ABSTRACT
Beta-propiolactone (BPL) is an inactivating agent that is widely used in the vaccine industry. However, its effects on vaccine protein antigens and its mechanisms of action remain poorly understood. Here we present cryo-electron microscopy (cryo-EM) structures of BPL-treated coxsackievirus A16 (CVA16) mature virions and procapsids at resolutions of 3.9 Å and 6.5 Å, respectively. Notably, both particles were found to adopt an expanded conformation resembling the 135S-like uncoating intermediate, with characteristic features including an opened 2-fold channel, the externalization of the N terminus of VP1 capsid protein, and the absence of pocket factor. However, major neutralizing epitopes are very well preserved on these particles. Further biochemical analyses revealed that BPL treatment impairs the abilities of CVA16 particles to bind to the attachment receptor heparan sulfate and to a conformation-dependent monoclonal antibody in a BPL dose-dependent manner, indicating that BPL is able to modify surface-exposed amino acid residues. Taken together, our results demonstrate that BPL treatment may induce alteration of the overall structure and surface properties of a nonenveloped viral capsid, thus revealing a novel mode of action of BPL.
IMPORTANCE Beta-propiolactone (BPL) is commonly used as an inactivating reagent to produce viral vaccines. It is recognized that BPL inactivates viral infectivity through modification of viral nucleic acids. However, its effect on viral proteins remains largely unknown. Here, we present high-resolution cryo-EM structures of BPL-treated coxsackievirus A16 (CVA16) mature virions and procapsids, which reveals an expanded overall conformation and characteristic features that are typical for the 135S-like uncoating intermediate. We further show that the BPL concentration affects the binding of inactivated CVA16 particles to their receptor/antibody. Thus, BPL treatment can alter the overall structure and surface properties of viral capsids, which may lead to antigenic and immunogenic variations. Our findings provide important information for future development of BPL-inactivated vaccines.
KEYWORDS: Beta-propiolactone, coxsackievirus A16, cryo-EM structure, structural alteration, surface modification
INTRODUCTION
Enterovirus 71 (EV71) and coxsackievirus A16 (CVA16) are the two major causative agents of hand, foot, and mouth disease (HFMD), which has been prevalent in young children in the Asia-Pacific region (1). According to a recent survey, EV71 and CVA16 were associated with 50.4% and 38.3% of the 266 laboratory-confirmed HFMD cases, respectively (2). Moreover, coinfection with EV71 and CVA16 may have led to the generation of recombinant strains currently circulating in China (3, 4). It is therefore of great importance to develop vaccines for EV71 and CVA16.
Both EV71 and CVA16 belong to the Enterovirus genus of the Picornaviridae family. They are small nonenveloped viruses of ∼30 nm in diameter with a single-stranded positive-sense RNA genome of ∼7.4 kb encapsulated in icosahedral capsids (5–8). Cell culture-derived CVA16 particles naturally exist in two forms, the mature virions (also termed full particles), which contain the infectious viral RNA genome, and the noninfectious procapsids with intact VP0 but no viral RNA (9). Mature CVA16 virions may undergo conformational changes upon cellular attachment and receptor binding to produce an uncoating intermediate termed the “135S-like particle,” which has been resolved to atomic detail by X-ray crystallography (10). The key features of 135S-like particles that differ from those of mature virions include an expanded capsid, lost pocket factor, no evidence of VP4, an extruded VP1 N terminus, and an enlarged 2-fold opening (10, 11). Recently, Ren et al. reported crystal structures of the unexpanded CVA16 mature virion, the procapsid, and a recombinant virus-like particle (VLP) (12). In addition, the structure of the insect cell-produced CVA16 VLP has been determined at 5.5 Å by cryo-electron microscopy (cryo-EM) single-particle analysis (13). These studies reveal that the capsid of CVA16 virions is similar to those of other enteroviruses, which is an arrangement of 60 copies of protomers each consisting of 4 subunits known as VP1, VP2, VP3, and VP4. For enteroviruses, in general VP1 to -3 share a jelly-roll-fold-like structure and form a quasi-T=3 symmetry on the virus surface, and VP4 is a small linear protein lying beneath the surface and interlacing with the VP1 to -3 N-terminal extensions to surround the RNA genome. The outer surface of enterovirus capsids has some characteristic features, including a mesa-like feature at the 5-fold symmetry axis, a three-blade propeller-like feature surrounding the 3-fold symmetry axis, and a depression called a “canyon” between the mesa and propeller (5, 6, 14–16).
Development of EV71 vaccines has advanced rapidly, with two inactivated whole-virus vaccine candidates having completed phase 3 clinical trials and one inactivated whole-virus vaccine approved in China (17). However, the development of inactivated whole-virus vaccines for CVA16 has proven challenging, and contradictory results were obtained from preclinical studies of a few experimental CVA16 vaccines developed by different groups. For example, Chong et al. reported that formaldehyde-inactivated CVA16 mature virions (also termed “R-particles” in that paper) was able to induce neutralizing antibodies in mice and rabbits, whereas sera from mice immunized with formaldehyde-treated CVA16 procapsids (also termed “P-particles” in that paper) exhibited no neutralization activity at all (9). Notably, in the same study, the neutralizing antibody titer toward CVA16 elicited by 3 doses of 2.5 μg of inactivated CVA16 “R-particles” was much lower than that against EV71 induced by the same regimen of inactivated EV71 counterpart in mice (128 versus 2,560) and in rabbits (256 versus 32,000) (9). Another research group showed that 3 doses (400 endotoxin units [EU] per dose) of a formaldehyde-inactivated CVA16 experimental vaccine (which contains both mature virions and procapsids as indicated by the presence of both VP0 and VP2 on SDS-PAGE and confirmed by electron microscopy) elicited neutralizing antibodies with relatively low titers (256 on average) in mice (18). Surprisingly, our recent study showed that 2 doses (1 or 2 μg per dose) of beta-propiolactone (BPL)-inactivated CVA16 experimental vaccine could efficiently induce neutralizing antibodies with titers up to 4,096 (19, 20). Although the methods used for antigen quantification in these studies are different, the apparently significant difference in immunogenicity between the formaldehyde- and BPL-inactivated CVA16 experimental vaccines leads us to hypothesize that their structures might be different.
To test this hypothesis, in this study we aimed to determine the structures of BPL-inactivated CVA16 mature virions and procapsids by high-resolution cryo-EM single-particle analysis. Furthermore, we examined the surface properties of the BPL-inactivated CVA16 particles by biochemical assays. Our results revealed that BPL treatment could induce conformational changes and alter the binding capacity of CVA16 particles. These findings should have strong implications for the development of inactivated whole-virus vaccines.
RESULTS
Cryo-EM reconstructions of BPL-inactivated CVA16 particles.
