ABSTRACT
Hepatitis B virus (HBV) infection may cause acute hepatitis B, chronic hepatitis B (CHB), liver cirrhosis, and hepatocellular carcinoma (HCC). However, the mechanisms by which HBV evades host immunity and maintains chronic infection are largely unknown. Here, we revealed that matrix metalloproteinase 9 (MMP-9) is activated in peripheral blood mononuclear cells (PBMCs) of HBV-infected patients, and HBV stimulates MMP-9 expression in macrophages and PBMCs isolated from healthy individuals. MMP-9 plays important roles in the breakdown of the extracellular matrix and in the facilitation of tumor progression, invasion, metastasis, and angiogenesis. MMP-9 also regulates respiratory syncytial virus (RSV) replication, but the mechanism underlying such regulation is unknown. We further demonstrated that MMP-9 facilitates HBV replication by repressing the interferon (IFN)/Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway, IFN action, STAT1/2 phosphorylation, and IFN-stimulated gene (ISG) expression. Moreover, MMP-9 binds to type I IFN receptor 1 (IFNAR1) and facilitates IFNAR1 phosphorylation, ubiquitination, subcellular distribution, and degradation to interfere with the binding of IFANR1 to IFN-α. Thus, we identified a novel positive-feedback regulation loop between HBV replication and MMP-9 production. On one hand, HBV activates MMP-9 in infected patients and leukocytes. On the other hand, MMP-9 facilitates HBV replication through repressing IFN/JAK/STAT signaling, IFNAR1 function, and IFN-α action. Therefore, HBV may take the advantage of MMP-9 function to establish or maintain chronic infection.
IMPORTANCE Hepatitis B virus (HBV) infection may cause chronic hepatitis B (CHB) and hepatocellular carcinoma (HCC). However, the mechanisms by which HBV maintains chronic infection are largely unknown. Matrix metalloproteinase 9 (MMP-9) plays important roles in the facilitation of tumor progression, invasion, metastasis, and angiogenesis. However, the effects of MMP-9 on HBV replication and pathogenesis are not known. This study reveals that MMP-9 expression is activated in patients with CHB, and HBV stimulates MMP-9 production in PBMCs and macrophages. More interestingly, MMP-9 in turn promotes HBV replication through suppressing IFN-α action. Moreover, MMP-9 interacts with type I interferon receptor 1 (IFNAR1) to disturb the binding of IFN-α to IFNAR1 and facilitate the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1. Therefore, these results discover a novel role of MMP-9 in viral replication and reveal a new mechanism by which HBV evades host immunity to maintain persistent infection.
KEYWORDS: hepatitis B virus, interferon/Janus kinase/signal transducers and activators of transcription pathway, matrix metalloproteinase 9, type I IFN receptor 1
INTRODUCTION
Two billion people have been infected with hepatitis B virus (HBV) worldwide; about 350 million infected patients develop chronic hepatitis B (CHB), and 600,000 die each year from HBV-related liver cirrhosis or hepatocellular carcinoma (HCC) (1). Adults infected with HBV can clear the virus by the host immune system in 95% of cases, while 90% of perinatal infections develop into CHB (2). Chronic HBV infection begins when the immune response fails to clear the virus, and CHB accounts for approximately 50% of HCC cases (3–5).
Interferons (IFNs) are classified into three types by their unique cell surface receptors. Type I IFNs act through ubiquitously expressed IFN-α/β receptors (type I IFN receptor 1 [IFNAR1] and IFNAR2), which are associated with tyrosine kinase 2 (TYK2) and Janus kinase 1 (JAK1), respectively (6). Once IFN binds to IFNAR, TYK2 and JAK1 are activated by transphosphorylation to phosphorylate IFNAR1/2. Signal transducer and activator of transcription 1 (STAT1) and STAT2 are recruited to the receptor complex and phosphorylated to form a new complex with IFN regulatory factor 9 (IRF9) and then translocate into the nucleus to activate IFN-stimulated genes (ISGs) (7).
