ABSTRACT
Segment reassortment and base mutagenesis of influenza A viruses are the primary routes to the rapid evolution of high-fitness virus genotypes. We recently described a predominant G57 genotype of avian H9N2 viruses that caused countrywide outbreaks in chickens in China during 2010 to 2013, which led to the zoonotic emergence of H7N9 viruses. One of the key features of the G57 genotype is the replacement of the earlier A/chicken/Beijing/1/1994 (BJ/94)-like M gene with the A/quail/Hong Kong/G1/1997 (G1)-like M gene of quail origin. We report here the functional significance of the G1-like M gene in H9N2 viruses in conferring increased infection severity and infectivity in primary chicken embryonic fibroblasts and chickens. H9N2 virus housing the G1-like M gene, in place of the BJ/94-like M gene, showed an early surge in viral mRNA and viral RNA (vRNA) transcription that was associated with enhanced viral protein production and with an early elevated release of progeny virus comprising largely spherical rather than filamentous virions. Importantly, H9N2 virus with the G1-like M gene conferred extrapulmonary virus spread in chickens. Five highly represented signature amino acid residues (37A, 95K, 224N, and 242N in the M1 protein and 21G in the M2 protein) encoded by the prevalent G1-like M gene were demonstrated to be prime contributors to enhanced infectivity. Therefore, the genetic evolution of the M gene in H9N2 virus increases reproductive virus fitness, indicating its contribution to the rising virus prevalence in chickens in China.
IMPORTANCE We recently described the circulation of a dominant genotype (genotype G57) of H9N2 viruses in countrywide outbreaks in chickens in China, which was responsible, through reassortment, for the emergence of H7N9 viruses that cause severe human infections. A key feature of the genotype G57 H9N2 virus is the presence of the quail-origin G1-like M gene, which had replaced the earlier BJ/94-like M gene. We found that H9N2 virus with the G1-like M gene, but not the BJ/94-like M gene, showed an early surge in progeny virus production and more severe pathology and extrapulmonary virus spread in chickens. Five highly represented amino acid residues in the M1 and M2 proteins derived from the G1-like M gene were shown to mediate enhanced virus infectivity. These observations enhance what we currently know about the roles of reassortment and mutations in virus fitness and have implications for assessing the potential of variant influenza viruses that can cause a rising prevalence in chickens.
KEYWORDS: chicken, H9N2 virus, M gene, reassortment, replication
INTRODUCTION
Avian H9N2 and H5N1 influenza A viruses are two major globally circulating subtypes in poultry populations (1, 2). Unlike highly pathogenic H5N1 viruses, the low-pathogenicity nature of H9N2 viruses had attracted less attention in disease management and public health controls (3). In 2013, a novel reassortant H7N9 virus carrying six internal genes from avian H9N2 influenza virus caused serious outbreaks in humans in China (4, 5), which led to intense scrutiny of the evolution of H9N2 viruses. We previously demonstrated that H9N2 viruses of genotype G57 have become predominant in chickens since 2010 and that genotype G57-type viruses, with enhanced infectivity and antigenic drift, had caused nationwide outbreaks in chicken flocks during 2010 to 2013 (6). The increased prevalence of H9N2 viruses in chickens has directly contributed to the emergence in humans of H7N9 and other novel reassortants with H9N2-like segments (6). Furthermore, prevalent chicken H9N2 isolates are able to preferentially bind to the human-type sialic acid receptor and to be transmitted between ferrets by respiratory droplets (3). Presently, H9N2 viruses continue to cause mild infections in humans in China and other countries based on etiological and serological evidence (7–14). In 2013 to 2016, 18 human cases were laboratory confirmed in China, while only 10 cases were reported during the previous 14 years of 1999 to 2012 (http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/). This evidence suggests that the prevalent H9N2 virus poses an increasing threat to human health.
H9N2 influenza viruses are enzootic in poultry in several Asian and Middle Eastern countries (http://www.who.int/influenza/vaccines/virus/characteristics_virus_vaccines/en/). Phylogenetic analysis revealed that multiple lineages of H9N2 viruses have been circulating, including A/chicken/Beijing/1/1994 (BJ/94)-like, A/quail/Hong Kong/G1/1997 (G1)-like, and A/duck/Hong Kong/Y439/1997 (Y439)-like viruses (15, 16). Since the first isolation of BJ/94-like and G1-like viruses in China in the mid-1990s, the two lineages have become predominant in chickens and quail, respectively, indicating relative host restriction (17, 18). During the cocirculation of BJ/94-like and G1-like H9N2 viruses in poultry, genetic reassortments between the two lineages were observed (15). Several virus segments, including PB2, PB1, PA, and M, from G1-like viruses have been introduced into BJ/94-like viruses, but only the G1-like M gene is firmly established in BJ/94-like viruses (6, 15). The stable replacement of the BJ/94-like M gene with the G1-like M gene since 2004 is one key change in the generation of the G57 genotype of H9N2 influenza viruses (6). Thus, the main question is whether the G1-like M gene confers a replication advantage to H9N2 viruses that contributes to their increasing prevalence in chickens in China since 2010.
The M gene performs multiple roles in the life cycle of influenza A virus through encoding the matrix protein (M1) and the proton channel protein (M2) (19, 20). M1 is the most abundant viral protein responsible for the structural shell of the virus linking the viral envelope with the nucleocapsid (19); it is involved in the shuttling of the viral ribonucleotide protein (vRNP) complex between the nucleus and cytoplasm during viral replication (21–23). M2 is an integral membrane protein inserted into the viral envelope and possesses proton channel activity (20); it is thought to function at an early stage of the virus life cycle (24–26). Once virions have undergone endocytosis, the M2 proton channel is believed to permit the flow of protons from the endosome into the virion interior, which promotes vRNP release into the cytoplasm (24). In addition, the M1 and M2 proteins play an important role in influenza virus assembly and budding and are key determinants of virus morphology (19). Changes in the M1 and M2 genes have been found to be critical for viral replication and the pathogenicity of avian H5N1, seasonal human H1N1 and H3N2, and pandemic H1N1/2009 viruses in avian or mammalian cells and hosts (27–29).
In this study, we examined the preferential selection of the M gene from the G1-like lineage in place of the BJ/94-like lineage in avian H9N2 viruses in chickens. H9N2 virus with the G1-like M gene, relative to the presence of the BJ/94-like M gene, showed early and enhanced replication in chicken embryo fibroblasts (CEFs) and chickens, extrapulmonary virus spread, and a changed viral morphology, which collectively indicate increased virus fitness. Five dominant amino acid residues in the M1 and M2 proteins encoded by G1-like M genes are identified as being critical contributors to enhanced virus replication.
RESULTS
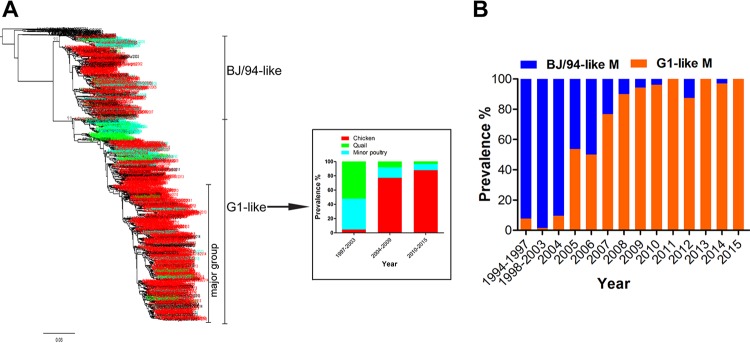
Increased prevalence of the G1-like M gene segment in chicken H9N2 viruses in China.