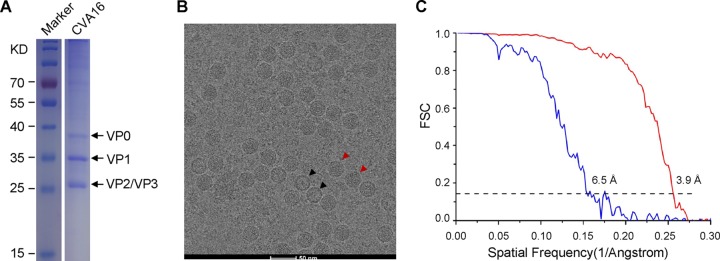
BPL-inactivated CVA16 was prepared as described in Materials and Methods. As revealed by the SDS-PAGE analysis, high-purity CVA16 was obtained (Fig. 1A). The structures of CVA16 viral particles were determined by cryo-EM single-particle analysis. Two types of viral particles were visualized in the cryo-EM micrographs; one was the mature virion (full particle), and the other was the procapsid (empty particle) (Fig. 1B). Separate reconstructions of the mature virion and procapsid of CVA16 were made.
FIG 1.
SDS-PAGE analysis, cryo-EM image, and resolution evaluation of CVA16 viral particles. (A) SDS-PAGE and Coomassie blue staining of purified BPL-inactivated CVA16. The lanes for the inactivated CVA16 sample and the protein ladder marker on the same gel were aligned for molecular mass estimation. (B) Representative cryo-EM micrograph of CVA16. The red and black arrowheads indicate mature virion and empty capsid, respectively. Scale bar, 50 nm. (C) Resolution evaluation of the cryo-EM maps of the BPL-inactivated CVA16 particles by Fourier shell correlation (FSC) at a 0.143 criterion. Red and blue curves represent the FSCs of the mature virion and the empty capsid, respectively.
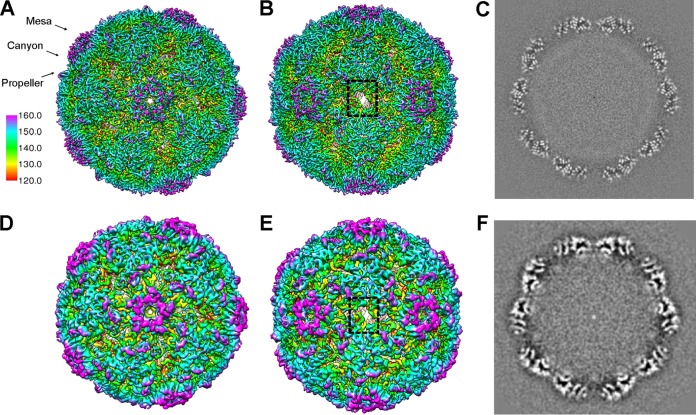
A total of 25,610 mature virions were extracted and further reconstructed to a 3.9-Å resolution cryo-EM density map, as judged by the Fourier shell correlation coefficient (FSC) of 0.143 (Fig. 1C). The map of the CVA16 mature virion depicted the characteristic features of picornaviruses, including a mesa at the 5-fold symmetry axis, a three blade propeller-like feature surrounding the 3-fold symmetry axis, and a “canyon” between the mesa and propeller (Fig. 2A to C). Surprisingly, we noticed that the 2-fold axis channel was opened in this structure (Fig. 2B).
FIG 2.
Cryo-EM density maps of BPL-inactivated CVA16 particles. (A and B) The overall structure of the BPL-inactivated CVA16 mature virion viewed along the 5-fold (A) or the 2-fold (B) axis of symmetry. The locations of the mesa, canyon, and propeller are indicated by black arrows. The 2-fold opening regions are indicated by a dashed black rectangle. (C) Central section of the cryo-EM map of BPL-inactivated CVA16 mature virion visualized along the 5-fold axis of symmetry to show the quality of the map. (D and E) The overall structure of BPL-inactivated CVA16 empty capsid viewed along a 5-fold (D) or 2-fold (E) axis of symmetry. The reconstructions are colored by radius as indicated by the scale bar. (F) Central section of the cryo-EM map of the BPL-inactivated CVA16 empty capsid visualized along the 5-fold axis of symmetry.
We also obtained the CVA16 procapsid map at a resolution of 6.5 Å from 1,642 empty particles (Fig. 1C and 2D to F). The overall architecture of the procapsid (Fig. 2D to F) is highly similar to that of the BPL-inactivated mature virion (Fig. 2A to C), except that no density corresponding to the viral genome was observed in this density map. The capsid portions of our two cryo-EM maps were nearly identical in terms of particle size and the dimensions in the 2-fold and 5-fold channels.
The BPL-inactivated CVA16 mature virion structurally resembles the expanded 135S-like particle.
We compared our cryo-EM density map of the BPL-inactivated mature virion with each of the crystal structures of the formaldehyde-inactivated CVA16 mature virion (PDB 5C4W), procapsid (PDB 5C9A) (12), the CVA16 135S-like uncoating intermediate (PDB 4JGY) (10), and the crystal structures of the EV71 (sharing about 80% sequence identify with CVA16) mature virion (PDB 3VBS) and procapsid (PDB 3VBU) (5). Surprisingly, among these crystal structures, the CVA16 135S-like structure and EV71 procapsid fit well into our cryo-EM map of the CVA16 mature virion, with cross correlation scores (CC) of 0.84 and 0.74, respectively, whereas the others yielded a much lower CC (less than 0.15) (Table 1).
TABLE 1.
Correlation between the BPL-inactivated CVA16 cryo-EM maps and the corresponding crystal structures of CVA16 and EV71
| Cryo-EM map | Correlation coefficient for crystal structure |
||||
|---|---|---|---|---|---|
| CVA16 |
EV71 |
||||
| Mature virion (PDB 5C4W) | Procapsid (PDB 5C9A) | 135S particle (PDB 4JGY) | Mature virion (PDB 3VBS) | Procapsid (PDB 3VBU) | |
| Mature viriona | 0.15 | 0.15 | 0.84 | 0.14 | 0.74 |
| Procapsidb | 0.19 | 0.2 | 0.81 | 0.19 | 0.86 |
Cryo-EM map of the BPL-inactivated CVA16 mature virion.
Cryo-EM map of the BPL-inactivated CVA16 procapsid.