Matrix metalloproteinase 9 (MMP-9) is expressed in normal leukocytes and transformed cells (8). Elevated serum levels of MMP-9 were detected in CHB and HCC patients (9, 10), and HBV upregulates MMP-9 in hepatocytes (11). MMP-9 plays important roles in the breakdown of the extracellular matrix and the facilitation of tumor progression, invasion, metastasis, and angiogenesis. It was reported previously that MMP-9 mediated respiratory syncytial virus (RSV) replication in vitro and in vivo (12) and promoted the multiplication of RSV (13). Another report showed that MMP-9 exerted antiviral activity against RSV by enhancing neutrophil recruitment to the lungs in mice (14). However, the role of MMP-9 in the replication of HBV is unknown.
Here, we revealed that MMP-9 levels are elevated in peripheral blood mononuclear cells (PBMCs) of CHB patients and that HBV upregulates MMP-9 in PBMCs and macrophages in vitro. Interestingly, MMP-9 in turn facilitates HBV replication in human hepatoma cells through suppressing IFN-α action. More importantly, MMP-9 interacts with IFNAR1 to disturb the binding of IFN-α to IFNAR1. Moreover, MMP-9 facilitates the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1. Therefore, we revealed a novel mechanism by which MMP-9 promotes virus replication through repressing IFN/JAK/STAT signaling and IFNAR1 activity.
RESULTS
HBV activates MMP-9 expression in PBMCs of CHB patients and in macrophages differentiated from THP-1 cells.
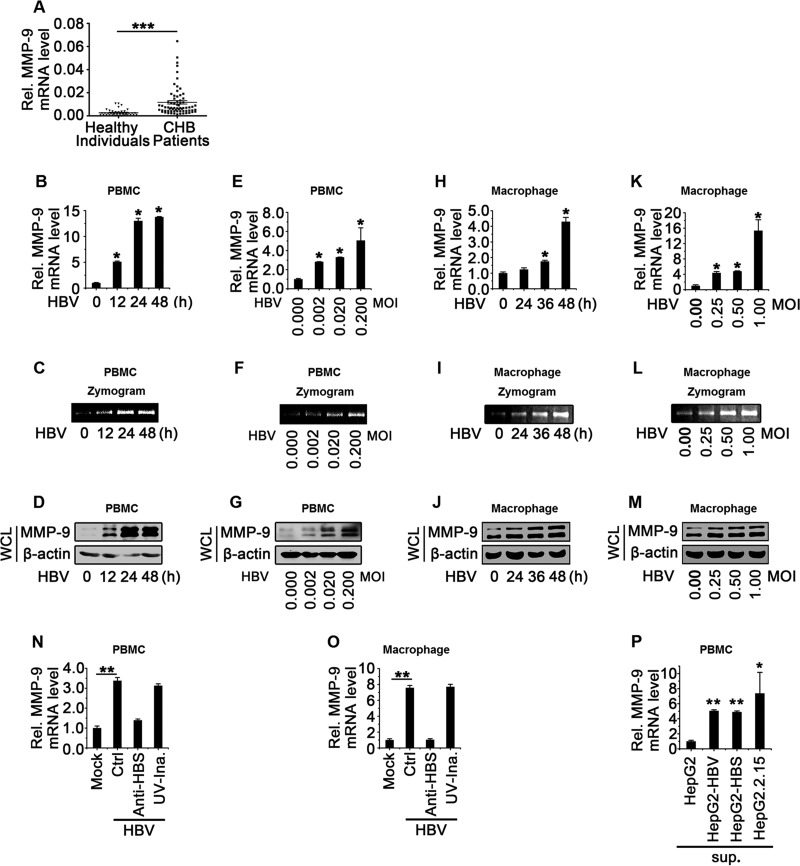
MMP-9 is produced mainly by leukocytes, and HBV activates MMP-9 in hepatocytes and in the serum of CHB patients (9, 11, 15, 16); meanwhile, HBV is recognized by liver macrophages upon infection of primary liver cells (17). Here, we determined whether HBV activates MMP-9 in leukocytes or macrophages in vivo and in vitro. Analyses of PBMCs isolated from CHB patients (n = 69) and healthy individuals (n = 40) revealed that MMP-9 mRNA levels were significantly higher in CHB patients than in healthy individuals (Fig. 1A), suggesting that MMP-9 is activated in PBMCs of CHB patients.
FIG 1.