Previous studies revealed that H9N2 viruses with G1-like M genes were present mainly in quails and to a lesser extent in other minor poultry, but few G1-like M genes were isolated from chicken H9N2 viruses (15, 16). To better understand the current circulation of G1-like M genes in different poultry populations in China, we performed a phylogenetic analysis of all available sequences from 1997 to 2015. As shown in Fig. 1A, H9N2 viruses with G1-like M genes were predominantly present (95.52%) in quail and other minor poultry in 1997 to 2003 but were uncommon (4.48%) in chickens. In 2004 to 2009, G1-like M genes had reassorted with BJ/94-like M genes in chicken H9N2 viruses (the hemagglutinin [HA] gene belonged to the BJ/94-like lineage), leading to the proportion of G1-like M genes increasing to 77.05%. Since 2010, G1-like M genes of H9N2 viruses from chickens have been detected in 87.79% of isolates, but those of quail and minor poultry isolates were found in only 12.21% of the H9N2 isolates with G1-like M genes; the G1-like M gene remained the predominant lineage in quail and minor poultry.
FIG 1.
Prevalence of the G1-like M gene in chicken, quail, and minor poultry (A) and prevalence of BJ/94-like and G1-like M genes in chicken H9N2 viruses (B) in China. The phylogenetic tree of the M gene was generated with all available M sequences from H9N2 viruses isolated from various hosts in China during the period of 1994 to 2015. In the tree, the names of viruses from chicken, quail, minor poultry, and others are shown in red, green, blue, and black, respectively. For each column from left to right, the actual numbers of virus isolates are 67, 183, and 475 (A) and 13, 141, 21, 41, 16, 30, 40, 53, 53, 64, 88, 189, 33, and 6 (along the x axis) (B). In panel B, all HA genes of chicken H9N2 viruses belonged to the BJ/94-like lineage; the G1-like M gene has become predominant in the BJ/94-like viruses through reassortment.
We further examined the dynamic prevalence of BJ/94-like and G1-like M genes in chicken H9N2 viruses. In 1994, H9N2 viruses with the BJ/94-like M gene segment were first isolated from chickens; this gene segment remained dominant at an ∼97% frequency in this host until 2004 (Fig. 1B). H9N2 viruses with G1-like M genes were found in some chickens in 1997 to 2004, but by 2005, the year after reassortment, the rate of detection of the G1-like M gene in chicken H9N2 viruses had increased sharply. Since 2007, the G1-like M gene-containing H9N2 virus replaced the BJ/94-like M gene-containing virus as the dominant (94.95%) genotype in chickens. These findings suggest that the segmental replacement of the BJ/94-like gene with the G1-like M gene could be a significant adaptation of H9N2 viruses that confers improved infection fitness in chickens.
The G1-like M gene confers early elevated levels of viral mRNA transcription, vRNA production, and protein expression.
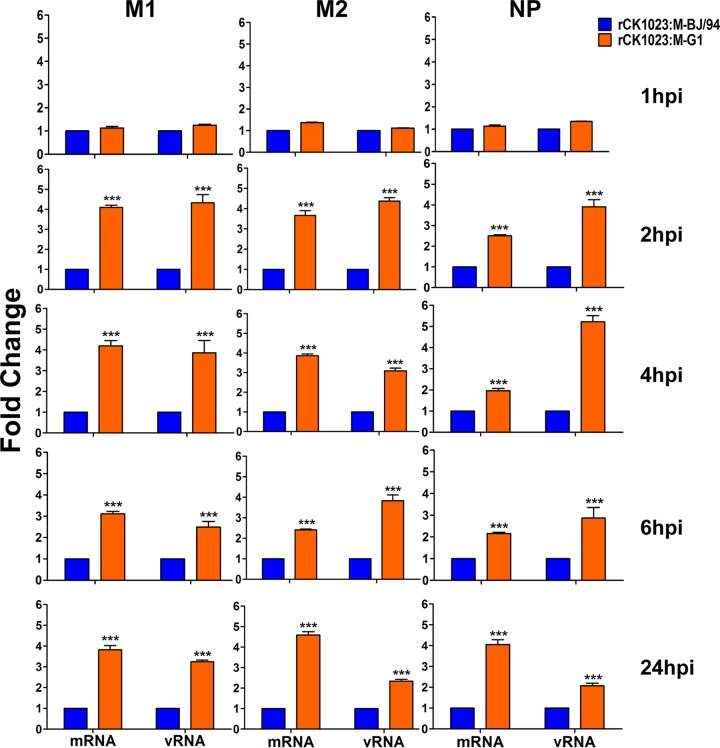
To address the viral fitness hypothesis, we first determined if the replacement of the BJ/94-like gene with the G1-like M gene affects viral infection in terms of viral mRNA transcription and viral RNA (vRNA) production. We produced the virus rCK1023:M-BJ/94 from a wild-type H9N2 isolate (A/chicken/Shandong/Lx1023/2007 [Lx1023]) that contained a BJ/94-like M gene and another virus, rCK1023:M-G1, based on the same H9N2 virus backbone with only the M gene replaced with the G1-like M segment from A/chicken/Jiangsu/TS/2010 (TS). Levels of viral transcription (mRNA) and genomic replication (vRNA) were determined in CEFs separately infected with the rCK1023:M-BJ/94 and rCK1023:M-G1 viruses for 1, 2, 4, 6, and 24 h by real-time PCR. As shown in Fig. 2, the rCK1023:M-G1 virus produced significantly higher levels of viral M1, M2, and nucleoprotein (NP) mRNAs and vRNA, from as early as 2 h postinoculation (hpi) onwards, than did the rCK1023:M-BJ/94 virus (P < 0.05). Thus, compared to the BJ/94-like M gene, the G1-like M gene in H9N2 virus enhanced early viral mRNA and vRNA transcription in CEFs.
FIG 2.
Relative expression levels of viral M1, M2, and NP mRNAs and vRNA of the rCK1023:M-G1 and rCK1023:M-BJ/94 H9N2 viruses in CEFs. CEFs were infected with the indicated H9N2 viruses at an MOI of 0.01 for 1, 2, 4, and 6 h or at an MOI of 0.001 for 24 h. mRNA and vRNA expression levels are presented as fold changes relative to the values for the rCK1023:M-BJ/94 virus. Data are presented as means ± standard deviations of results from three independent experiments. Statistical significance was based on two-way ANOVA (***, P < 0.001).