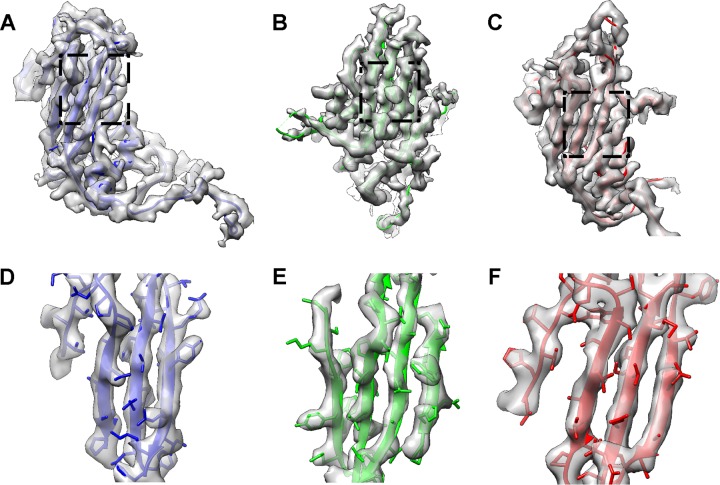
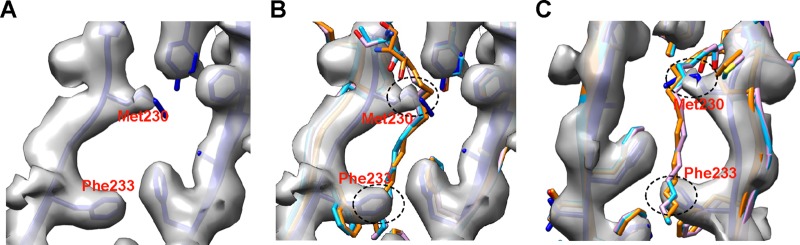
Fitting of the CVA16 135S-like crystal structure into the capsid density of our mature virion map showed that all the main chains could be fit unambiguously (Fig. 3). VP1, VP2, and VP3 share an eight-strand β-barrel-like topology, and all of these strands were separated and the distinctive bulky side chains were discerned in the cryo-EM map, demonstrating that the resolution is indeed around 4 Å. In the 135S-like crystal structure, some residues in the VP1 to -3 GH loop are missing, including F211 to N218 in VP1, G137 to N141 in VP2, and H180 to G184 in VP3 (10). These residues were also missing in our structure (Fig. 3), suggesting intrinsic flexibility in those regions. In addition, the densities corresponding to T98 to Q101 and D219 to N228 of VP1, as well as A3 to V13 and E142 to S144 of VP2, were not very well resolved in our cryo-EM map. These residues were resolved in the crystal structure but with relative high b-factors of more than 100 A2 (10), indicating a highly dynamic nature of these regions. It is noteworthy that no density corresponding to VP4 and pocket factor was found in the BPL-inactivated CVA16 mature virion. Instead, the pocket collapsed, with the pocket factor binding site occupied by the side chains of M230 and F233 in VP1 (Fig. 4).
FIG 3.
Fitting of the 135S-like crystal structure into the cryo-EM map of the BPL-inactivated mature virion. (A to C) The crystal structure of the CVA16 135S-like particle (PDB 4JGY) fits well into the corresponding density map. VP1, VP2, and VP3 are shown in blue, green, and red, respectively. The same color scheme is followed throughout. (D to F) Expanded views of panels A to C (indicated by the dashed rectangles), illustrating the high-resolution structure features, including separation of beta strands as well as the majority of bulky side chains.
FIG 4.
Collapsed hydrophobic pocket in VP1 of BPL-inactivated CVA16. (A) In the hydrophobic pocket of the BPL-inactivated CVA16 mature virion, the side chains of Met230 and Phe233 in VP1 partially occupy the binding position for pocket factor. The positions of cryo-EM densities for the side chains of Met230 and Phe233 were determined according to the fitted CVA16 135S crystal structure (PDB 4JGY) (blue). (B) The structures in panel A were superimposed with the crystal structures of the EV71 mature virion (PDB 3VBS) (orange), the CVA16 mature virion (PDB 5C4W) (cyan), and the CVA16 empty capsid (PDB 5C9A) (pink), which all have a pocket factor, so as to illustrate where a pocket factor supposed to reside in the pocket. (C) Panel B viewed from the back. The potential clashes between pocket factor and the side chains of Met230 and Phe233 in VP1 are indicated by black dashed ovals.
Previous studies have shown that some picornaviruses can “breathe,” which is accompanied by transient reversible externalization of the capsid protein VP4 and the N terminus of VP1 (21, 22). Upon binding to its cellular receptor, the virus undergoes a series of irreversible conformational changes to form an expanded 135S entry intermediate with its VP1 N terminus and VP4 penetrated into cell membrane, which are postulated to facilitate viral entry by anchoring the entry intermediate to the cell membrane and forming a translocation pore for viral RNA (10, 11, 23, 24). In our cryo-EM map, the VP1 N terminus appeared to extrude from the base of the canyon at the VP1-VP3 interface, with the density corresponding to N62 to L70 traversing across the capsid (Fig. 5A and B), whereas the N-terminal 61 residues of VP1 were not resolved in our structure (Fig. 5A and B), probably due to high dynamics. These observations are in line with the finding for the 135S-like crystal structure (10).
FIG 5.
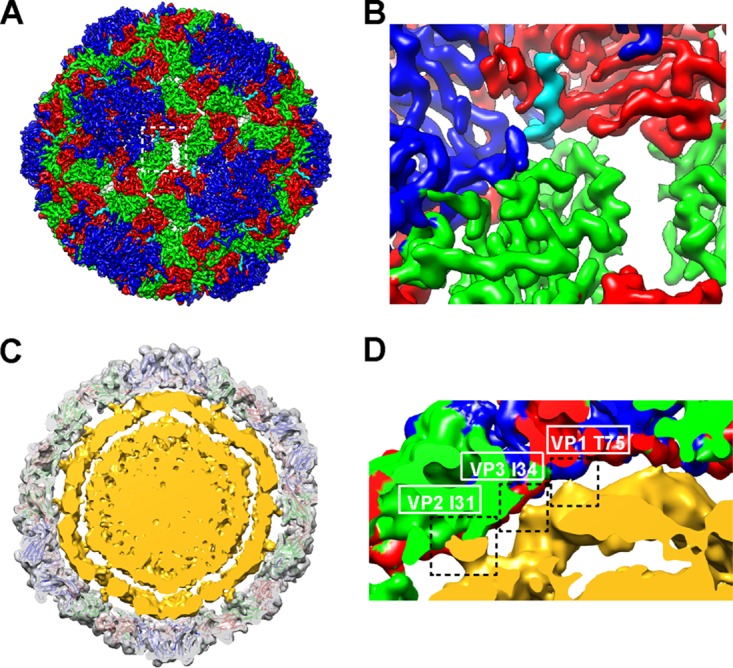
Features of the cryo-EM structure of the BPL-inactivated CVA16 mature virion. (A) Externalization of the VP1 N terminus from the base of canyon. The density for residues 62 to 70 of VP1 is rendered in cyan. Densities for other parts of VP1, VP2, and VP3 are rendered in blue, green, and red, respectively. (B) Expanded view of panel A. (C and D) Cross section of BPL-inactivated CVA16 virion structure that was low-pass filtered to 10-Å resolution. The capsid is rendered in transparent gray with the fitted atomic model. The RNA shell and core are rendered in yellow. (D) Expanded view of panel C. Three contacts between the capsid and the RNA shell are indicated by dashed rectangles, and the residues that contributed to the contacts are labeled.