MMP-9 is upregulated in PBMCs of CHB patients and activated by HBV in PBMCs and macrophages in vitro. (A) MMP-9 mRNA levels in PBMCs of CHB patients (n = 69) and healthy individuals (n = 40) were measured by qPCR. Points represent MMP-9 mRNA levels of each sample. (B to G) PBMCs (1 × 106) obtained from healthy individuals were incubated with supernatants isolated from HepG2 cultures (without HBV) or HepG2.2.15 cultures (containing HBV) at an MOI of 0.5 for different times (B to D) or for 24 h at different MOIs (E to G). The MMP-9 mRNA level was measured by qPCR and normalized to the GAPDH mRNA level. (B and E) Results are standardized to a value of 1 for HepG2 supernatant-treated cells. (C and F) MMP-9 proteinase activity in the supernatants was determined by gelatin zymography assays. (D and G) The MMP-9 protein level in WCLs was determined by Western blotting. (H to M) Macrophages (1 × 106) differentiated from THP-1 cells were incubated with supernatants isolated from HepG2 cultures or HepG2.2.15 cultures at an MOI of 0.5 for different times (H to J) or at different MOIs for 48 h (K to M). The MMP-9 mRNA level was measured by qPCR and normalized to the GAPDH mRNA level. (H and K) Results are standardized to a value of 1 for HepG2 supernatant-treated cells. (I and L) MMP-9 proteinase activity in the supernatants was determined by gelatin zymography assays. (J and M) The MMP-9 protein level in WCLs was determined by Western blotting. (N) PBMCs (1 × 106) isolated from healthy individuals were incubated with supernatants of HepG2 cultures, supernatants of HepG2.2.15 cultures, anti-HBsAg antibody-pretreated supernatants of HepG2.2.15 cultures, or UV-inactivated HepG2.2.15 supernatants. The MMP-9 mRNA level was measured by qPCR. (O) Macrophages (1 × 106) differentiated from THP-1 cells were incubated with supernatants of HepG2 cultures, supernatants of HepG2.2.15 cultures, anti-HBsAg antibody-pretreated supernatants of HepG2.2.15 cultures, or UV-inactivated HepG2.2.15 supernatants. The MMP-9 mRNA level was measured by qPCR. (P) PBMCs (1 × 106) isolated from healthy individuals were incubated with supernatants of HepG2 cultures, supernatants of cultures of HepG2 cells transfected with pHBV1.3, supernatants of cultures of HepG2 cells transfected with pCMV-HBS, or supernatants of HepG2.2.15 cultures. The MMP-9 mRNA level was measured by qPCR. Graphs show means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further determine the role of HBV in the regulation of MMP-9, PBMCs isolated from healthy individuals were incubated with the supernatants of HepG2 cells (without HBV) or HepG2.2.15 cell cultures (with HBV). Quantitative PCR (qPCR), Western blot, and gelatin zymography assays revealed that MMP-9 mRNA, enzyme activity, and protein levels were upregulated by HBV in time-dependent fashions (Fig. 1B to D) and dose-dependent manners (Fig. 1E to G) in PBMCs. In addition, macrophages differentiated from human acute monocytic leukemia cells (THP-1) were incubated with the supernatants of HepG2 or HepG2.2.15 cell cultures. Similarly, qPCR, gelatin zymography assays, and Western blotting revealed that MMP-9 mRNA, enzyme activity, and protein levels were upregulated by HBV in time-dependent fashions (Fig. 1H to J) and dose-dependent manners (Fig. 1K to M) in macrophages.
The effect of HBV on the expression of MMP-9 and the mechanism underlying such regulation were then determined. PBMCs isolated from healthy individuals and THP-1-differentiated macrophages were incubated with the supernatants of HepG2 cells, the supernatants of HepG2.2.15 cells, the supernatants of HepG2.2.15 cells pretreated with neutralizing antibody (Ab) (anti-hepatitis B virus s antigen [anti-HBsAg]), or the supernatants of HepG2.2.15 cells inactivated with UV light. MMP-9 mRNA was upregulated by HBV, and such activation was attenuated by treatment with anti-HBsAg antibody but was not affected by treatment with UV light (Fig. 1N and O). In addition, PBMCs isolated from healthy individuals were incubated with the supernatants of HepG2 cells, the supernatants of HepG2 cells transfected with pHBV1.3, the supernatants of HepG2 cells transfected with pCMV-HBS, or the supernatants of HepG2.2.15 cells. The level of MMP-9 mRNA was upregulated by the supernatants of HepG2 cells transfected with pHBV1.3 or with pCMV-HBS and activated by the supernatants of HepG2.2.15 cells (Fig. 1P). These results revealed that HBV activates MMP-9 expression in PBMCs and macrophages and implicated the involvement of the HBs protein, but not viral replication, in the HBV-mediated activation of MMP-9. Our results are consistent with data from previous studies that showed that the HBV envelope protein induces NF-κB in nonparenchymal liver cells (17), and MMP-9 expression is induced by the activation of NF-κB signaling (11). Taken together, we demonstrated that HBV activates MMP-9 expression in PBMCs of CHB patients and in macrophages differentiated from THP-1 cells.
