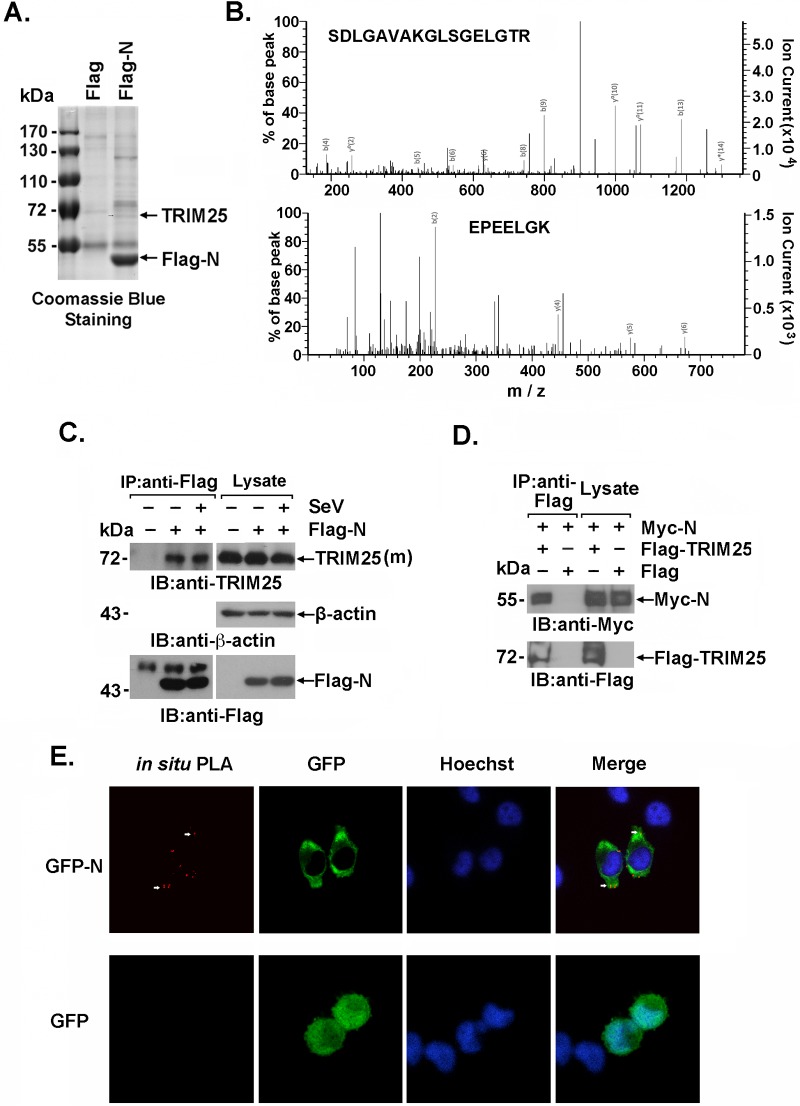
To investigate the cellular interaction partners of the SARS-CoV N protein in humans, lysates from 293T cells that exogenously expressed the Flag-N protein or Flag-vector control were immunoprecipitated with an anti-Flag antibody. The immunoprecipitates were subjected to SDS-PAGE and Coomassie brilliant blue staining (Fig. 1A). Stained bands that appeared in the Flag-N but not the control immunoprecipitates were subjected to trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Two TRIM25 peptides in the indicated band were identified by a Mascot search (Matrix Science, Boston, MA). These results suggested that TRIM25, the E3 ligase for RIG-I ubiquitination that plays a vital role in the innate immune response to RNA virus infection, was present in the Flag-N immunoprecipitates and that is might associate with the SARS-CoV N protein (Fig. 1B). The interaction between the SARS-CoV N protein and TRIM25 was verified by immunoblotting. Endogenous TRIM25 was coimmunoprecipitated with Flag-N but not with the Flag-vector control, with or without Sendai virus (SeV) infection (Fig. 1C). Moreover, an interaction between exogenous Flag-TRIM25 and the Myc-N protein was observed by immunoblotting anti-Flag immunoprecipitates with the anti-Myc antibody (Fig. 1D). Further, to demonstrate the association between the SARS-CoV N protein and endogenous TRIM25 in situ, HeLa cells expressing green fluorescent protein (GFP)-N or GFP alone were subjected to a Duolink-based in situ proximity ligation assay (PLA) followed by confocal microscopy. PLA-positive signals (red points) indicated that colocalization of the SARS-CoV N protein (but not GFP) and endogenous TRIM25 was exclusively in the cytoplasm. This distribution was similar to that observed for the SARS-CoV N protein (Fig. 1E). These data collectively showed that TRIM25 was an association partner of the SARS-CoV N protein in human cells.
FIG 1.
SARS-CoV N protein interacts with TRIM25. (A) Cell extracts prepared from 293T cells transfected with Flag-SARS N protein (Flag-N) or Flag vector were subjected to anti-Flag immunoprecipitation. The immunoprecipitates were resolved using SDS-PAGE electrophoresis and Coomassie blue staining. (B) The stained bands marked by arrows in Fig. 1A were digested with trypsin and analyzed by LC-MS/MS. Two peptides (SDLGAVAKGLSGELGTR and EPEELGK) matching the TRIM25 protein sequence were identified. (C and D) 293T cells transfected with the indicated plasmids were infected with or without SeV (C), and the cell lysates were subjected to anti-Flag immunoprecipitation (IP). The immunoprecipitates were analyzed by immunoblotting (IB) with anti-TRIM25 (C) or anti-Myc (D). (E) HeLa cells transfected with GFP-N were fixed with 4% paraformaldehyde and incubated with anti-TRIM25 and anti-GFP antibodies. An in situ PLA assay was conducted as described and imaged with a confocal microscope at ×100 magnification. Red dots indicate the interaction.
The C terminus of the SARS-CoV N protein interacts with the TRIM25 SPRY domain.