Apart from the capsid in the CVA16 mature virion map, we were also enabled to visualize the density beneath attributed to viral RNA. Low-pass filtering of this map to 10-Å resolution revealed a condensed RNA core surrounded by a looser RNA shell (Fig. 5C and D). The gaps between the capsid and the outer RNA shell and between the outer RNA shell and the inner RNA core were about 13 Å and 15 Å, respectively. Three contacts between the capsid and the RNA shell were clearly seen at the locations of VP1 T75, VP3 I34, and VP2 I31 (Fig. 5C and D).
We should point out that the direct observation of extrusion of the VP1 N terminus, the absence of VP4 and pocket factor, and the presence of RNA genome in the map of the BPL-treated mature virion strongly suggest that BPL had triggered formation of the 135S-like particle.
The BPL-treated CVA16 procapsid is expanded and broadly similar to the 135S-like particle.
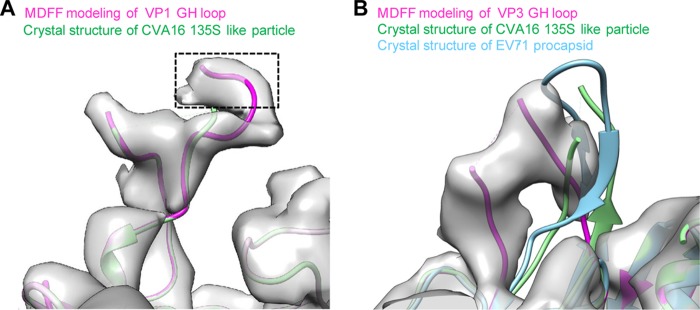
The crystal structure of the CVA16 135S-like particle also fits into our cryo-EM density map of BPL-treated procapsid (CC = 0.81) better than the crystal structure of the formaldehyde-inactivated CVA16 procapsid or mature virion (CC = 0.2 and 0.19, respectively) (Table 1). Comparison of the BPL-treated procapsid cryo-EM map with the CVA16 135S-like crystal structure showed that the densities for the N terminus of VP1 (residues 1 to 72) and the N terminus of VP0 (residues 1 to 98) were missing beneath the capsid in our map. Interestingly, extra density corresponding to the VP1 GH loop (residues 210 to 220), which was missing in the crystal structures, was resolved in our map (Fig. 6A).
FIG 6.
Structure comparison. (A) Extra density corresponding to the VP1 GH loop was found in our cryo-EM map of the BPL-treated CVA16 procapsid. Using molecular dynamics flexible fitting (MDFF) software, we built residues 216 to 220 of this loop (extra purple model portion compared to the green model), which is missing in the crystal structure of the CVA16 135S-like particle (PDB 4JGY) (green). (B) The VP3 GH loop in our map of the CVA16 procapsid adopts a different conformation than in the crystal structures of the EV71 procapsid (PDB 3VBU) (cyan) and CVA16 135S-like particle (PDB 4JGY) (green). Using MDFF software, we built part of this loop against the density of our CVA16 procapsid map (purple).
In addition, the crystal structures of EV71 procapsid (PDB 3VBU) but not the mature virion (PDB 3VBS) (5) could fit into our CVA16 procapsid map reasonably well (CC = 0.86) (Table 1). Most parts of the crystal structures of the CVA16 135S-like particle and EV71 procapsid matched our CVA16 procapsid map, except for the VP3 GH loop region (residues T175 to T191) (Fig. 6B). The significant conformational changes of the VP3 GH loop have been previously reported in expanded particles of EV71 and CVA16 (5, 10), which indicates that it may play a significant role in the viral entry process, such as receptor binding or virus uncoating. Overall, the cryo-EM density map of the BPL-treated procapsid is nearly identical to that of the BPL-treated mature virion, with only a few differences attributed to the VP0 N-terminal and VP1 N-terminal regions as well as the VP1 and VP3 GH loops (data not shown).
Neutralizing epitopes are well preserved in BPL-treated CVA16 particles.
Six linear neutralizing epitopes of CVA16, namely, PEP32, PEP37, PEP55, PEP63, PEP71, and PEP91, have been previously identified (25). We located these epitopes on our cryo-EM maps of the BPL-treated CVA16 mature virion and procapsid. All of these epitopes were distributed around the canyon (Fig. 7), suggesting that the canyon region is a predominant site targeted by neutralizing antibodies. Our cryo-EM maps assigned PEP32, PEP37, PEP55, and PEP71 to the surface-exposed BC, CD, EF, and GH loops of VP1, respectively (Fig. 7B and D). Part of PEP55 (T98 to T103) and most of PEP71 (F211 to C225) were missing in the cryo-EM map of the mature virion, likely due to a high degree of dynamics in these regions. PEP91 (orange), which resides in the C-terminal loop of VP1, appeared to be surface exposed as well, whereas PEP63 (light green) was less exposed than the other five epitopes on both particles (Fig. 7). Taken together, these data demonstrate that the important linear neutralizing epitopes are well preserved in the BPL-treated CVA16 particles. In addition, we showed that BPL-treated CVA16 particles could be recognized by the conformation-dependent neutralizing monoclonal antibody 9B5 (Fig. 8), suggesting the presence of conformational neutralizing epitopes on the surface of these particles.
FIG 7.

Epitope location on the cryo-EM structures of BPL-inactivated CVA16 capsids. (A) A single asymmetric unit of our cryo-EM map of the BPL-treated CVA16 mature virion. Densities for VP1, VP2, and VP3 are rendered in blue, green, and red, respectively. (B) Previously identified linear neutralizing epitopes of CVA16 in the asymmetric unit shown in panel A. (C) A single asymmetric unit of our cryo-EM map of the BPL-treated CVA16 procapsid. (D) Previously identified linear neutralizing epitopes of CVA16 in the asymmetric unit shown in panel C. Epitopes are colored as follows: PEP32, residues 94 to 108 of VP1, pink; PEP37, residues 109 to 123 of VP1, cyan; PEP55, residues 163 to 177 of VP1, magenta; PEP63, residues 187 to 201 of VP1, light green; PEP71, residues 211 to 225 of VP1, yellow; PEP91, residues 271 to 285 of VP1, orange.
FIG 8.