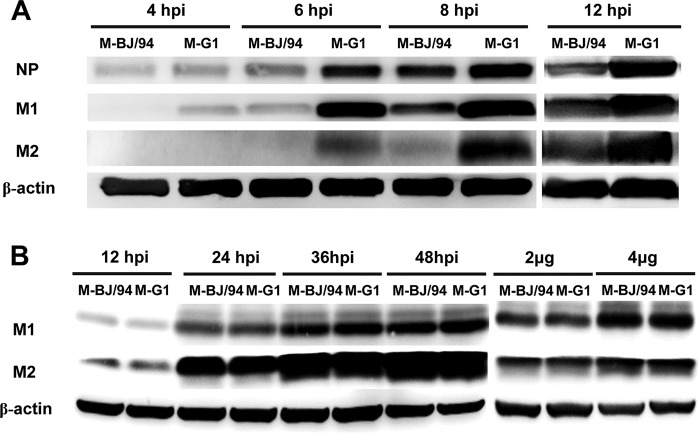
We next examined viral protein expression in CEFs separately infected with the rCK1023:M-BJ/94 and rCK1023:M-G1 viruses at 4, 6, 8, and 12 hpi (Fig. 3A). The M1 and M2 proteins of rCK1023:M-G1 were detected at 4 and 6 hpi, respectively, while those of rCK1023:M-BJ/94 were found 2 h later. At each time point, rCK1023:M-G1 produced higher levels of the M1, M2, and NP proteins than did rCK1023:M-BJ/94. Thus, the accumulation of the M1, M2, and NP proteins was earlier and higher during infection with the rCK1023:M-G1 virus than with the corresponding rCK1023:M-BJ/94 virus. To examine if differences between the G1-like and BJ/94-like M genes alone could affect protein production, 293T cells were transfected with individual M gene expression plasmids derived from BJ/94-like and G1-like M gene segments. Western blotting at 12, 24, 36, and 48 h posttransfection or at 36 h posttransfection with a plasmid dose of 2 μg or 4 μg showed no significant difference in M1 and M2 protein expression levels between the two M genes (Fig. 3B). Taken together, the enhanced viral mRNA transcription conferred by the G1-like M gene segment was translated into earlier and higher-level accumulation of H9N2 viral proteins.
FIG 3.
Replacement of the BJ/94-like M gene with the G1-like M gene in avian H9N2 virus increases viral protein expression. (A) CEFs were infected with the rCK1023:M-BJ/94 or rCK1023:M-G1 virus at an MOI of 0.1. Cells were harvested at the indicated time points, and Western blotting was performed on cell lysates to detect the viral NP, M1, and M2 proteins. The panels showing viral protein expression at 4, 6, and 8 hpi and that at 12 hpi are photos taken from different gels. (B) 293T cells were separately transfected with M gene expression plasmids derived from field H9N2 isolates Lx1023 and TS, which housed BJ/94-like and G1-like M gene segments, respectively. At 12, 24, 36, and 48 h posttransfection, cell lysates were harvested for Western blotting. Comparative M gene protein expression analysis was conducted at 36 h posttransfection with 2 μg or 4 μg of individual plasmids.
The G1-like M gene results in an increased frequency of spherical virions released from CEFs.
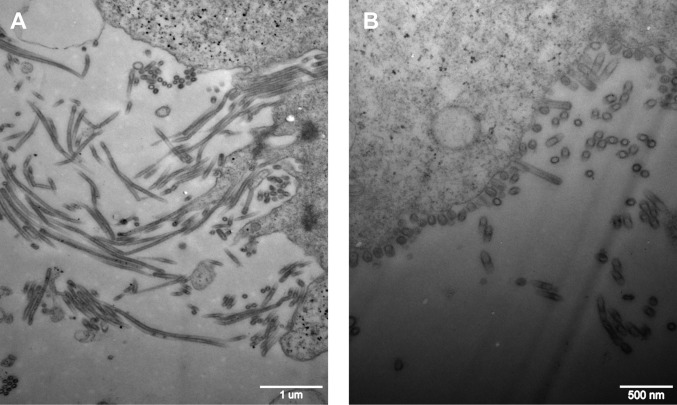
The influenza virus M1 and M2 proteins have been implicated in the determination of variation in virus morphology (19). Shorter spherical virus particles have been associated with a higher output of avian progeny virus (30). We compared the morphologies of the nascent rCK1023:M-BJ/94 and rCK1023:M-G1 viruses from CEFs by transmission electron microscopy (Fig. 4). About 61.90% of rCK1023:M-BJ/94 virus particles were morphologically filamentous, whereas rCK1023:M-G1 virions were predominantly spherical or ovoid (96.05%), with only 3.95% filamentous particles. Thus, the G1-like M gene segment specified a spherical morphological phenotype, which might have facilitated virus replication and shedding of the rCK1023:M-G1 virus from CEFs.
FIG 4.
Transmission electron micrographs of negatively stained H9N2 virus particles housing the BJ/94-like (A) or G1-like (B) M gene. CEFs were infected at an MOI of 3.0 for 15 h. rCK1023:M-BJ/94 progeny were mainly filamentous, and rCK1023:M-G1 particles were primarily spherical/ovoid.
The G1-like M gene in H9N2 virus confers early replication in CEFs and increased virus output.
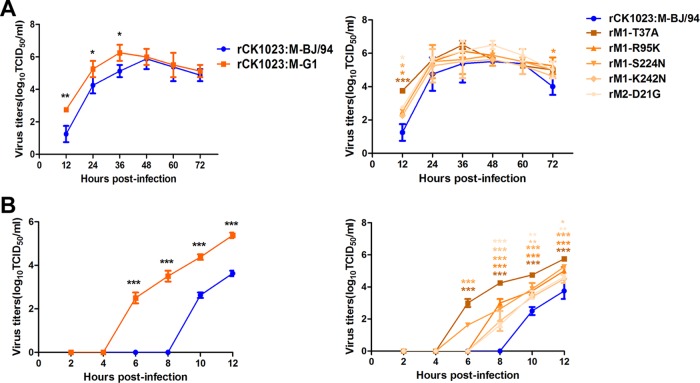
To compare the effects of different M gene lineages on viral replication in vitro, multistep replication kinetics assays of the rCK1023:M-BJ/94 and rCK1023:M-G1 viruses were performed with a multiplicity of infection (MOI) of 0.001 in CEFs over a duration of 72 h. The level of rCK1023:M-G1 virus production was up to 10-fold higher than that for the rCK1023:M-BJ/94 virus from 12 to 36 hpi (P < 0.05) (Fig. 5A, left). We further conducted single-replication-cycle kinetics analyses at an MOI of 0.01 over 12 h of infection (Fig. 5B, left). The rCK1023:M-G1 virus generated progeny at levels that were around 60-fold higher than those produced by the rCK1023:M-BJ/94 virus from 6 to 12 hpi. Notably, the rCK1023:M-G1 progeny virus output was detected earlier, by 6 hpi, about 4 h ahead of the detection of rCK1023:M-BJ/94 progeny virus. Taken together, the G1-like M gene in H9N2 virus conferred an earlier release and a higher overall output of progeny virus from CEFs.
FIG 5.
Virus output of rCK1023:M-G1, rCK1023:M-BJ/94, and M gene point mutants of rCK1023:M-BJ/94 H9N2 viruses from infected CEFs. (A) Multistep growth curves of H9N2 viruses from CEFs inoculated at an MOI of 0.001. (B) Single replication cycle of H9N2 viruses in CEFs inoculated at an MOI of 0.01. Virus titers were determined from supernatants collected at the indicated time points. Statistical significance was based on two-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The G1-like M gene in H9N2 virus confers increased severity and early onset of infection in chickens.