MMP-9 plays a stimulatory role in the activation of HBV replication in hepatocytes.
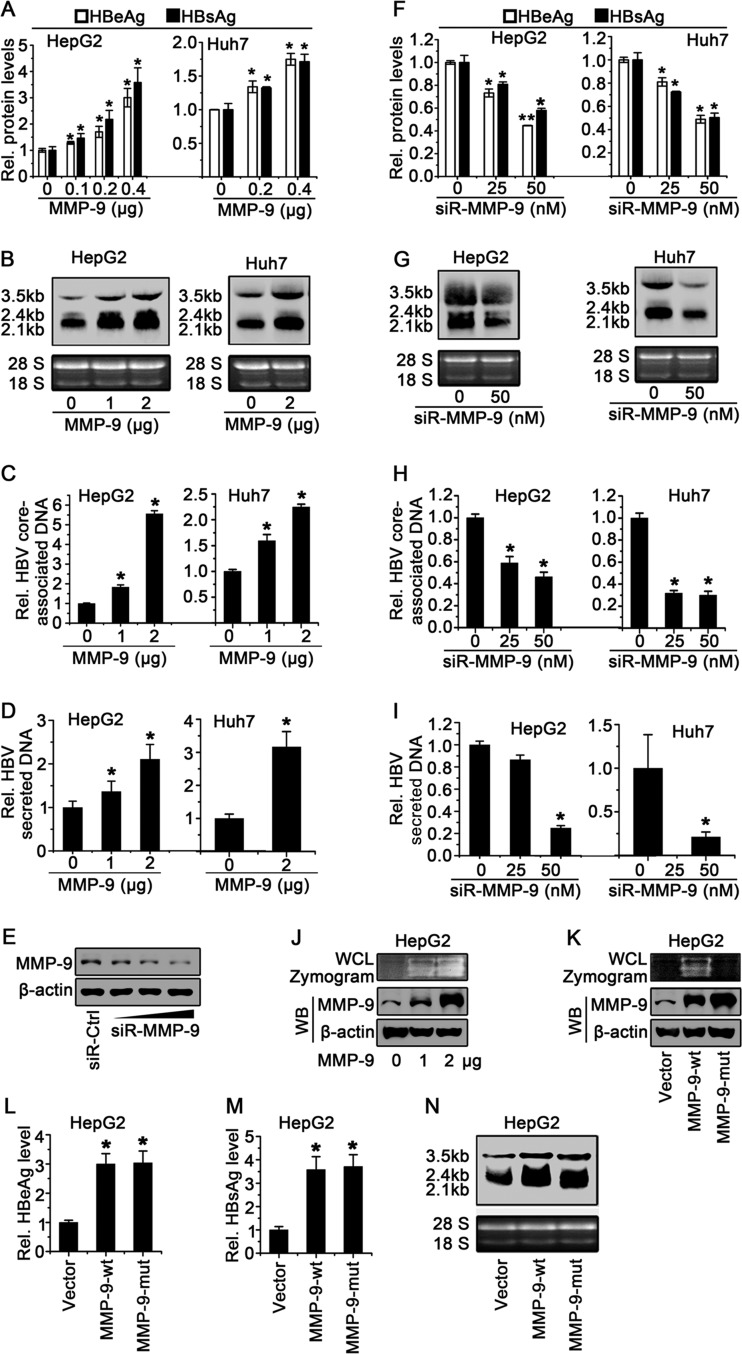
MMP-9 plays important roles in wound healing, arthritis, angiogenesis, and tumor metastasis (18), but the effect of MMP-9 on HBV replication has not been reported. Here, we initially determined the role of MMP-9 in the regulation of HBV replication in HepG2 cells and Huh7 cells cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9. Hepatitis B virus e antigen (HBeAg) and HBsAg were upregulated by MMP-9 in dose-dependent manners in HepG2 cells (Fig. 2A, left) and Huh7 cells (Fig. 2A, right). In addition, HBV RNAs (3.5-kb, 2.4-kb, and 2.1-kb RNAs) were enhanced by MMP-9 in HepG2 cells (Fig. 2B, left) and Huh7 cells (Fig. 2B, right). Moreover, HBV core-associated DNA was facilitated by MMP-9 (Fig. 2C). Finally, secreted HBV core-associated DNA was stimulated by MMP-9 (Fig. 2D). These results revealed that the overexpression of MMP-9 upregulates HBV replication.