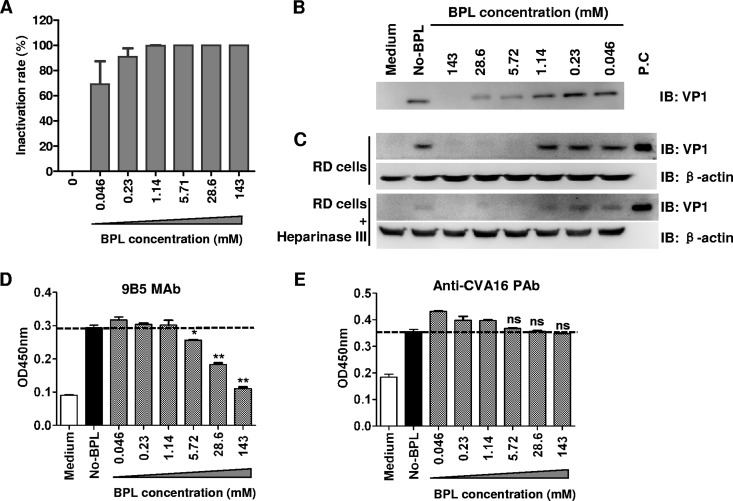
BPL treatment alters surface properties of CVA16 particles. CVA16 was treated with different concentrations of BPL as indicated for virus inactivation. (A) Inactivation rates were determined as the percentage of the TCID50 of BPL-treated samples in comparison to that of CVA16 without BPL treatment. (B) The interaction of BPL-treated CVA16 with heparin-conjugated agarose was determined by pulled down assay. (C) The attachment of BPL-treated CVA16 to RD cells with or without heparinase III treatment was determined as described in Materials and Methods. β-Actin served as and internal control. The virus was detected with rabbit anti-VP1 polyclonal antibodies by Western blotting assay. The positive control (P.C) was purified CVA16. (D and E) The reactivities of BPL-treated CVA16 with monoclonal antibody 9B5 (D) and anti-CVA16 VLP polyclonal antibodies (E) were determined by ELISA and compared to that of CVA16 without BPL treatment. The ELISA results are shown as means ± standard deviations of optical densities at 450 nm (OD450) for triplicate wells. The dotted line indicates signal generated from virus without BPL treatment. Representative results from two independent experiments are shown. Statistical significance between BPL-treated and nontreated virus was determined by the Student t test using GraphPad Prism version 4 and is indicated as follows: n.s., P > 0.05; *, P < 0.05; **, P < 0.01.
BPL treatment alters surface property of CVA16 particles in a dose-dependent manner.
Our structural study described above corroborated that BPL inactivation can induce conformational changes of CVA16 particles. We further investigated whether the surface properties of CVA16 could be affected by the BPL treatment. CVA16 was treated with different concentrations of BPL, and then the treated samples were measured for infectivity. Figure 8A shows that the inactivation of CVA16 infectivity was BPL dose dependent. Heparan sulfate glycosaminoglycans (HSPGs) are identified attachment receptors for EV71 (26). HSPGs also serve as attachment receptors for CVA16, and the virus could interact with heparin in vitro and be pulled down by heparin-conjugated agarose (our unpublished data). We then performed pulldown assays to examine the binding of inactivated CVA16 to heparin. As shown in Fig. 8B, CVA16 samples treated with higher concentrations of BPL exhibited reduced heparin binding activities. We further determined the binding of BPL-inactivated CVA16 on RD cells (which are HSPG positive) with or without heparinase III treatment by attachment assay. As shown in Fig. 8C, CVA16 samples inactivated by higher concentrations of BPL exhibited reduced amounts of attached virus on RD cells, and the attached virus on RD cells treated with heparinase III generated a reduced overall signal compared to that of RD cells without treatment. We then analyzed the ability of BPL-treated CVA16 to bind to a conformation-dependent monoclonal antibody, 9B5, by enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 8D, the binding of BPL-treated CVA16 samples to the monoclonal antibody was impaired in a BPL concentration-dependent manner; in contrast, as a control, the reactivity of anti-CVA16 polyclonal antibodies to treated CVA16 was not significantly affected by corresponding BPL concentrations (Fig. 8E). These results suggest that some specific conformations or surface accessibility of the CVA16 particles was altered following BPL treatment.
DISCUSSION
Formaldehyde and BPL are the two most commonly used inactivating reagents in the production of inactivated viral vaccines. It is generally recognized that formaldehyde inactivates viruses through cross-linking of viral surface proteins (27, 28), whereas BPL modifies mainly viral DNA or RNA through acylation or alkylation for virus inactivation (29–31). Hence, BPL-inactivated viruses may maintain a more authentic surface structure and higher immunogenicity than those treated with formaldehyde/formalin (32). However, a few recent studies show that BPL can also react with peptides or proteins (33, 34), resulting in altered function of key viral proteins, such as reduced membrane fusion activity of influenza virus hemagglutinin (HA) protein (35). In the present study, for the first time we provide evidence that BPL treatment can negatively affect binding of CVA16 particles. Specifically, the abilities of CVA16 particles to bind heparan sulfate and the target cells as well as a conformation-dependent antibody were impaired by BPL treatment in a dose-dependent manner (Fig. 8). This functional alteration is likely due to chemical modification of the surface-exposed residues on CVA16 particles by BPL. It has been demonstrated that BPL can react strongly with cysteine, methionine, and histidine and, to a lesser degree, with aspartic acid, glutamic acid, tyrosine, lysine, serine, and threonine (arginine was not tested in the study), yielding alkylated and acylated products with extended side chains (33). The exact residues of BPL modification on CVA16 particles remain to be determined.
The most striking finding in the present study is the overall structural alteration of CVA16 capsids following BPL inactivation. Our high-resolution (up to 3.9 Å) cryo-EM maps of the BPL-inactivated CVA16 particles reveal that both the BPL-inactivated CVA16 mature virion and procapsid adopt an expanded conformation resembling that of the uncoating intermediate 135S-like particle. Expanded forms of procapsid have been reported for a number of enteroviruses. For example, poliovirus procapsid could convert into an expanded form (which was termed H-antigenic procapsids) at pH 8.4 or 8.5 (36). In addition, the EV71 procapsid was found to be expanded and to possess different antigenic properties than the mature virion (5, 37). Several characteristics of the 135S-like particle were evident in our cryo-EM structure of CVA16 mature virion, including the opening of the 2-fold channel, the externalization of N-terminal VP1 protein, and the lack of pocket factor. These findings are vastly different from the structural features of formaldehyde-inactivated CVA16 mature virions and procapsids, both of which manifested as unexpanded forms (12). The specific mechanism underlying the BPL-induced conformational shift of CVA16 capsids from their native unexpanded forms to the expanded 135S-like uncoating intermediate remains unclear. Usually, expulsion of a lipid pocket factor, which lies at the bottom of the “canyon” on enterovirus capsids, is the prelude to conformation shift of virions from 160S to 135S (38, 39). It has been shown that pocket factors play an important role in stabilizing enteroviruses in the native state (40). In our study, we did not observe density corresponding to the pocket factor at its expected binding site, indicating the loss of pocket factor in the BPL-treated CVA16 particles either with or without genomes. In contrast, it was recently reported that both formaldehyde-inactivated CVA16 mature virion and procapsid contain a sphingosine-like pocket factor (12). Therefore, it is likely that BPL may directly or indirectly deplete the pocket factor, thus triggering the conformational shift for both the mature virion and procapsid. Further experimentation is required to verify this hypothesis and unravel the underlying mechanism.