Typically, the avian H9N2 virus is mildly pathogenic in chickens; its replication is confined largely to the upper respiratory tract, causing little or no overt clinical signs, especially in specific-pathogen-free (SPF) chickens (6). The pathogenicity and replication of the rCK1023:M-BJ/94 and rCK1023:M-G1 H9N2 viruses were evaluated in SPF chickens. All chickens infected with the rCK1023:M-BJ/94 or rCK1023:M-G1 virus showed no overt clinical signs. Lung and kidney tissues were collected at 3 days postinoculation (dpi) for histopathological examination. rCK1023:M-BJ/94 virus-infected lungs showed mild inflammatory changes and bronchitis (Fig. 6A). The rCK1023:M-G1 virus, on the other hand, showed evidence of more severe inflammation, with interstitial pneumonia and bronchopneumonia, characterized by alveolar interstitial consolidation, extensive infiltration of inflammatory cells, and sloughing of the mucous epithelial lining (Fig. 6B). The kidneys from rCK1023:M-BJ/94 virus-infected chickens appeared normal (Fig. 6C). However, renal congestion was evident in rCK1023:M-G1 virus-infected chickens (Fig. 6D). Expectedly, viral NP was extensively detected in the lungs (bronchioles, terminal bronchioles, and alveoli) of chickens infected with each H9N2 virus type (Fig. 6E and F). However, NP was detected only in the tubular renal epithelial cells of chickens infected with the rCK1023:M-G1 virus (Fig. 6G and H), indicating extrapulmonary infection.
FIG 6.
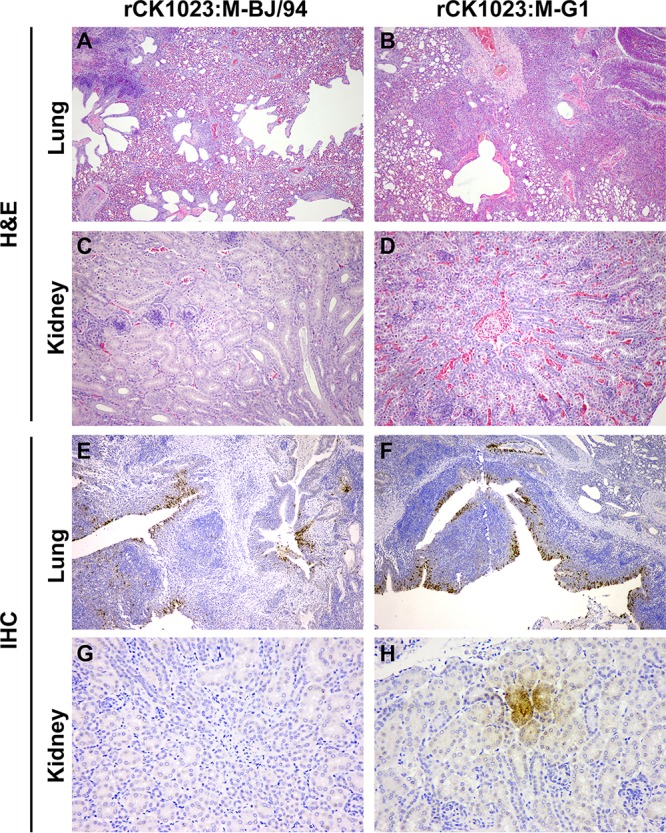
Histological examination of lungs and kidneys from chickens infected with H9N2 virus housing the BJ/94-like or G1-like M gene. Shown are representative H&E (A to D) and immunohistochemical (IHC) (E to H) stainings of lung and kidney sections at 3 dpi. (A and C) rCK1023:M-BJ/94 virus infection causes mild bronchitis and inflammatory changes (A) and no apparent pathological changes in the kidney (C). (B and D) rCK1023:M-G1 virus infection causes severe interstitial pneumonia and bronchopneumonia (B) and hyperemic renal congestion (D). (E and F) There is extensive viral NP distribution in the pulmonary tissues with each virus type. (G and H) However, viral NP is detected in the kidney only during rCK1023:M-G1 virus infection. Magnifications, ×100 (A, B, E, and F), ×200 (C and D), and ×400 (G and H).
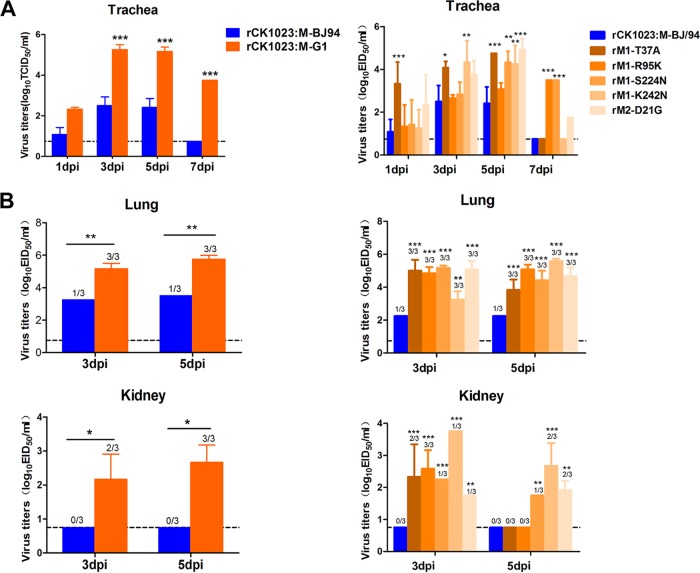
At 1, 3, 5, and 7 dpi, tracheal and cloacal virus titers were determined for three chickens per virus group; virus titers from lung and kidney tissues were ascertained at 3 and 5 dpi. At each time point of tracheal sampling, the rCK1023:M-G1 virus produced more progeny than did the rCK1023:M-BJ/94 virus; from 3 to 7 dpi, the difference in virus output was over 500-fold (P < 0.001) (Fig. 7A, left). rCK1023:M-G1 virus shedding from the trachea lasted for at least 7 days, while rCK1023:M-BJ/94 virus shedding lasted for 5 days (Fig. 7A, left). The rCK1023:M-G1 virus also showed cloacal virus shedding in one infected chicken, while no virus was found in the cloacae of H9N2-M-BJ/94-infected chickens (data not shown). There was a high level of recovery of the rCK1023:M-G1 virus from the lungs of all infected chickens at both 3 and 5 dpi (Fig. 7B, left). The level of rCK1023:M-BJ/94 virus recovery, however, was lower (up to 100-fold lower; P < 0.05), and recovery was successful in only 1 out of 3 infected chickens at each time point. A similar contrast of high-level rCK1023:M-G1 virus recovery but no rCK1023:M-BJ/94 virus detection in the kidneys of infected chickens was observed (Fig. 7B, left). Collectively, the G1-like M gene in H9N2 virus conferred a greater severity of infection, more progeny virus, and extrapulmonary virus spread in chickens.
FIG 7.
Virus titers of rCK1023:M-G1, rCK1023:M-BJ/94, and M gene point mutants of rCK1023:M-BJ/94 H9N2 viruses recovered from chicken trachea (A) and lung and kidney (B). Nine 6-week-old SPF White Leghorn chickens per group were inoculated with 106 EID50 of the indicated viruses; tracheal swabs from three chickens per group were taken at 1, 3, 5, and 7 dpi; and lungs and kidneys were harvested from three chickens per group at 3 and 5 dpi for virus titration. Virus titers are means ± standard deviations. Dashed lines indicates the lower limit of detection. Statistical significance was based on two-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Highly represented amino acid residues encoded by prevalent G1-like M genes from chicken H9N2 viruses.