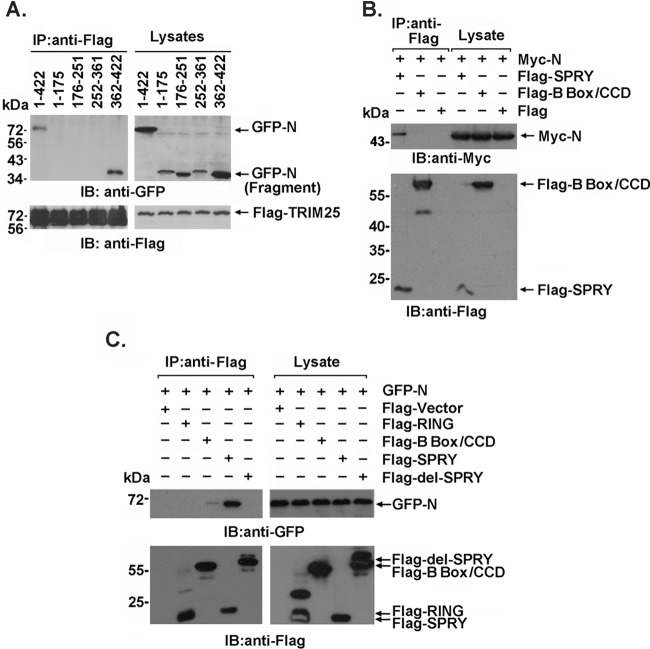
To investigate the interaction between TRIM25 and the SARS-CoV N protein, GFP-tagged truncated fragments of the SARS-CoV N protein (aa 1 to 175, 176 to 251, 252 to 361, and 362 to 422) were cotransfected with Flag-TRIM25 into 293T cells. The C terminus of SARS-CoV N (aa 362 to 422), but not the other truncated regions, was responsible for the TRIM25 association (Fig. 2A). Reciprocally, we also produced Flag-tagged TRIM25 truncated proteins containing the SPRY domain, B box/central coiled-coil domain (CCD), or RING domain and found that only the SPRY domain was involved in the SARS-CoV N association (Fig. 2B and C). Furthermore, a Flag-tagged SPRY domain with deleted TRIM25 (Flag-del-SPRY) failed to interact with the N protein (Fig. 2C). These findings clearly defined the interaction domains that contributed to the association of the two proteins.
FIG 2.
The interaction domain of the SARS-CoV N protein and TRIM25. (A) Anti-Flag immunoprecipitates prepared from lysates of 293T cells expressing Flag-TRIM25 and GFP-tagged full-length or truncated SARS CoV N protein were analyzed by immunoblotting with anti-Flag and anti-GFP. (B) Anti-Flag immunoprecipitates prepared from lysates of 293T cells expressing Myc-N protein and the Flag-B box/CCD or Flag-SPRY domain were analyzed by immunoblotting with anti-Flag and anti-Myc. (C) Anti-Flag immunoprecipitates prepared from lysates of 293T cells expressing the GFP-N or Flag-tagged TRIM25 domain were analyzed by immunoblotting with anti-Flag and anti-GFP.
The SARS-CoV N protein interferes with the TRIM25–RIG-I interaction.
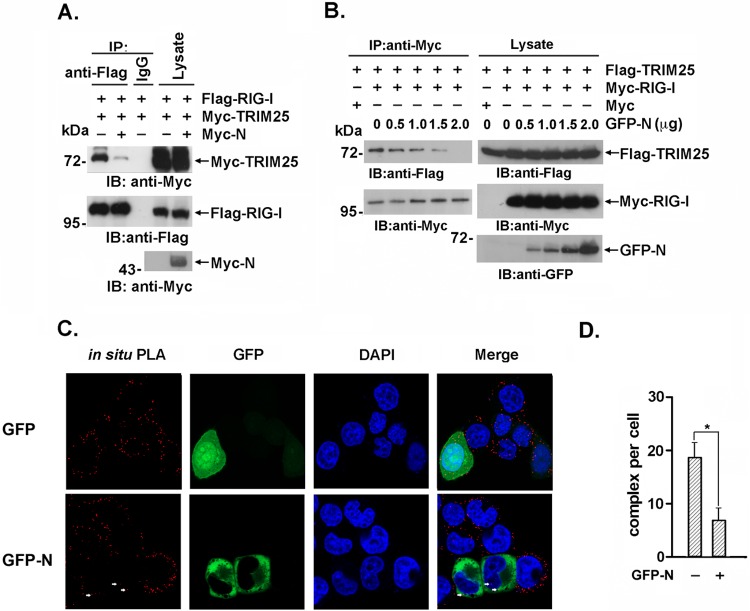
The TRIM25 SPRY and RING domains have been reported to be involved in the interaction with the RIG-I N-terminal CARDs and E2 ubiquitin-conjugating enzymes, respectively (22). Due to the association of the TRIM25 SPRY domain with the SARS-CoV N protein, we hypothesized that SPRY domain-mediated RIG-1 binding might be competitively regulated by the SARS-CoV N protein. To investigate this hypothesis, Flag–RIG-I and Myc-TRIM25 were coexpressed with or without Myc-N in 293T cells. The interaction between RIG-I and TRIM25 was substantially attenuated by coexpression of the SARS-CoV N protein in a dose-dependent manner (Fig. 3A and B). Additionally, the in situ PLA analysis demonstrated that the interaction between endogenous RIG-I and TRIM25 was decreased by exogenous SARS-CoV N expression but not by GFP expression (Fig. 3C and D). These results collectively indicated that the SARS-CoV N protein competitively bound to the TRIM25 SPRY domain and interfered with the association between TRIM25 and its ubiquitination substrate RIG-I.
FIG 3.
The interaction between TRIM25 and RIG-I is inhibited by the SARS-CoV N protein. (A) 293T cells were transfected with Flag–RIG-I and Myc-TRIM25 with or without Myc-N. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Flag or anti-Myc. (B) 293T cells were transfected with Flag-TRIM25, Myc-RIG-I, and the indicated amount of GFP-N. Anti-Flag immunoprecipitates were analyzed by immunoblotting with anti-Flag, anti-Myc, or anti-GFP. (C) HeLa cells expressing the GFP-N or GFP plasmid were fixed with 4% paraformaldehyde and incubated with anti-TRIM25 and anti-RIG-I antibodies. The in situ PLA assay was conducted as described in the text, and the results were imaged with a confocal microscope at magnification of ×100. Red dots indicate the interaction. (D) Red dots indicating positive PLA signals were counted in 30 randomly selected cells. The data are expressed as the means ± SD (*, P < 0.05).
The SARS-CoV N protein suppresses RIG-I ubiquitination.