It is believed that acylation or alkylation of the viral genome is the primary (if not the sole) mechanism for BPL-mediated inactivation of CVA16. In the present study, we demonstrate that the overall structure and heparan sulfate binding capacity of CVA16 particles were altered following BPL treatment. It is possible that these changes might have influenced viral infectivity. Previous studies showed that the expanded uncoating intermediates (also termed “A-particles”) of some picornaviruses cannot infect cells efficiently because they have lost the ability to bind the receptor (23, 41, 42). Therefore, the structural and functional alterations of CVA16 particles observed in the present study might also contribute to virus inactivation, likely due to a decrease or even loss of receptor binding ability. Interestingly, preclinical studies from us and other groups showed that BPL-inactivated CVA16 vaccines are more immunogenic than the formaldehyde-inactivated ones with regard to neutralizing antibody titers (9, 18–20). Current results suggest that the 135S-like uncoating intermediate represents a form of antigen superior to native viral particles for the development of CVA16 vaccine as well as vaccines for other related enteroviruses.
In summary, by high-resolution cryo-EM single-particle analysis coupled with biochemical assays, we demonstrated that BPL treatment could induce structural alteration and surface modification of CVA16 capsids. These findings provide new insight into the working mechanism of BPL as a widely used inactivating agent and therefore may have broad implication for the development of inactivated whole-virus vaccines.
MATERIALS AND METHODS
Cells, virus, and antibodies.
RD and Vero cells were maintained as previously described (8). CVA16-SZ05 (GenBank accession number EU262658) was grown on Vero cells. Rabbit polyclonal antibodies against the CVA16 VP1 protein were described previously (8). CVA16 VLP was produced as described previously (43). To generate rabbit polyclonal antibodies against CVA16 VLP, a rabbit was subcutaneously immunized with 100 μg of CVA16 VLP emulsified in complete Freund's adjuvant (Sigma) and boosted twice with 50 μg of VLP plus incomplete Freund's adjuvant (Sigma) at 3 and 6 weeks postpriming. At 2 weeks after the last immunization, sera were collected from the immunized rabbit, titrated, and served as the stock of anti-CVA16 polyclonal antibodies. The animal studies were approved by the Institutional Animal Care and Use Committee at the Institut Pasteur of Shanghai, and the animals were cared for in accordance with the institutional guidelines. The 9B5 anti-CVA16 monoclonal antibodies were generated in-house from mice immunized with inactivated CVA16 as described previously (44).
CVA16 preparation.
Inactivated CVA16 was as prepared as described previously (20) with minor modifications. Briefly, the CVA16/SZ05 strain was amplified in Vero cells and inactivated with BPL. The inactivated virus was first purified by 20% sucrose cushion and 10 to 50% sucrose gradient ultracentrifugation and then further purified by size exclusion chromatography with a Sephacryl S500 column (GE Healthcare) in phosphate-buffered saline (PBS). The purified virus was concentrated to 3.22 mg/ml by ultrafiltration with a filter with a 100-kDa cutoff (Millipore). The purified samples were subjected to SDS-PAGE and Coomassie blue staining.
Cryo-EM imaging.
An aliquot of 2 μl of sample was deposited onto a glow-discharged holey carbon Quantifoil Cu grid (R1.2x1.3, 200 mesh; Quantifoil), which was covered with a thin layer of homemade continuous carbon film. After 2 s of blotting to remove extra sample, the grid was plunge-frozen into liquid ethane using an FEI Mark IV Vitrobot. Specimens were examined under low-dose conditions at 300 kV with an FEI Titan Krios transmission electron microscope equipped with a Cs corrector. Images were recorded on a Falcon II direct electron detector in the 18-frame movie mode. The electron dose rate was set to ∼25 e−/Å2 · s, and the exposure time was 1.1 s. All the images were recorded at a nominal magnification of 57,000, corresponding to a pixel size of 1.1 Å/pixel, and with the defocus ranging from 1.5 to 2.5 μm.
Cryo-EM single-particle and 3D reconstruction.
To correct the drift and beam-induced motion, the 18 frames in each movie were aligned and averaged to a single micrograph using Motioncorr (45). Particles were boxed semiautomated using e2boxer.py from EMAN2.1 (46). CTF fitting was automatically performed using the fitctf2.py program in jspr (47) and then visually adjusted using the EMAN1.9 ctfit program (48). After CTF correction, the particles were separated into two halves, and on each half of the data we performed the reference-free two-dimensional (2D) analysis and initial model building utilizing EMAN2.1. In the following 3D reconstruction process, the gold standard procedure was followed using the jspr package (49). The further refinements of defocus, astigmatism, and magnification were all carried out in jspr (49). The map resolutions were assessed using gold standard criteria of a 0.143 FSC cutoff.
Structure analysis.
After reconstruction, the cryo-EM density maps were sharpened with structure factors to boost the density map Fourier amplitudes and then low-pass filtered to the assessed resolution using e2proc3d.py in EMAN2.1 (46). The central section image along the 5-fold symmetrical axis was generated using EMAN1.9 (48). To calculate the correlation coefficient between our cryo-EM density maps and the related crystal structures, we first fitted the crystal structures (including formaldehyde-inactivated CVA16 mature virion [PDB 5C4W], procapsid [PDB 5C9A] [12], and 135S-like uncoating intermediate [PDB 4JGY] [10], as well as native EV71 mature virion [PDB 3VBS] and procapsid [PDB 3VBU] [5]) into our cryo-EM maps using the fit in map module in UCSF Chimera (50). The fitted crystal structure was then converted into a density map in the same resolution as our cryo-EM map using the pdb2mrc program in EMAN1.9 (48). Afterwards, the correlation coefficient for the generated map and our cryo-EM map was calculated using the fit in map module in UCSF Chimera (50).
The linear neutralizing epitopes of CVA16 were identified as described previously (25). The location of the epitope in our cryo-EM map of mature CVA16 was determined according to the fitted crystal structure of the CVA16 135S particle (PDB 4JGY), and the epitope residues were chosen and colored. After that, the cryo-EM density within 3.5 Å of the epitope residues was depicted using the color zone module in UCSF Chimera. A similar procedure was adopted to determine the location of the epitope in our cryo-EM map of the CVA16 procapsid, other than that the crystal structure of the native EV71 procapsid (PDB 3VBU) was used as the reference.
Flexible fitting on the models of residues 216 to 220 in the VP1 GH loop (missing in the crystal structure of the CVA16 135S-like particle), as well as part of the VP3 GH loop (resolved in our CVA16 empty-capsid cryo-EM map but in a conformation different from that of the crystal structures of the EV71 E-particle [PDB 3VBS] and the CVA16 135S-like particle [PDB: 4JGY]) was carried out with the restraint of the cryo-EM density map of the BPL-treated CVA16 procapsid, using molecular dynamics flexible fitting (MDFF) (51).
Infectivity assay.