We sought to identify critical amino acid residues represented in the M1 and M2 proteins derived from prevalent G1-like M genes that could explain the increased severity and early onset of production of H9N2 virus in chickens. In the M phylogenetic tree (Fig. 1A), the prevalent M genes circulating in chickens since 2010 are mostly from the major group of the G1-like lineage, while outside the major group, the M genes are mainly from the earlier period. We compared the amino acid sequences of the M1 and M2 proteins between the major group and the nonmajor group of chicken-origin M genes within the G1-like lineage (Fig. 1A). The alignment identified five amino acid residues as being highly represented in the major group of the G1-like lineage, M1-37A, M1-95K, M1-224N, M1-242N, and M2-21G (Table 1); these residues were also present in the rCK1023:M-G1 virus. They were uncommon in the M genes of the G1-like nonmajor group (0.96 to 5.77%) and in the BJ/94-like lineage (0 to 2.23%). Therefore, the five amino acid residues are distinct molecular markers of G1-like M genes of prevalent H9N2 viruses.
TABLE 1.
Highly represented amino acid residues in prevalent G1-like M genes from chicken H9N2 viruses in China
| Mutation | Time of emergence (yr) | Prevalence in chicken H9N2 influenza viruses (%)a |
||
|---|---|---|---|---|
| With G1-like M gene from major group | With G1-like M gene from nonmajor group | With BJ/94-like M gene | ||
| M1-37A | 2007 | 73.54 | 5.77 | 1.34 |
| M1-95K | 2008 | 96.25 | 4.81 | 0.45 |
| M1-224N | 2008 | 93.44 | 0.96 | 0.45 |
| M1-242N | 2006 | 96.02 | 4.81 | 0 |
| M2-21G | 2007 | 85.48 | 2.38 | 2.23 |
The total numbers of the viruses with either the G1-like M gene from the major group, the G1-like M gene from the nonmajor group, or the BJ/94-like M gene are 427, 104, and 224, respectively.
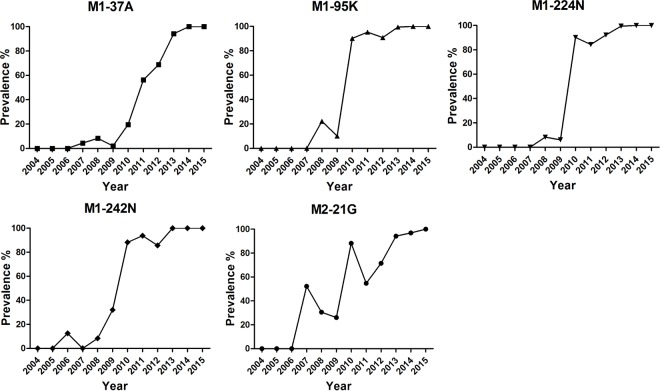
We further investigated the dynamic prevalence of the five amino acid residues of the G1-like M genes of chicken H9N2 viruses in China. As shown in Fig. 8 and Table 1, all five residues first emerged in 2006 to 2008, followed by a sharp increase in their detection in the subsequent years that coincided with the countrywide H9N2 virus chicken outbreaks of 2010 to 2013. This finding suggests that the five distinct residues encoded by the prevalent G1-like M gene could be involved in the recent increase in the reproductive fitness of the H9N2 virus in chickens.
FIG 8.
Prevalence of the five distinct amino acid residues encoded by G1-like M genes of chicken H9N2 viruses in China. Shortly after 2004, the G1-like M segment was introduced into and established in the BJ/94-like H9N2 viruses in chickens. G1-like M gene residues M1-37A, M1-95K, M1-224N, M1-242N, and M2-21G were first detected in 2007, 2008, 2008, 2006, and 2007, respectively. All M1 and M2 sequences were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genomes/FLU). The total numbers of virus isolates in each year from 2006 to 2015 were 6, 20, 35, 52, 51, 63, 77, 189, 32, and 6, respectively.
Dominant G1-like M gene amino acid residues confer increased and early-onset virus replication in CEFs and chickens.
We constructed five virus mutants (rM1-T37A, rM1-R95K, rM1-S224N, rM1-K242N, and rM2-D21G), each with a single amino acid substitution in the M gene of the rCK1023:M-BJ/94 virus. Compared with the rCK1023:M-BJ/94 virus, mutant viruses exhibited higher-level (Fig. 5A, right) and earlier (Fig. 5B, right) production of progeny virus from CEFs. The rM1-T37A mutant virus, in particular, showed significantly greater virus output at between 6 and 12 hpi and earlier virus output, about 4 h ahead, than the parental rCK1023:M-BJ/94 virus (P < 0.001).
SPF chickens infected with the five mutant H9N2 viruses variously showed elevated virus output from tracheal, lung, and kidney samples relative to that of the parental rCK1023:M-BJ/94 virus (Fig. 7A and B, right). The rM1-R95K, rM1-K242N, and rM2-D21G mutants but not the parental virus remained detectable in tracheal samples at 7 dpi (Fig. 7A, right). The rM1-T37A mutant consistently showed elevated tracheal release of virus at 1, 3, and 5 dpi (Fig. 7A, right). All chickens inoculated with each of the five mutant viruses significantly produced higher virus loads in the lung and kidney than did those infected with the parental virus (Fig. 7B, right). For both sampling time points (3 and 5 dpi), virus recovery from the lungs of all chickens infected with each mutant virus was successful, but virus recovery from the lung was successful in only 1 in 3 infections with the parental virus at each time point (Fig. 7B, right). All five mutant viruses were also isolated from the kidneys of infected chickens at increased levels at 3 dpi albeit with differing frequencies. At 5 dpi, 3 out of 5 mutant viruses were still isolated from the kidneys of infected chickens at differing frequencies (Fig. 7B, right). No virus was recovered from kidneys of chickens infected with the parental virus. Collectively, these results demonstrated that each of the five amino acid residues, identified in the major group of G1-like M genes, introduced into the parental rCK1023:M-BJ/94 H9N2 virus was able to increase virus replication, elicit an earlier onset of virus release, and confer extrapulmonary spread in chickens.
DISCUSSION
In the present study, our combined in vitro and in vivo findings clearly demonstrated that the recent evolution of the M gene through reassortment and mutations has significantly contributed to the fitness of the H9N2 virus in chickens, which could account for its increased prevalence in chicken flocks in China during 2010 to 2013. We found that the G1-like M gene in the H9N2 virus was able to confer increased infectivity and severity of infection in primary CEFs and chickens. Crucially, the H9N2 virus housing the G1-like M gene, but not the BJ/94-like M gene, exhibited an early surge (detected by 2 hpi) in viral mRNA and vRNA synthesis that was associated with enhanced viral protein production and with an early and elevated release of progeny virus from infected cells (up to 4 h earlier) and chickens. Five signature amino acid residues (37A, 95K, 224N, and 242N in the M1 protein and 21G in the M2 protein) were demonstrated to be functionally important for the enhanced virus fitness effect of the G1-like M gene.