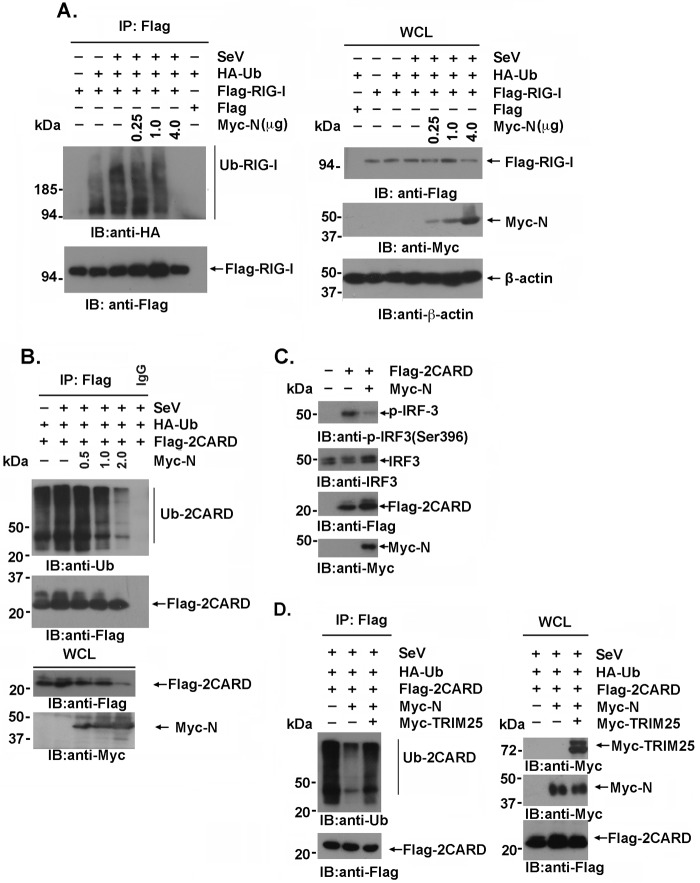
Next, we investigated whether TRIM25-mediated RIG-I ubiquitination at the N-terminal CARD was regulated by the SARS-CoV N protein. Lysates prepared from 293T cells cotransfected with Flag–RIG-I, hemagglutinin (HA)-ubiquitin, and the indicated amounts of the Myc-N expression plasmids were immunoprecipitated with an anti-Flag antibody and analyzed by immunoblotting with anti-HA or anti-Flag. As previously reported, TRIM25-mediated RIG-I ubiquitination was potentiated by SeV infection but was substantially suppressed by increasing SARS-CoV N protein expression (Fig. 4A) in a dose-dependent manner. In agreement with this result, the ubiquitination of the two CARDs of RIG-I was similarly inhibited by exogenous N protein expression (Fig. 4B). Phosphorylation of IRF3(Ser 396) plays an essential role in IRF-3 activation responses to virus infection (23). In concert, IRF3(S396) phosphorylation induced by the overexpression of the two CARD domains of RIG-I was suppressed by the SARS-CoV N protein (Fig. 4C). Notably, the suppression of RIG-I ubiquitination by the SARS-CoV N protein could be partially rescued by TRIM25 overexpression (Fig. 4D). These results indicated that competitive binding of TRIM25 by the SARS-CoV N protein interfered with TRIM25's association with its ubiquitination substrate, thereby inhibiting RIG-I ubiquitination and activation in a SARS-CoV N dose-dependent manner.
FIG 4.
TRIM25-mediated RIG-I ubiquitination is suppressed by the SARS-CoV N protein. (A, B, and D) 293T cells transfected with the indicated plasmids for 36 h were infected with or without SeV at an MOI of 2 for 12 h. The anti-Flag immunoprecipitates prepared from the cell extracts were analyzed by immunoblotting with the indicated antibodies. (C) 293T cells were transfected with the vector, Flag-2CARD, or Myc-N. At 24 h posttransfection, whole-cell lysates were subjected to immunoblotting with anti-IRF3 and anti-p-IRF3(S396) antibodies.
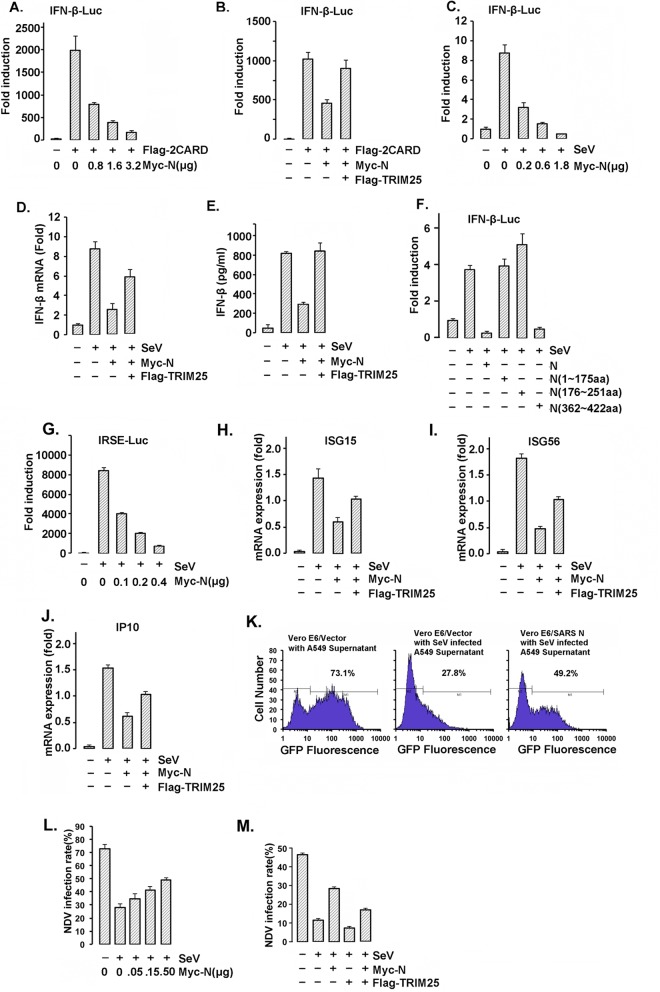
The SARS-CoV N protein suppresses type I IFN production.
RIG-I is activated by TRIM25-mediated ubiquitination, which plays a vital role in virus-induced type I IFN production. To examine the regulation of RIG-I activity by the SARS-CoV N protein, we constructed a luciferase reporter under the control of the IFN-β promoter (IFN-β–Luc) to quantify IFN-β promoter activation. In concert with the inhibition of RIG-I ubiquitination by the N protein, IFN-β promoter activation induced by RIG-I CARD domain overexpression was significantly inhibited by SARS-CoV N expression in a dose-dependent manner (Fig. 5A). However, coexpression of TRIM25 with SARS-CoV N counteracted the inhibition caused by the N protein (Fig. 5B). Similarly, IFN-β promoter activation induced by SeV infection was suppressed by the SARS-CoV N protein in a dose-dependent manner (Fig. 5C). Both the IFN-β transcription and expression levels could be suppressed by the N protein and rescued by TRIM25 overexpression (Fig. 5D and E). Next, we investigated whether the inhibition of IFN production by the N protein was dependent on the TRIM25 association. Only full-length N and the fragment from aa 362 to 422 that could interact with TRIM25, but not the other truncated fragments, were responsible for the suppression of IFN-β production (Fig. 5F). Accordingly, the transcriptional activity of the IFN-responsive sequence element (IRSE) and the transcription of IFN-stimulated gene 15 (ISG15), ISG56, and IP10, which greatly depend on the cellular IFN level, were similarly inhibited by the SARS-CoV N protein and partially recovered by TRIM25 overexpression (Fig. 5G to J).
FIG 5.