CVA16/SZ05 (3.89 × 108 50% tissue culture infective doses [TCID50]/ml) was incubated with BPL (Wako, Japan) at different final concentrations (143, 28.6, 5.72, 1.14, 0.23, and 0.046 mM) overnight at 4°C. The virus was then incubated for 2 h at 37°C to hydrolyze the remaining BPL. Virus without added BPL served as a control. The pH of inactivated virus was adjusted to 7.0 to match that of the control, and then the infectivity of inactivated virus was titrated by TCID50 assay on RD cells as described previously (8). To determine the inactivation rates with different BPL treatments, their corresponding TCID50 values were normalized to that of the control sample without BPL addition.
Pulldown assay.
Pulldown assays were performed to determine the influence of BPL inactivation on the virus interacting with heparin. Briefly, 200 μl of the viruses inactivated by different concentrations of BPL were incubated with 20 μl of heparin-conjugated agarose (Sigma) and 200 μl of Dulbecco modified Eagle medium (DMEM) containing 2% fetal bovine serum (FBS) and incubated overnight at 4°C with rotation. The mixtures were subjected to centrifugation at 3,000 rpm for 3 min. The supernatants were carefully removed, and the beads were washed three times with DMEM. Virus without BPL inactivation and DMEM containing 2% FBS served as controls. The virus-bound beads were analyzed by SDS-PAGE and Western blotting with rabbit polyclonal antibodies against the VP1 protein of CVA16.
Virus attachment assay.
RD cells were seeded into 24-well plates (1 × 105 cells/well) 1 day before the experiment. The cells were treated with or without 0.0125 IU/ml of heparinase III (Yuanye Biotech, Shanghai) in PBS at 37°C for 1 h. The plate was washed with cold PBS to remove heparinase and incubated with 250 μl/well of BPL-treated CVA16 or medium at 4°C for 1 h for virus attachment. The plate was washed three times to remove unattached virus, and then the cells were collected and subjected to Western blot assay. Purified CVA16 served as a positive control. The virus was detected with rabbit anti-VP1 polyclonal antibodies. β-Actin was detected using an anti-β-actin monoclonal antibody (Sigma) and served as an internal control.
ELISA.
The antigenicity of BPL-inactivated CVA16 was assessed by sandwich ELISA. Briefly, ELISA plates were coated overnight at 4°C with 50 μl/well of 1/5,000-diluted rabbit anti-CVA16 VLP serum and blocked with 5% nonfat milk in PBS–Tween 20 (PBST) for 2 h at 37°C. The BPL-treated virus samples were added to the plate and incubated at 37°C for 2 h. Virus without BPL treatment and DMEM containing 2% FBS served as controls. The plate was then incubated with 50 μl/well of 9B5 monoclonal antibody (0.2 μg/ml) or 1/1,000-diluted anti-CVA16 VLP polyclonal antibodies at 37°C for 2 h, followed by incubation with a horseradish peroxidase-conjugated secondary antibody for 1 h. All the samples were diluted with PBST plus 1% nonfat milk. After each incubation step, the plates were washed five times with PBST. After color development, the absorbance at 450 nm was determined on a 96-well plate reader.
Accession number(s).
The cryo-EM density maps of the CVA16 mature virion and premature procapsid have been deposited in the Electron Microscopy Data Bank under accession codes EMD-8324 and EMD-8325, respectively. The structures of the VP1 and VP3 subunits were deposited in PDB under accession numbers 5TSK and 5TSL, respectively.
ACKNOWLEDGMENTS
We thank Xia Jin of the Institute Pasteur of Shanghai for critical reading of the manuscript. We thank Junrui Li and Mi Cao from the Electron Microscopy facility and staff from the Data Base & Computation facility of the National Center for Protein Science Shanghai for their assistance with the EM instruments and the parallel computing.
This work was supported by grants from the National Natural Science Foundation of China (31370930, 31670754, and 31500153), the Science and Technology Commission of Shanghai Municipality (13431900600 and 15XD1524900), CAS Pilot Strategic Science and Technology Projects B (XDB08030201), the National Basic Research Program of China (2013CB910401), the CAS-Shanghai Science Research Center (CAS-SSRC-YH-2015-01), and the STS Program of the Chinese Academy of Sciences (KFJ-EW-STS-098).
REFERENCES
- 1.Xing W, Liao Q, Viboud C, Zhang J, Sun J, Wu JT, Chang Z, Liu F, Fang VJ, Zheng Y, Cowling BJ, Varma JK, Farrar JJ, Leung GM, Yu H. 2014. Hand, foot, and mouth disease in China, 2008-12: an epidemiological study. Lancet Infect Dis 14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang F, Zhang T, Hu Y, Wang X, Du J, Li Y, Sun S, Sun X, Li Z, Jin Q. 2011. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol J 8:508. doi: 10.1186/1743-422X-8-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yip CC, Lau SK, Zhou B, Zhang MX, Tsoi HW, Chan KH, Chen XC, Woo PC, Yuen KY. 2010. Emergence of enterovirus 71 “double-recombinant” strains belonging to a novel genotype D originating from southern China: first evidence for combination of intratypic and intertypic recombination events in EV71. Arch Virol 155:1413–1424. doi: 10.1007/s00705-010-0722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Zhu Z, Yang W, Ren J, Tan X, Wang Y, Mao N, Xu S, Zhu S, Cui A, Zhang Y, Yan D, Li Q, Dong X, Zhang J, Zhao Y, Wan J, Feng Z, Sun J, Wang S, Li D, Xu W. 2010. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol J 7:94. doi: 10.1186/1743-422X-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol 19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F, Liu Q, Cai Y, Leng Q, Huang Z. 2011. Construction and characterization of an infectious clone of coxsackievirus A16. Virol J 8:534. doi: 10.1186/1743-422X-8-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Ku Z, Cai Y, Sun B, Leng Q, Huang Z. 2011. Detection, characterization and quantitation of coxsackievirus A16 using polyclonal antibodies against recombinant capsid subunit proteins. J Virol Methods 173:115–120. doi: 10.1016/j.jviromet.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Chong P, Guo MS, Lin FH, Hsiao KN, Weng SY, Chou AH, Wang JR, Hsieh SY, Su IJ, Liu CC. 2012. Immunological and biochemical characterization of coxsackie virus A16 viral particles. PLoS One 7:e49973. doi: 10.1371/journal.pone.0049973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat Commun 4:1929. doi: 10.1038/ncomms2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J Virol 79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren J, Wang X, Zhu L, Hu Z, Gao Q, Yang P, Li X, Wang J, Shen X, Fry EE, Rao Z, Stuart DI. 2015. Structures of coxsackievirus A16 capsids with native antigenicity: implications for particle expansion, receptor binding, and immunogenicity. J Virol 89:10500–10511. doi: 10.1128/JVI.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong M, Zhu H, Zhou J, Yang C, Feng J, Huang X, Ji G, Xu H, Zhu P. 2014. Cryo-electron microscopy study of insect cell-expressed enterovirus 71 and coxsackievirus a16 virus-like particles provides a structural basis for vaccine development. J Virol 88:6444–6452. doi: 10.1128/JVI.00200-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 15.Hogle JM, Chow M, Filman DJ. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 16.Butan C, Filman DJ, Hogle JM. 2014. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol 88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong P, Liu CC, Chow YH, Chou AH, Klein M. 2015. Review of enterovirus 71 vaccines. Clin Infect Dis 60:797–803. doi: 10.1093/cid/ciu852. [DOI] [PubMed] [Google Scholar]