The establishment of the G1-like M gene, in place of the BJ/94-like M gene, in chicken H9N2 viruses has important evolutionary implications. During the 1990s, two distinct H9N2 virus lineages (BJ/94-like and G1-like) were established in chickens and quail, respectively, in China (16). Subsequently, phylogenetic analysis revealed two-way transmissions of BJ/94-like and G1-like H9N2 viruses between chickens and quails in this country (15, 16, 31). Over the past 20 years, the G1-like M gene is the only segment from quail G1-like viruses to be established in chickens. Based on epidemiological predictions by Xu and colleagues (16), two-way transmissions between different types of poultry can increase the risk of transmission of H9N2 virus mutants to humans by direct infection or indirectly through the contribution of their internal genes to the promotion of novel subtypes (16). Our present findings along with the findings of others on the emergence of H7N9 viruses (4, 6, 32) have in part corroborated this prediction. Namely, through viral transmission from quails to chickens, the G1-like M gene was reassorted into the chicken H9N2 virus to generate the G57 genotype, which subsequently provided six internal genes to the novel H7N9 viruses causing severe outbreaks in humans. The ability of the G1-like M gene to increase and initiate early virus replication would be enormously advantageous to virus genotype dominance.
Reassortment and subsequent mutation are important strategies of influenza viruses in host adaptation (33). Here, we identified five highly represented amino acid residues in the M1 and M2 proteins encoded by prevalent G1-like M genes from chicken H9N2 viruses in China. With the exception of M1-37A, the remaining four residues (M1-95K, M1-224N, M1-242N, and M2-21G) are located at or close to known functional domains. M1-224 and M1-242 are located in the binding site of the M1 protein for vRNP (34). M1-95 is located close to the M1 nuclear localization signal sequence (22), which is also a potential binding site for nuclear export protein (NEP) (35). M2-21 is located close to the transmembrane domain of the M2 proton channel protein (26). Each of the five amino acid residues (M1-37A, M1-95K, M1-224N, M1-242N, and M2-21G) introduced into the parental rCK1023:M-BJ/94 H9N2 virus was variously able to increase virus replication, elicit an earlier onset of virus release, and confer extrapulmonary spread in chickens. These mutations may function through the above-described domain-mediated effects, especially through the effects on M2 proton channel activity and shuttling of vRNP between the nucleus and cytoplasm, to facilitate the early production of the viral genome, proteins, and virus particles. We surmise that these residues in concert could have a greater and more consistent promotional impact on virus propagation.
The M1 and M2 proteins play important roles at later stages of the virus life cycle through participating in virus assembly and the budding of nascent virions. The influenza virus virion is pleomorphic, forming spherical and filamentous virions, but little is known about the functional significance of influenza virus morphology (19). It was thought that human infection produces predominantly filamentous virions (36, 37). Several groups demonstrated that the M segment of the 2009 pandemic influenza virus confers increased filamentous morphology and efficient contact transmissibility in mammalian hosts (29, 38). Interestingly, serial passage of filamentous isolates of influenza virus in eggs caused a loss of filamentous morphology (39, 40) and led to improved growth in eggs (39). Low-pathogenicity avian H2N3 virus-infected CEFs were found to produce mainly spherical virions, whereas the same infection in duck embryonic fibroblasts (which are inherently more resistant to virus replication) generated largely filamentous virions (30). In our study, CEFs infected with H9N2 virus housing the G1-like M gene produced mainly spherical/ovoid virus particles, whereas H9N2 virus harboring the BJ/94-like M gene generated mainly filamentous virions. The amino acid change from K to R at position 95 in the M1 protein has a critical role in filamentous particle formation (41, 42). The G1-like M1 protein possesses a reverse mutation, R95K, which may account for the increased prevalence of spherical particles of the corresponding H9N2 virus. In summary, the change from a filamentous to a spherical morphology correlated with more efficient infection by the rCK1023:M-G1 virus in CEFs and chickens, suggesting that spherical virions are better adapted to virus replication.
Relative to other lineages of H9N2, G1-like H9N2 viruses or viruses with G1-like genes appear to show increased infectivity to humans (7, 17, 43, 44). We found that all of the human H9N2 virus isolates with available M gene sequences from databases have G1-like M genes (data not shown). The H9N2 virus subtype has contributed its internal genes, including the G1-like M gene, to H5N1, H7N9, and H10N8 subtypes, with ensuing human infections (4, 17, 45). The H9N2 virus-origin PB2, M, and NP genes are key virulence genes in human cases of H7N9 virus infection (46). Experimentally, H9N2 viruses have been shown to have extensive reassortment compatibility with pandemic H1N1 (pH1N1) (47, 48), human H3N2 (49), or avian H5N1 (50) viruses. Therefore, the threat of H9N2 viruses harboring the G1-like M gene segment in chickens to human health should be taken seriously.
Our data indicate that the early virus replication and more severe infection acquired by genetic reassortment are critical in conferring virus fitness to better counter host defenses and increasingly cause outbreaks in avian or mammalian populations.
MATERIALS AND METHODS
Ethical approval.
All animal studies were performed in compliance with recommendations of the guide for the care and use of laboratory animals of China Agricultural University (CAU) (approval SKLAB-B-2010-003) and with the approval of the Beijing Association for Science and Technology of China (approval SYXK, Beijing, 2007-0023).
Phylogenetic analysis.
All available M gene sequences of H9N2 viruses isolated from various hosts in China during 1994 to 2015 were downloaded from the Influenza Virus Resource at the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genomes/FLU). Multiple-sequence alignment was carried out by using MUSCLE (51), guided by amino acid sequence alignment. The final alignment covered 804 nucleotides (positions 79 to 882) (reference strain A/turkey/California/189/66) and contained 1,145 sequences. The aligned sequences were then used to generate a maximum likelihood (ML) tree by employing FastTree (version 2.1.7) (52). The generalized time-reversible (GTR+) gamma model was used, and phylogenetic uncertainty was assessed by the Shimodaira-Hasegawa (SH) test for each split in the tree and was resampled 1,000 times. The final tree was viewed and edited in FigTree (version 1.4.2) (http://tree.bio.ed.ac.uk/software/figtree/) and rooted by using A/turkey/California/189/66.
Viruses, plasmids, and cells.
The use of wild-type H9N2 viruses Lx1023 and TS was previously described (6, 53). Lx1023 and TS house the BJ/94-like M gene and the G1-like M gene, respectively. M gene expression plasmids derived from H9N2 virus strains Lx1023 and TS were generated by separately inserting each M gene coding sequence into the pcDNA3.1 vector. CEFs were isolated from 10-day-old embryonated chicken eggs by trypsin digestion (54). Human embryonic kidney (293T) cells, Madin-Darby canine kidney (MDCK) cells, and CEFs were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml of penicillin, and 100 μg/ml of streptomycin at 37°C in a 5% CO2 atmosphere.
Generation of recombinant and mutant H9N2 viruses by reverse genetics.