The SARS-CoV N protein inhibits TRIM25-mediated RIG-I activation and interferon production. (A, B, C, and F) 293T cells were cotransfected with the IFN-β-Luc firefly luciferase reporter plasmid, the Renilla luciferase control reporter plasmid pRL and the indicated plasmids for 36 h. The transfected cells were infected with or without SeV at an MOI of 2 for 12 h (C and F). The luciferase activity of the cell lysates was analyzed with the dual luciferase reporter assay system (Promega) and was measured on a Monolight 2010 luminometer. (D and E) A549 cells transfected with Myc-N and Flag-TRIM25 were infected with or without SeV at an MOI of 2 for 36 h. The mRNA was extracted, and the IFN-β mRNA expression level was determined by RT-PCR (D). The IFN-β concentrations in the cell culture supernatants were measured using a VeriKine human IFN-β enzyme-linked immunosorbent assay (ELISA) kit (E). (G) 293T cells cotransfected with the IRSE-Luc reporter plasmid, pRL, and the other indicated plasmids were infected with or without SeV at an MOI of 2 for 24 h. The luciferase activity of the cell lysates was analyzed with the dual luciferase reporter assay system and was measured using a Monolight 2010 luminometer. (H to J) Total RNA was extracted from A549 cells expressing the indicated plasmids. The ISG15, ISG56, and IP10 mRNA expression levels were determined by RT-PCR. All results in panels A to I are expressed as the means ± SD from three independent experiments (ns, nonsignificant; *, P < 0.05; **, P < 0.01, ***, P < 0.001). (K to M) A549 cells were transfected with 0.5 μg of Myc-vector (left and middle) or Myc-N (right) for 24 h. The cells were infected with SeV at an MOI of 2 for 16 h (middle and right) or not infected (left). Cell culture supernatants were harvested and incubated with Vero E6 cells for 2 h prior to infection of the Vero E6 cells with NDV-GFP at an MOI of 2. After 24 h, the GFP fluorescence of the Vero E6 cells was detected by flow cytometry. The cell culture supernatants were harvested and incubated with Vero E6 cells for 2 h prior to infection of the Vero E6 cells with NDV-GFP at an MOI of 2. The rate of NDV infection of the Vero E6 cells was measured by flow cytometry after 24 h of infection.
Because the SARS-CoV N protein suppressed the production of IFN induced by SeV, we next investigated whether viral infection and proliferation were regulated by the N protein. First, A549 cells exogenously expressing Myc-N or Myc-vector were infected (or not, in the control) with SeV for 16 h to induce IFN production. In accordance with the previous finding, SeV-induced IFN-β transcription in A549 cells was suppressed by the SARS-CoV N protein. IFN-β-sensitive Vero E6 cells were preincubated with the above-mentioned A549 cell culture supernatants for 4 h and then infected with GFP-expressing Newcastle disease virus (NDV-GFP). The proliferation of NDV-GFP in Vero E6 cells was determined by the GFP-positive cell number and the fluorescence intensity detected by flow cytometry. As shown in Fig. 5K, Vero E6 cells preincubated with SeV-infected A549 cell culture supernatants showed a significantly decreased NDV-GFP infection rate compared with Vero E6 cells preincubated with the normal A549 cell culture supernatant without SeV infection (∼73.1% versus ∼27.8%). This difference was due to the antiviral activity of the IFN-β released by the SeV-infected A549 cells (Fig. 5K, left and middle). In accordance with the finding that the N protein inhibits IFN production (Fig. 5A to E), a much higher NDV-GFP infection rate was observed in Vero E6 cells preincubated with the supernatant collected from SeV-infected A549 cells expressing the SARS-CoV N protein (∼49.2% versus 27.8%) (Fig. 5K). Additionally, the NDV infection rate was increased in an N protein dose-dependent manner (Fig. 5L) and was compromised when TRIM25 was coexpressed with the N protein (Fig. 5M). These results demonstrated that the SARS-CoV N-protein-mediated inhibition of IFN production relied on its host cell association partner TRIM25.
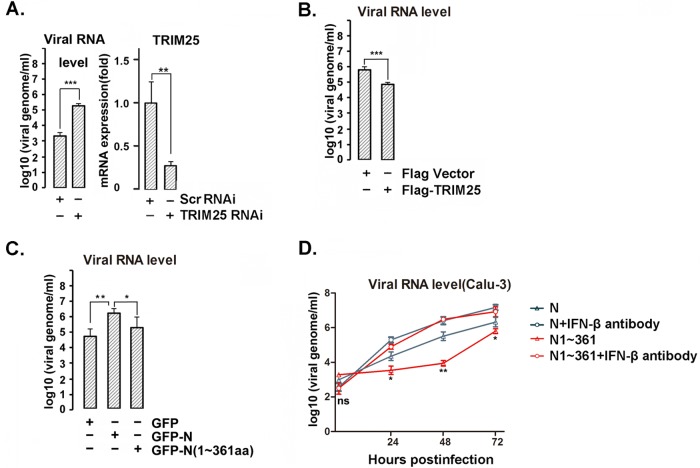
N protein promotes virus replication by inhibiting TRIM25-mediated IFN-β production.
Next, we investigated the function of TRIM25 in SARS-CoV infection and proliferation in human cells using the de novo-synthesized recombinant SARS-CoV strain v2163 (24). A549 cells were transfected with a TRIM25-targeting small interfering RNA (siRNA) or scrambled siRNA for 48 h, resulting in ∼70% TRIM25 knockdown (Fig. 6A). The siRNA-transfected cells were infected with recombinant SARS-CoV at a dose of 0.05 PFU per cell for 24 h. The virus particles released into the cell culture supernatant were collected, and the genomic RNA copy numbers were determined by quantitative PCR (qPCR). Compared with the scrambled siRNA, the TRIM25-targeting siRNA resulted in a significantly higher SARS-CoV titer in A549 cells (Fig. 6A). TRIM25 overexpression reduced SARS-CoV release in A549 cells (Fig. 6B). These results suggested that higher TRIM25 levels may compensate for the activity inhibition of RIG-I ubiquitination mediated by the viral N protein because the N protein suppressed RIG-I ubiquitination in a dose-dependent manner. Thus, a higher N protein/TRIM25 ratio should contribute more suppression. Further, A549 cells were transfected with full-length N protein or the N terminus (aa 1 to 361); 24 h later, cells were infected with SARS-CoV. At 24 h postinfection, the viral genomic RNA copy numbers were determined by qPCR. Overexpression of full-length N protein led to a higher viral genomic copy number than with the N protein N-terminal version in A549 cells (Fig. 6C). SARS-CoV can productively infect the human bronchial epithelium Calu-3 cell line and shows more sensitivity to IFN treatment (25, 26). To further study the effect of the N protein on SARS-CoV replication, Calu-3 cells were transfected with full-length N protein or its N-terminal truncation (aa 1 to 361), which do not interact with TRIM25. At 24 h posttransfection, cells were infected with SARS-CoV for the indicated times in the presence or absence of IFN-β-specific neutralization antibody at a concentration of 2.7 μg/ml. Compared with the N terminus of the N protein, the full-length N protein showed little, if any, effect on viral replication in the presence of the IFN-β-specific neutralization antibody, while a much higher viral titer was observed in the cells overexpressing full-length N protein in the absence of IFN-β antibody, suggesting that the overexpressed N protein promoted viral replication by suppressing IFN production (Fig. 6D). Unexpectedly, a lower viral titer was observed (data not shown) in the cells expressing the C-terminal N protein, possibly due to the incorporation of this protein into a nucleocapsid that may block the formation of functionally infectious virus, since the C-terminal N protein plays vital roles in N protein polymerization (14, 27).