- 18.Yang E, Cheng C, Zhang Y, Wang J, Che Y, Pu J, Dong C, Liu L, He Z, Lu S, Zhao Y, Jiang L, Liao Y, Shao C, Li Q. 2014. Comparative study of the immunogenicity in mice and monkeys of an inactivated CA16 vaccine made from a human diploid cell line. Hum Vaccin Immunother 10:1266–1273. doi: 10.4161/hv.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Ku Z, Liu Q, Leng Q, Huang Z. 2014. A combination vaccine comprising of inactivated enterovirus 71 and coxsackievirus A16 elicits balanced protective immunity against both viruses. Vaccine 32:2406–2412. doi: 10.1016/j.vaccine.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Cai Y, Liu Q, Huang X, Li D, Ku Z, Zhang Y, Huang Z. 2013. Active immunization with a coxsackievirus A16 experimental inactivated vaccine induces neutralizing antibodies and protects mice against lethal infection. Vaccine 31:2215–2221. doi: 10.1016/j.vaccine.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Cheng NQ, Chow M, Filman DJ, Steven AC, Hogle JM, Belnap DM. 2011. An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles. J Virol 85:9974–9983. doi: 10.1128/JVI.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Lee LY, Roivainen M, Filman DJ, Hogle JM, Belnap DM. 2012. Structure of the Fab-labeled “breathing” state of native poliovirus. J Virol 86:5959–5962. doi: 10.1128/JVI.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fricks CE, Hogle JM. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol 64:1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panjwani A, Strauss M, Gold S, Wenham H, Jackson T, Chou JJ, Rowlands DJ, Stonehouse NJ, Hogle JM, Tuthill TJ. 2014. Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore. PLoS Pathog 10:e1004294. doi: 10.1371/journal.ppat.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi JP, Huang XL, Liu QW, Huang Z. 2013. Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine 31:2130–2136. doi: 10.1016/j.vaccine.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 26.Tan CW, Poh CL, Sam IC, Chan YF. 2013. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol 87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metz B, Kersten GF, Baart GJ, de Jong A, Meiring H, ten Hove J, van Steenbergen MJ, Hennink WE, Crommelin DJ, Jiskoot W. 2006. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug Chem 17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 28.Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ, Jiskoot W. 2004. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem 279:6235–6243. [DOI] [PubMed] [Google Scholar]
- 29.Colburn NH, Richardson RG, Boutwell RK. 1965. Studies of the reaction of beta-propiolactone with deoxyguanosine and related compounds. Biochem Pharmacol 14:1113–1118. doi: 10.1016/0006-2952(65)90040-7. [DOI] [PubMed] [Google Scholar]
- 30.Mate U, Solomon JJ, Segal A. 1977. In vitro binding of beta-propiolactone to calf thymus DNA and mouse liver DNA to form 1-(2-carboxyethyl) adenine. Chem Biol Interact 18:327–336. doi: 10.1016/0009-2797(77)90018-7. [DOI] [PubMed] [Google Scholar]
- 31.Roberts JJ, Warwick GP. 1963. The reaction of beta-propiolactone with guanosine, deoxyguanylic acid and RNA. Biochem Pharmacol 12:1441–1442. doi: 10.1016/0006-2952(63)90216-8. [DOI] [PubMed] [Google Scholar]
- 32.Budimir N, Huckriede A, Meijerhof T, Boon L, Gostick E, Price DA, Wilschut J, de Haan A. 2012. Induction of heterosubtypic cross-protection against influenza by a whole inactivated virus vaccine: the role of viral membrane fusion activity. PLoS One 7:e30898. doi: 10.1371/journal.pone.0030898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uittenbogaard JP, Zomer B, Hoogerhout P, Metz B. 2011. Reactions of beta-propiolactone with nucleobase analogues, nucleosides, and peptides: implications for the inactivation of viruses. J Biol Chem 286:36198–36214. doi: 10.1074/jbc.M111.279232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.She YM, Cheng K, Farnsworth A, Li X, Cyr TD. 2013. Surface modifications of influenza proteins upon virus inactivation by beta-propiolactone. Proteomics 13:3537–3547. doi: 10.1002/pmic.201300096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnafous P, Nicolai MC, Taveau JC, Chevalier M, Barriere F, Medina J, Le Bihan O, Adam O, Ronzon F, Lambert O. 2014. Treatment of influenza virus with beta-propiolactone alters viral membrane fusion. Biochim Biophys Acta 1838:355–363. doi: 10.1016/j.bbamem.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 36.Rombaut B, Vrijsen R, Boeye A. 1987. A pH-dependent dissociation of poliovirus procapsids. Virology 157:245–247. doi: 10.1016/0042-6822(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 37.Liu CC, Guo MS, Lin FH, Hsiao KN, Chang KH, Chou AH, Wang YC, Chen YC, Yang CS, Chong PC. 2011. Purification and characterization of enterovirus 71 viral particles produced from vero cells grown in a serum-free microcarrier bioreactor system. PLoS One 6:e20005. doi: 10.1371/journal.pone.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergelson JM, Coyne CB. 2013. Picornavirus entry. Adv Exp Med Biol 790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann MG, Bella J, Kolatkar PR, He Y, Wimmer E, Kuhn RJ, Baker TS. 2000. Cell recognition and entry by rhino- and enteroviruses. Virology 269:239–247. doi: 10.1006/viro.2000.0258. [DOI] [PubMed] [Google Scholar]
- 40.Hadfield AT, Diana GD, Rossmann MG. 1999. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci U S A 96:14730–14735. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Sena J, Mandel B. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554–566. [DOI] [PubMed] [Google Scholar]
- 42.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. 2014. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol 88:5755–5765. doi: 10.1128/JVI.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Liu Q, Ku Z, Hu Y, Ye X, Zhang Y, Huang Z. 2016. Coxsackievirus A16-like particles produced in Pichia pastoris elicit high-titer neutralizing antibodies and confer protection against lethal viral challenge in mice. Antiviral Res 129:47–51. doi: 10.1016/j.antiviral.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Ku Z, Liu Q, Ye X, Cai Y, Wang X, Shi J, Li D, Jin X, An W, Huang Z. 2014. A virus-like particle based bivalent vaccine confers dual protection against enterovirus 71 and coxsackievirus A16 infections in mice. Vaccine 32:4296–4303. doi: 10.1016/j.vaccine.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Jiang W, Guo F, Liu Z. 2012. A graph theory method for determination of cryo-EM image focuses. J Struct Biol 180:343–351. doi: 10.1016/j.jsb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 49.Guo F, Jiang W. 2014. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol 1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. 2009. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]