All eight gene segments of Lx1023 and the M gene of TS (G1-like M gene) were amplified by reverse transcription-PCR (RT-PCR) and individually cloned into a dual-promoter plasmid, pHW2000 (47). The reverse genetic virus rCK1023:M-BJ/94, containing all eight genes from Lx1023, and the reassortant virus rCK1023:M-G1, with the M gene from the TS virus and the remaining seven genes from the Lx1023 virus, were generated in 293T cells as previously described (47). In the backbone of rCK1023:M-BJ/94, mutations T37A, R95K, S224N, and K242N were separately introduced into the M1 protein, and the D21G mutation was introduced into the M2 protein, by using a QuikChange site-directed mutagenesis kit (Agilent, Santa Clara, CA) according to the manufacturer's instructions. Primer sequences are available upon request. The rescued viruses possessing a single mutation were designated rM1-T37A, rM1-R95K, rM1-S224N, rM1-K242N, and rM2-D21G. All viruses were propagated in 9-day-old SPF chicken embryos and sequence verified prior to use.
Quantitative real-time PCR.
Levels of mRNA and vRNA were determined in CEFs infected with different H9N2 viruses at an MOI of 0.01 or 0.001. Total RNA was extracted from infected CEF cells by using TRIzol reagent according to the manufacturer's instructions (Invitrogen). For the detection of mRNA and vRNA, the oligo(dT) primer and the uni-12 primer (5′-AGCAAACGACC-3′), respectively, were used to generate cDNAs by reverse transcription with 1 μg of total RNA per sample using Superscript III first-strand synthesis SuperMix (Invitrogen). The quantitative real-time PCR (qRT-PCR) mixture for each reaction sample consisted of 10 μl of 2× SYBR green PCR master mix (Applied Biosystems), 7 μl of nuclease-free water, 0.5 μl of each primer, and 2 μl of a cDNA template (diluted 1:100). mRNA and vRNA levels of the M1, M2, and NP genes as well as β-actin mRNA levels were quantified by using the 7500 real-time PCR system (Applied Biosystems) with the following program: 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Expression values for each gene, relative to values for β-actin, were calculated by using the 2−ΔΔCT method. Each experiment comprised three technical replicates for each sample, and two experimental replicates were performed. Primers for the amplification of the β-actin, M1, M2, and NP genes are as follows: β-actin forward primer 5′-AGAGCTACGAGCTGCCTGAC-3′ and reverse primer 5′-CGTGGATGCCACAGGACT-3′, M1 forward primer 5′-CCATCAGGCCCCCTCAAAGCCGAGA-3′ and reverse primer 5′-ACGGTGAGCGTGAACACGAACCCTA-3′, M2 forward primer 5′-TTTCTTCAAATGCATTTATCGTCGC-3′ and reverse primer 5′-AAAATGACCATCGTCAACATCCACA-3′, and NP forward primer 5′-AGAGACGGAAAATGGGTGAGAGAGC-3′ and reverse primer 5′-GGATCCATTCCAGTACGCACGAGAG-3′.
Western blotting.
Total cell protein lysates were extracted from transfected 293T cells or infected CEFs with radioimmunoprecipitation assay (RIPA) lysis buffer, and the total protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit (Beyotime, China). Protein samples derived from cell lysates were heated at 100°C for 10 min, separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, USA), and subsequently incubated with an appropriate primary antibody. Primary antibodies were specific for β-actin (Beyotime, China) and influenza A virus M1 (GeneTex, USA), M2 (Thermo Fisher Scientific, USA), and NP (Biorbyt, UK). Horseradish peroxidase (HRP)-conjugated anti-rabbit or -mouse secondary antibody was used (Beyotime, China). The presence of HRP was detected by using a Western Lightning chemiluminescence kit (Amersham, USA) according to the manufacturer's protocol.
Virus titration and replication kinetics.
Fifty percent tissue culture infectious dose (TCID50) assays were performed on MDCK cells inoculated with 10-fold serially diluted viruses and incubated at 37°C in a 5% CO2 atmosphere for 72 h. TCID50 values were calculated according to the Reed-Muench method (55). Multistep replication kinetics were determined by infecting CEFs at an MOI of 0.001. After 1 h of incubation at 37°C, the cells were washed twice and further incubated in serum-free DMEM containing 0.5 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin. Supernatants were sampled at 12, 24, 36, 48, 60, and 72 hpi. Single-replication-cycle kinetics assays were similarly conducted except with starting virus inoculations at an MOI of 0.01. Supernatants were sampled at 2, 4, 6, 8, 10, and 12 hpi.
Chicken challenge study.
Nine 6-week-old SPF White Leghorn chickens were inoculated intranasally with 106 50% egg infective doses (EID50) of each stock virus. Three chickens per group were euthanized at 3 and 5 dpi, and lungs and kidneys were collected for virus titration and histopathological examination. Tracheal and cloacal swabs from the remaining three chickens of each group were collected at 1, 3, 5, and 7 dpi. The virus titer detection limit was 0.75 log10 EID50/ml.
Histopathology and immunohistochemistry.
Lungs and kidneys collected at 3 dpi were fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). The tissue sections were also immunostained for viral NP with a monoclonal primary antibody (AA5H; Abcam, Hong Kong, China). The secondary antibody (Millipore, Billerica, MA, USA) used was conjugated to HRP, and the color reaction was based on the use of an HRP reaction kit (diaminobenzidine tetrahydrochloride; Sigma, St. Louis, MO, USA). Two independent pathologists scored all slides from experimental groups in a blind manner.
Transmission electron micrographs.
For imaging of virions, samples were prepared as previously described, with some modifications (39). Briefly, CEFs were infected at an MOI of 3.0. At 15 hpi, cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 to 3 h at room temperature or overnight at 4°C. Cells were then embedded in Eponate 12 resin, cut into 80-nm sections, and stained with 5% uranylacetate and 2% lead citrate. After sample preparation, grids were imaged at 75 kV by using a JEOL1200EX transmission electron microscope.
Statistical analysis.