FIG 6.
N protein promotes virus replication by inhibiting TRIM25-mediated IFN-β production. (A) A549 cells were transfected with TRIM25 siRNA or scramble siRNA. At 24 h posttransfection, the cells were infected with recombinant SARS-CoV at an MOI of 0.05 PFU per cell for 24 h. The virus particles released into the cell culture supernatants were determined by viral genomic RNA level as assayed by qPCR. The TRIM25 mRNA expression level was determined by RT-PCR (right). (B) A549 cells transfected with Flag-TRIM25 or Flag vector were infected with recombinant SARS-CoV as described for panel A. The virus particles released into the cell culture supernatants were quantified by viral genomic RNA level as assayed by qPCR. (C) A549 cells transfected with GFP vector, GFP-N, or GFP-N(1–361) were infected with SARS-CoV as described for panel A. The virus particles released into the cell culture supernatants were quantified by viral genomic RNA level as assayed by qPCR. (D) Calu-3 cells were transfected with full-length N protein or N(1–361), and 24 h after transfection, the cells were infected with mouse-adapted SARS-CoV at an MOI of 0.05 PFU per cell in the presence or absence of IFN-β-specific neutralizing antibody. Viral RNA from viral particles in the supernatant was quantified by qPCR. The results are expressed as the means ± SD from three independent experiments (ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). The significance of results for cells not treated with the antibody from panel D was analyzed using the t test.
MERS-CoV N protein interaction with TRIM25.
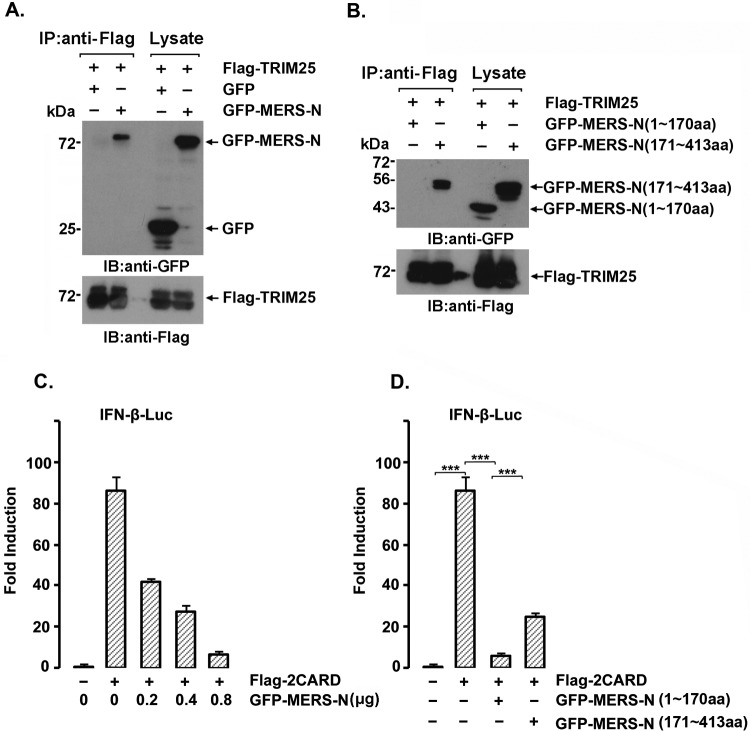
Middle East respiratory syndrome CoV (MERS-CoV) is a new human coronavirus, discovered in the Middle East in 2012, with high lethality and that is comparable to SARS-CoV in many aspects (28). The SARS-CoV N shares only 20 to 30% homology with the N proteins of other known coronaviruses (29) except SARS-like bat viruses, including HKU4, HKU5, and MERS-CoV (40 to 50%) (30, 31). To test whether MERS-CoV could interact with TRIM25 and suppress IFN-β expression, lysates from cells expressing Flag-TRIM25 and GFP-MERS-N or GFP as a control were subjected to immunoprecipitation with anti-Flag, and the immunoprecipitates were immunoblotted with anti-Flag and anti-GFP. GFP-MERS-N, but not GFP, coprecipitated with Flag-TRIM25 (Fig. 7A). Similar to the SARS-CoV N protein, the C-terminal part of the MERS-CoV N protein interacted with TRIM25 (Fig. 7B). The Flag-2CARD of RIG-I-induced IFN production was suppressed by the MERS-N protein in a dose-dependent manner (Fig. 7C). Unlike the case for the SARS-CoV N protein, both the N- and C-terminal portions of the MERS-CoV N protein suppressed RIG-I-induced IFN production (Fig. 7D), suggesting that the MERS-CoV N protein interferes with IFN production by interacting with TRIM25, similar to SARS-CoV and other unidentified pathways.
FIG 7.
The MERS-CoV N protein interacts with TRIM25 and inhibits RIG-I interferon signaling. (A and B) 293T cells were transfected with 2 μg of the indicated plasmids for 36 h. Whole-cell lysates were subjected to anti-Flag immunoprecipitation and immunoblotting with anti-Flag or anti-GFP antibodies. (C and D) 293T cells were cotransfected with the IFN-β-Luc firefly luciferase reporter plasmid, the Renilla luciferase control reporter plasmid pRL, Flag-2CARD, and increasing amounts of the GFP-MERS-N plasmid (C) or GFP-MERS-N (aa 1 to 170) or GFP-MERS-N (aa 171 to 413) (D) for 36 h. The luciferase activity of the cell lysates was analyzed with the dual luciferase reporter assay system (Promega) and was measured with a Monolight 2010 luminometer. The results are expressed as the means ± SD from three independent experiments.
DISCUSSION
Most human coronaviruses cause mild infections, but the outbreaks of SARS in 2003 and MERS in 2012 represented highly lethal coronavirus epidemics. SARS-CoV and MERS-CoV have similar genome structures, viral growth kinetics, and clinical presentations (26). Therefore, understanding the virulence mechanisms and microbe-host interactions of these pathogens will be beneficial for the development of effective therapies to control outbreaks of similar coronaviruses in the future (30, 32, 33).
Inhibition of IFN production and function plays key roles in virus escape from the innate immunity. Previous studies have shown that both SARS-CoV and MERS-CoV failed to elicit strong IFN expression during the innate immune response. Viral infection failed to induce transcription of type I interferon in peripheral blood mononuclear cells (PBMCs) derived from SARS patients and in MERS-CoV-infected ex vivo respiratory tissues (34, 35). Elderly patients, especially those who were immunocompromised, had bad outcomes from SARS and MERS infection (36). Several SARS-CoV-encoded proteins, such as nsp1, nsp7, nsp15, PLpro, M, open reading frame 3b (ORF3b), ORF6, and ORF9b, were found to antagonize IFN production by different pathways (37). The MERS-CoV 4a protein suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral responses (38, 39), and its M protein, ORF4a, ORF4b, and ORF5 inhibit the nuclear trafficking and activation of IRF3 and the activity of NF-κB and the ISRE promoter (40). Another well-studied coronavirus, the murine coronavirus mouse hepatitis virus (MHV), could induce type I interferon expression dependent on RIG-I recognition. Notably, the nucleocapsid protein of MHV, which shares ∼34% homology with that of SARS-CoV, is also regarded as a type I IFN antagonist by interfering with RNase L activity associated with the induction of 2′,5′-oligoadenylate synthetase (2′,5′-OAS (41).