All statistical analyses were performed by using GraphPad Prism software version 5.00 (GraphPad Software Inc., San Diego, CA, USA). Statistically significant differences between experimental groups were determined by analysis of variance (ANOVA). Differences were considered statistically significant at a P value of <0.05.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31430086), the National Key Research and Development Program (2016YFD0500204 and 2016YFD0500201), the National Key Technology Research and Development Program of China (2015BAD12B01). Y.B. is supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
REFERENCES
- 1.Neuman G, Chen H, Gao GF, Shu Y, Kawaoka Y. 2010. H5N1 influenza viruses: outbreaks and biological properties. Cell Res 20:51–61. doi: 10.1038/cr.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su S, Bi Y, Wong G, Gray GC, Gao GF, Li S. 2015. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol 89:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam TY, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung YH, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 6.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. 2015. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A 112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Liu J. 2015. H9N2 influenza virus in China: a cause of concern. Protein Cell 6:18–25. doi: 10.1007/s13238-014-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair PJ, Putnam SD, Krueger WS, Chum C, Wierzba TF, Heil GL, Yasuda CY, Williams M, Kasper MR, Friary JA, Capuano AW, Saphonn V, Peiris M, Shao H, Perez DR, Gray GC. 2013. Evidence for avian H9N2 influenza virus infections among rural villagers in Cambodia. J Infect Public Health 6:69–79. doi: 10.1016/j.jiph.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coman A, Maftei DN, Krueger WS, Heil GL, Friary JA, Chereches RM, Sirlincan E, Bria P, Dragnea C, Kasler I, Gray GC. 2013. Serological evidence for avian H9N2 influenza virus infections among Romanian agriculture workers. J Infect Public Health 6:438–447. doi: 10.1016/j.jiph.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Gray GC, Ferguson DD, Lowther PE, Heil GL, Friary JA. 2011. A national study of US bird banders for evidence of avian influenza virus infections. J Clin Virol 51:132–135. doi: 10.1016/j.jcv.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okoye J, Eze D, Krueger WS, Heil GL, Friary JA, Gray GC. 2013. Serologic evidence of avian influenza virus infections among Nigerian agricultural workers. J Med Virol 85:670–676. doi: 10.1002/jmv.23520. [DOI] [PubMed] [Google Scholar]
- 12.Uyeki TM, Nguyen DC, Rowe T, Lu X, Huprimmer J, Huynh LP, Hang NLK, Katz JM. 2012. Seroprevalence of antibodies to avian influenza A (H5) and A (H9) viruses among market poultry workers, Hanoi, Vietnam, 2001. PLoS One 7:e43948. doi: 10.1371/journal.pone.0043948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Ju L, Liu P, Zhou J, Lv X, Li L, Shen H, Su H, Jiang L, Jiang Q. 2015. Serological and virological surveillance of avian influenza A virus H9N2 subtype in humans and poultry in Shanghai, China, between 2008 and 2010. Zoonoses Public Health 62:131–140. doi: 10.1111/zph.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawar SD, Tandale BV, Raut CG, Parkhi SS, Barde TD, Gurav YK, Kode SS, Ac M. 2012. Avian influenza H9N2 seroprevalence among poultry workers in Pune, India, 2010. PLoS One 7:e36374. doi: 10.1371/journal.pone.0036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Pu J, Jiang Z, Tao G, Xia Y, Qi X, Liu L, Bo M, Tian F, Brown EG, Liu J. 2010. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol 146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Xu K, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang J, Li K, Fan X, Webster RG, Chen H, Peiris JS, Guan Y. 2007. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol 81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Y, Shortridge KF, Krauss S, Webster RG. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A 96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol 74:9372–9380. doi: 10.1128/JVI.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross TA, Dong H, Sharma M, Busath DD, Zhou HX. 2012. M2 protein from influenza A: from multiple structures to biophysical and functional insights. Curr Opin Virol 2:128–133. doi: 10.1016/j.coviro.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao S, Liu X, Yu M, Li J, Jia X, Bi Y, Sun L, Gao GF, Liu W. 2012. A nuclear export signal in the matrix protein of influenza A virus is required for efficient virus replication. J Virol 86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z, Robinson D, Wagner RR. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J Virol 69:1964–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130. doi: 10.1016/0092-8674(91)90576-K. [DOI] [PubMed] [Google Scholar]
- 24.Helenius A. 1992. Unpacking the incoming influenza virus. Cell 69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 25.Sugrue RJ, Hay AJ. 1991. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol 76:1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nao N, Kajihara M, Manzoor R, Maruyama J, Yoshida R, Muramatsu M, Miyamoto H, Igarashi M, Eguchi N, Sato M, Kondoh T, Okamatsu M, Sakoda Y, Kida H, Takada A. 2015. A single amino acid in the M1 protein responsible for the different pathogenic potentials of H5N1 highly pathogenic avian influenza virus strains. PLoS One 10:e0137989. doi: 10.1371/journal.pone.0137989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown EG, Liu H, Kit LC, Baird S, Nesrallah M. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc Natl Acad Sci U S A 98:6883–6888. doi: 10.1073/pnas.111165798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakdawala SS, Lamirande EW, Suguitan AL Jr, Wang W, Santos CP, Vogel L, Matsuoka Y, Lindsley WG, Jin H, Subbarao K. 2011. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog 7:e1002443. doi: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Mubarak F, Daly J, Christie D, Fountain D, Dunham SP. 2015. Identification of morphological differences between avian influenza A viruses grown in chicken and duck cells. Virus Res 199:9–19. doi: 10.1016/j.virusres.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Xu K, Li K, Smith GJ, Li J, Tai H, Zhang J, Webster RG, Peiris JS, Chen H, Guan Y. 2007. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol 81:2635–2645. doi: 10.1128/JVI.02316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, Liu X, Shu Y, Peng D, Jiang T. 2013. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14:446–452. doi: 10.1016/j.chom.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Neverov AD, Lezhnina KV, Kondrashov AS, Bazykin GA. 2014. Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes. PLoS Genet 10:e1004037. doi: 10.1371/journal.pgen.1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baudin F, Petit I, Weissenhorn W, Ruigrok RWH. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- 35.Akarsu H, Burmeister WP, Petosa C, Petit I, Müller CW, Ruigrok RWH, Baudin F. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J 22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu CM, Dawson IM, Elford WJ. 1949. Filamentous forms associated with newly isolated influenza virus. Lancet i:602. [DOI] [PubMed] [Google Scholar]
- 37.Kilbourne ED, Murphy JS. 1960. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J Exp Med 111:387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell PJ, Danzy S, Kyriakis CS, Deymier MJ, Lowen AC, Steel J. 2014. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J Virol 88:3802–3814. doi: 10.1128/JVI.03607-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seladischulman J, Steel J, Lowen AC. 2013. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J Virol 87:13343–13353. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choppin PW, Tamm I. 1960. Studies of two kinds of virus particles which comprise influenza A2 virus strains. III. Morphological characteristics: independence to morphological and functional traits. J Exp Med 112:945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourmakina S, Garcia-Sastre A. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J Gen Virol 84:517–527. doi: 10.1099/vir.0.18803-0. [DOI] [PubMed] [Google Scholar]
- 42.Elleman CJ, Barclay WS. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144–153. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster RG, Cox N, Hay A. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A 97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS, Guan Y. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 46.Bi Y, Xie Q, Zhang S, Li Y, Xiao H, Jin T, Zheng W, Li J, Jia X, Sun L, Liu J, Qin C, Gao GF, Liu W. 2015. Assessment of the internal genes of influenza A (H7N9) virus contributing to high pathogenicity in mice. J Virol 89:2–13. doi: 10.1128/JVI.02390-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Qin K, Wang J, Pu J, Tang Q, Hu Y, Bi Y, Zhao X, Yang H, Shu Y, Liu J. 2011. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci U S A 108:4164–4169. doi: 10.1073/pnas.1019109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimble JB, Sorrell E, Shao H, Martin PL, Perez DR. 2011. Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model. Proc Natl Acad Sci U S A 108:12084–12088. doi: 10.1073/pnas.1108058108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. 2009. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A 106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao X, Hu J, Wang J, Xu J, Cheng H, Xu Y, Li Q, He D, Liu X, Wang X, Gu M, Hu S, Xu X, Liu H, Chen S, Peng D, Liu X. 2016. Reassortant H5N1 avian influenza viruses containing PA or NP gene from an H9N2 virus significantly increase the pathogenicity in mice. Vet Microbiol 192:95–101. doi: 10.1016/j.vetmic.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bi Y, Lu L, Jing L, Yin Y, Yi Z, Gao H, Qin Z, Zeshan B, Liu J, Lei S, Liu W. 2011. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol J 8:505. doi: 10.1186/1743-422X-8-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Zou T, Hu X, Hong J. 2015. Type III interferon gene expression in response to influenza virus infection in chicken and duck embryonic fibroblasts. Mol Immunol 68:657–662. doi: 10.1016/j.molimm.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Reed LJ, Muench H. 1937. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]