Our work demonstrated that the N proteins of SARS-CoV could inhibit IFN-β production by interaction with TRIM25, thereby suppressing TRIM25-mediated RIG-I activation by ubiquitination, which provides in-depth insight into the mechanism of SARS-CoV-induced innate immune suppression. The MERS-CoV N protein was also demonstrated to interact with TRIM25 and to inhibit IFN-β promoter activity, which suggests that the N protein of MERS may be involved in IFN production in a way similar to that for SARS-CoV.
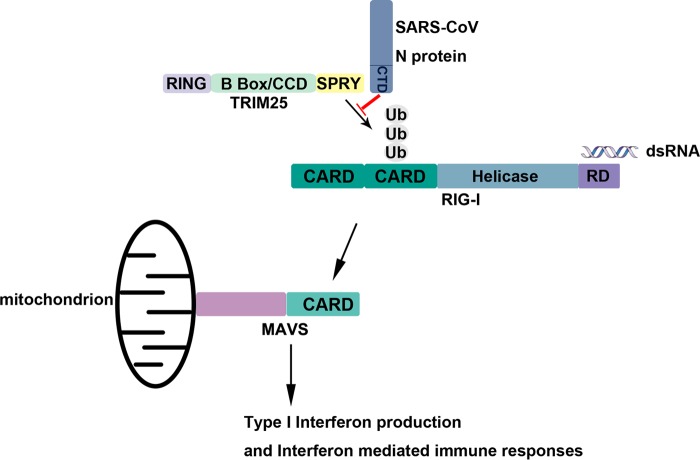
TRIM family proteins contain at least three domains (an N-terminal RING domain, one or two B boxes, and a central coiled-coil domain [CCD]). Most human TRIM proteins also contain a SPRY domain, which is involved in broad biological processes, including diverse cellular functions and antiviral and antimicrobial activity (42–44). As an ubiquitin E3 ligase, TRIM25 plays a crucial role in the RIG-I-mediated activation of the type I IFN pathway (22). TRIM25 acts as an E3 ubiquitin ligase in both MAVS ubiquitination and degradation (45). TRIM25 is also required for full NF-κB activation by ubiquitinating TRAF6 (46). Targeting TRIM25 is an efficient approach to inhibit the IFN signaling pathway. The NS1 protein of influenza A virus was reported to inhibit RIG-I activation by targeting the TRIM25 B box and CCD (4). In addition to the interactions of viral proteins, the subgenomic flavivirus RNA (sfRNA) of PR-2B serotype 2 (dengue virus serotype 2 [DENV-2]) binds to TRIM25, prevents its deubiquitylation, and results in the inhibition of RIG-I activation and, thus, IFN expression (47). Previous studies have suggested that the SARS-CoV N protein is capable of inhibiting IFN-β production by blocking the RIG-I signaling pathway, and the N protein of SARS-CoV was found to inhibit IRF3 activation induced by SeV (21, 48). Antagonism of IFN induction by the N protein was further mapped to the C terminus of the protein, while the N protein was shown not to interact with RIG-I and MDA5 (49). In this study, we show that the N protein of SARS-CoV suppressed IFN-β production by targeting the SPRY domain of TRIM25 and disturbing interaction between this SPRY domain and the N-terminal CARDs of RIG-I, resulting in the inhibition of RIG-I ubiquitination and downstream signaling and, ultimately, the production of type I interferon (Fig. 8). Considering that the inhibition of interferon production is mediated by the association of TRIM25 with N protein, abolition of inhibition should be observed after infection with a recombinant virus bearing a point mutant of the N protein that does not bind TRIM25, but it was not possible since no virus with such an N protein mutant could be successfully rescued because of the impaired functions.
FIG 8.
Schematic of N protein inhibiting type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination.
Proteins of SARS-CoV, including the N protein, suppress innate immunity by inhibiting IFN production other than by blocking IFN responses, suggesting that type I interferon might be employed for the treatment of early stage SARS infection. Administration of type I interferon in BALB/c mice before or at the initial stage of SARS-CoV infection resulted in a reduced virus titer, a limited inflammatory response, and mild clinical disease (50). Type I interferon treatment also improves the outcome of SARS-CoV and MERS-CoV infection in nonhuman primate models (51, 52) and has also been used in several clinical practices, together with other drugs (53–55).
In conclusion, our results provide evidence for the mechanism by which the SARS-CoV (and also MERS-CoV) N proteins inhibit RIG-I ubiquitination and thus suppress the release of type I IFN. The results help us to understand the pathogenicity of highly dangerous coronaviruses.
MATERIALS AND METHODS
Cell lines and viruses.
Cells (293T, HeLa, A549, Veros, and Calu-3) were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% (or 20% for Calu-3) heat-inactivated fetal bovine serum (FBS) (Gibco, Life Technologies, USA), 100 IU/ml of ampicillin, and 100 μg/ml of streptomycin. The cells were cultured in 5% CO2 and a humidified atmosphere at 37°C. The 293T and A549 cells were transfected using Vigofect reagent (Vigorous, Beijing, China) and jetPRIME reagent (Polyplus, New York, NY, USA) according to the manufacturer's instructions.
Sendai virus (SeV) and Newcastle disease virus with the green fluorescent protein gene incorporated into the genome (NDV-GFP) were amplified in 9- to 11-day embryonated specific-pathogen-free (SPF) eggs. The 50% tissue culture infectious dose (TCID50) in Vero cells was determined as previously described (56, 57).
DNA constructs.
Flag- or hemagglutinin (HA)-tagged protein genes were cloned into the pcDNA3-based expression vector (Invitrogen, Carlsbad, CA, USA). Enhanced GFP (EGFP)- or Myc-tagged protein genes were cloned into the pCMV-Myc expression vector (Clontech, Mountain View, CA, USA).
siRNAs targeting TRIM25 (siTRIM25) (sense, 5′-GCAAAUGUUCCCAGCACAATT-3′; antisense, 5′-UUGUGCUGGGAACAUUUGCTT-3′) and a nontarget siRNA (si-control) were purchased from Genepharm Technologies (Shanghai, China). All siRNA transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative RT-PCR.
RNA was extracted using an RNeasy minikit (Qiagen, Valencia, CA, USA), and 1 μg of RNA was used to synthesize cDNA using the ReverTra Ace qPCR RT reverse transcription (RT) master mix with genomic DNA (gDNA) remover (Toyobo, Osaka, Japan). Quantitative RT-PCR was performed using SYBR green Supermix (iQ Supermix; Bio-Rad, Hercules, CA, USA) with an iQ5 multicolor real-time PCR detection system (Bio-Rad). The primer sequences are shown in Table 1, and relative gene expression levels were calculated using the 2−ΔΔCT method (58).
TABLE 1.
Primers used in real-time PCR
| Gene product | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| β-Actin | TGACGTGGACATCCGCAAAG | CTGGAAGGTGGACAGCGAGG |
| IFN-α | CTGAATGACTTGGAAGCCTG | ATTTCTGCTCTGACAACCTC |
| IFN-β | GTCAGAGTGGAAATCCTAAG | ACAGCATCTGCTGGTTGAAG |
| IP10 | TCCCATCACTTCCCTACATG | TGAAGCAGGGTCAGAACATC |
| ISG15 | TCCTGGTGAGGAATAACAAGGG | CTCAGCCAGAACAGGTCGTC |
| ISG56 | TCGGAGAAAGGCATTAGATC | GACCTTGTCTCACAGAGTTC |
| TRIM25 | GACCACGGCTTTGTCATCTTC | AAAGTCCACCCTGAACTTATACATCA |
Immunoprecipitation and immunoblotting analysis.
Cell lysates were prepared in lysis buffer containing 1% Nonidet P-40 (59). Soluble proteins were subjected to immunoprecipitation with an anti-Flag antibody (M2, F2426; Sigma, St. Louis, MO, USA) or mouse IgG (A0910; Sigma). An aliquot of the total lysate (5%, vol/vol) was included as a control. The immunoblotting analysis was performed with anti-GFP (HRP-66002; Proteintech, Chicago, IL, USA), anti-TRIM25 (ab167154; Abcam, Cambridge, MA, USA), anti-Myc (SC-40; Santa Cruz, Santa Cruz, CA, USA), anti-Flag horseradish peroxidase (HRP) (A8592; Sigma), anti-β-actin (SC-1616; Santa Cruz), antiubiquitin (SC-8017; Santa Cruz), anti-HA (H9658; Sigma), anti-IRF3 (ab68481; Abcam), or anti-pIRF3(S396) (b138449; Abcam) antibodies. The antigen-antibody complexes were visualized using ECL chemiluminescence (Amersham Biosciences/GE Healthcare, Buckinghamshire, UK). On average, three independent experiments were performed, with standard deviations (SD) (error bars in figures) and P values determined by the t test.
LC-MS/MS analysis.
Plasmids expressing the Flag-tagged SARS-CoV N protein (Flag-N) were transiently transfected into 293T cells. Twenty-four hours after transfection, cell lysates were prepared and subjected to anti-Flag immunoprecipitation, SDS-PAGE, and Coomassie brilliant blue staining. Protein bands that coprecipitated with Flag-N but not control IgG were excised, digested with trypsin, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Micromass, Inc.) analysis to identify interacting proteins. The data were compared against Swiss-Prot using the Mascot search engine (Matrix Science) for protein identification.
NDV-GFP infection assay.
The GFP-expressing Newcastle disease virus (NDV-GFP) was used to infect cells for the previously described infection efficiency analysis (57). First, A549 cells transfected with or without SARS-CoV N protein were infected with SeV to induce IFN production. Vero cells were incubated with supernatants from the A549 cell culture and were then infected with NDV-GFP at the given viral titers. At 16 h postinfection, the number of GFP-positive cells and the GFP expression levels were analyzed by flow cytometry (BD FACSCalibur).
Duolink assay and confocal microscopy.
The Duolink in situ proximity ligation assay (PLA) (Duolink detection kit; Olink Bioscience, Uppsala, Sweden) was used to detect interactions between endogenous TRIM25 and the N protein or endogenous TRIM25 and RIG-I in the cells. Briefly, HeLa cells plated on glass coverslips were transfected with the GFP-N-expressing plasmid. After fixation with 4% formaldehyde, the cells were incubated with rabbit anti-RIG-I (4200; Millipore, Billerica, MA, USA) or rabbit anti-GFP (ab183734; Abcam, Cambridge, MA, USA) and mouse anti-TRIM25 (ab88669; Abcam) primary antibodies or with anti-TRIM25 antibody alone as a control. The Duolink system provides oligonucleotide-labeled secondary antibodies (PLA probes) for each of the primary antibodies. In combination with a DNA amplification-based reporter system, these antibodies generate a signal only when the two primary antibodies are in close proximity (∼10 nm). According to the manufacturer's instructions, the signal from each detected pair of primary antibodies was visualized as a fluorescent spot. Slides were evaluated using an LSM 510 Meta confocal microscope (Carl Zeiss). The cell images were exported in TIF format using the Zeiss LSM Image Browser (Carl Zeiss) for analysis. The number of interactions per cell was determined with the Duolink image tool developed by Olink Biosciences, and interactions were counted in at least three fields. Quantifications are given as the means ± SD. Representative results from experiments repeated three times are shown.
Dual luciferase assays.
To evaluate expression, 293T cells were seeded into 24-well plates and cotransfected with control plasmids or expression plasmids together with the luciferase reporter plasmid. A 10-ng sample of the pRL Renilla luciferase control reporter vector (pRL) was included as the control. Twenty-four hours after transfection, the cells were infected with Sendai virus (SeV) at a multiplicity of infection (MOI) of 2 for 16 h. The cell lysates were subjected to luciferase activity analysis using the dual luciferase reporter assay system (Promega). Total light production was measured using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA, USA). The results are expressed as the means ± SD from three independent experiments.
Virus replication assays.
A549 cells were seeded at a density of 2 × 106 cells per 25-cm2 flask (Corning, Corning, NY), and Vero and Calu-3 cells were seeded in 24-well plates (Corning, Corning, NY). After 24 h, the cells were washed with 1× phosphate-buffered saline (PBS) and incubated with medium containing SARS-CoV at an MOI of 0.05 PFU per cell (in each cell type) in a 1-ml volume for 45 min at 37°C. The medium was removed, the cells were washed twice with 1× PBS, and medium containing 2% FBS was added. Anti-IFN-β antibody (ab6979; Abcam, Cambridge, MA, USA) was used to neutralize IFN-β in Calu-3 cells at a concentration of 2.7 μg/ml. At the indicated time points postinfection, supernatant from each flask was subjected to RNA extraction using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcription using the ReverTra Ace qPCR RT master mix (FSQ-201; Toyobo) with the reverse primer given below. The Tiangen SuperReal premix (Probe) was used for quantitative nucleic acid amplification with specific primers in a Bio-Rad IQ5 optical system. Threshold cycle (CT) values were compared to those of a series of dilutions of standard samples. The primers used for quantifying SARS-CoV subgenomic mRNA 5 (M gene) are as follows: forward primer, 5′-CTCTTCTGAAGGAGTTCCTGAT-3′; reverse primer, 5′-GACAGCAGCAAGCACAAAACAA-3′; and 6-carboxyfluorescein (FAM) probe, 5′-GGCTCTTGTGGCCAGTAACACTT-3′. Viral titers in the supernatants were calculated as previously described using RT-PCR (60, 61).
All experiments with SARS-CoV were performed in a biosafety level 3 laboratory.
ACKNOWLEDGMENTS
This investigation was supported by grants 2016YFC1202400 and 2012CB518900 awarded by the Ministry of Science and Technology of China and by grants 30871240 and 81550001 awarded by the National Natural Science Foundation of China.
We declare no conflicts of interest.
Yong Hu, Wei Li, Ting Gao, and Yan Cui performed the experiments. Qingjun Ma, Xuan Liu, and Cheng Cao designed and supervised the research. Yanwen Jin and Ping Li developed the protocols and provided reagents.
REFERENCES
- 1.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh C-S, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 2.Chan YK, Gack MU. 2016. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 4.Gack MU, Albrecht RA, Urano T, Inn K-S, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, García-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang F, Ramanathan A, Miller MT, Tang G-Q, Gale M, Patel SS, Marcotrigiano J. 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katze MG, He Y, Gale M Jr. 2002. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 10.Lai MM. 2003. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci 10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, Pickles RJ, Corti D, Johnston RE, Baric RS, Denison MR. 2008. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci U S A 105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, Hsiao CD, Huang TH. 2007. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol 368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 14.Surjit M, Liu B, Kumar P, Chow VT, Lal SK. 2004. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem Biophys Res Commun 317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He R, Leeson A, Ballantine M, Andonov A, Baker L, Dobie F, Li Y, Bastien N, Feldmann H, Strocher U, Theriault S, Cutts T, Cao J, Booth TF, Plummer FA, Tyler S, Li X. 2004. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res 105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatakeyama S, Matsuoka Y, Ueshiba H, Komatsu N, Itoh K, Shichijo S, Kanai T, Fukushi M, Ishida I, Kirikae T, Sasazuki T, Miyoshi-Akiyama T. 2008. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology 380:99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, Hao Q, Mu Y, Timani KA, Ye L, Zhu Y, Wu J. 2006. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int J Biochem Cell Biol 38:1417–1428. doi: 10.1016/j.biocel.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Wang Q, Li C, Zhang Q, Wang T, Li J, Guan W, Yu J, Liang M, Li D. 2010. Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42. Virol J 7:99. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frieman M, Heise M, Baric R. 2008. SARS coronavirus and innate immunity. Virus Res 133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CK, Hsu YL, Chang YH, Chao FA, Wu MC, Huang YS, Hu CK, Huang TH. 2009. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J Virol 83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 23.Servant MJ, Grandvaux N, ten Oever BR, Duguay D, Lin R, Hiscott J. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem 278:9441–9447. doi: 10.1074/jbc.M209851200. [DOI] [PubMed] [Google Scholar]
- 24.Day CW, Baric R, Cai SX, Frieman M, Kumaki Y, Morrey JD, Smee DF, Barnard DL. 2009. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology 395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumaki Y, Day CW, Wandersee MK, Schow BP, Madsen JS, Grant D, Roth JP, Smee DF, Blatt LM, Barnard DL. 2008. Interferon alfacon 1 inhibits SARS-CoV infection in human bronchial epithelial Calu-3 cells. Biochem Biophys Res Commun 371:110–113. doi: 10.1016/j.bbrc.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, Becker S, Weber F. 2013. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol 87:5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Chen J, Chen K, Shen X, Jiang H. 2006. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry 45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 28.Durai P, Batool M, Shah M, Choi S. 2015. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 30.Lu G, Liu D. 2012. SARS-like virus in the Middle East: a truly bat-related coronavirus causing human diseases. Protein Cell 3:803–805. doi: 10.1007/s13238-012-2811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu B, Ge X, Wang LF, Shi Z. 2015. Bat origin of human coronaviruses. Virol J 12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, Snijder EJ, Canard B, Imbert I. 2014. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda-Okubo Y, Barnard D, Ong CH, Peng BH, Tseng CT, Petrovsky N. 2015. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J Virol 89:2995–3007. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reghunathan R, Jayapal M, Hsu LY, Chng HH, Tai D, Leung BP, Melendez AJ. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol 6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, Guan Y, Poon LL, Baric RS, Nicholls JM, Peiris JS. 2013. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol 87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham RL, Donaldson EF, Baric RS. 2013. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. 2013. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One 8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PC, Yuen KY, Jin DY. 2014. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J Virol 81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munir M. 2010. TRIM proteins: another class of viral victims. Sci Signal 3:jc2. [DOI] [PubMed] [Google Scholar]
- 43.Nisole S, Stoye JP, Saib A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 44.Ozato K, Shin DM, Chang TH, Morse HC III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10:44. doi: 10.1186/1741-7007-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee NR, Kim HI, Choi MS, Yi CM, Inn KS. 2015. Regulation of MDA5-MAVS antiviral signaling axis by TRIM25 through TRAF6-mediated NF-kappaB activation. Mol Cell 38:759–764. doi: 10.14348/molcells.2015.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, van Amerongen G, van Riel D, de Jong T, Itamura S, Chan KH, Tashiro M, Osterhaus AD. 2004. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med 10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan JF, Yao Y, Yeung ML, Deng W, Bao L, Jia L, Li F, Xiao C, Gao H, Yu P, Cai JP, Chu H, Zhou J, Chen H, Qin C, Yuen KY. 2015. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, Deif H, LaMere EA, Chang M, Kain KC, Farcas GA, Ferguson P, Latchford M, Levy G, Dennis JW, Lai EK, Fish EN. 2003. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Z, Zhang F, Xu M, Huang K, Zhong W, Cai W, Yin Z, Huang S, Deng Z, Wei M, Xiong J, Hawkey PM. 2003. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J Med Microbiol 52:715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 55.Al-Tawfiq JA, Momattin H, Dib J, Memish ZA. 2014. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis 20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bitzer M, Lauer U, Baumann C, Spiegel M, Gregor M, Neubert WJ. 1997. Sendai virus efficiently infects cells via the asialoglycoprotein receptor and requires the presence of cleaved F0 precursor proteins for this alternative route of cell entry. J Virol 71:5481–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol 77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Cao C, Leng Y, Kufe D. 2003. Catalase activity is regulated by c-Abl and Arg in the oxidative stress response. J Biol Chem 278:29667–29675. doi: 10.1074/jbc.M301292200. [DOI] [PubMed] [Google Scholar]
- 60.Mossel EC, Wang J, Jeffers S, Edeen KE, Wang S, Cosgrove GP, Funk CJ, Manzer R, Miura TA, Pearson LD, Holmes KV, Mason RJ. 2008. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gillim-Ross L, Taylor J, Scholl DR, Ridenour J, Masters PS, Wentworth DE. 2004. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J Clin Microbiol 42:3196–3206. doi: 10.1128/JCM.42.7.3196-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]