FIG 2.
MMP-9 enhances HBV replication in hepatocytes. (A to D) HepG2 cells (left) and Huh7 cells (right) were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 at different concentrations for 48 h. (A) HBeAg and HBsAg in the supernatants of transfected cells were assayed by an ELISA. (B) HBV RNAs (3.5-kb, 2.4-kb, and 2.1-kb RNAs) were detected by Northern blot analyses, and 18S and 28S rRNAs were used as loading controls. (C and D) Levels of HBV core-associated DNAs in cell cultures (C) or supernatants (D) were measured by qPCR. (E) HepG2 cells were transfected with control siRNA (siR-Ctrl) or siR-MMP-9 at different concentrations for 48 h. Levels of MMP-9 and β-actin proteins expressed in treated cells were determined by Western blot analyses using the corresponding antibodies. (F to I) HepG2 cells (left) and Huh7 cells (right) were cotransfected with pHBV1.3 and siR-MMP-9 at different concentrations for 48 h. (F) HBeAg and HBsAg levels in the supernatants were assayed by an ELISA. (G) HBV RNAs were detected by Northern blot analyses, and 18S and 28S rRNAs were used as loading controls. (H and I) Levels of HBV core-associated DNAs in cell cultures (H) or supernatants (I) were measured by qPCR. (J) HepG2 cells were transfected with pCMV-Tag2B or pCMV-Tag2B-MMP-9 for 48 h. MMP-9 enzyme activity in WCLs was determined by a gelatin zymography assay (top), and the MMP-9 protein level was determined by Western blotting (WB) (bottom). (K) HepG2 cells were transfected with pCMV-Tag2B, pCMV-Tag2B-MMP-9-wt, or pCMV-Tag2B-MMP-9-mut for 48 h. MMP-9 enzyme activity in WCLs was determined by a gelatin zymography assay (top), and the MMP-9 protein level was determined by Western blotting (bottom). (L to N) HepG2 cells were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 or pCMV-Tag2B-MMP-9-mut. (L and M) HBeAg (L) and HBsAg (M) in the supernatants were assayed by an ELISA. (N) HBV RNAs were detected by Northern blot analyses. A total of 5 × 105 cells were used to determine HBeAg/HBsAg expression levels, and 2 × 106 cells were used for analyses of HBV DNA/RNA levels. Results show means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01.
We then determined the effect of the knockdown of MMP-9 on the regulation of HBV replication in HepG2 cells and Huh7 cells cotransfected with pHBV1.3 and small interfering RNA (siRNA) specific for MMP-9 (siR-MMP-9). The MMP-9 protein was attenuated by siR-MMP-9 in a dose-dependent manner (Fig. 2E), indicating that siR-MMP-9 is effective. HBeAg and HBsAg were downregulated by siR-MMP-9 in dose-dependent manners in HepG2 cells (Fig. 2F, left) and Huh7 cells (Fig. 2F, right). In addition, HBV RNAs were attenuated by siR-MMP-9 in HepG2 cells (Fig. 2G, left) and Huh7 cells (Fig. 2G, right). Moreover, HBV core-associated DNA was repressed by siR-MMP-9 (Fig. 2H). Furthermore, the level of secreted HBV core-associated DNA was reduced by siR-MMP-9 (Fig. 2I). These results suggested that the knockdown of MMP-9 downregulates HBV replication.
Since MMP-9 is a matrix metalloproteinase, the effect of MMP-9 enzyme activity on HBV replication was evaluated in HepG2 cells transfected with pCMV-Tag2B-MMP-9. A gelatin zymography assay showed that MMP-9 enzyme activity was increased in HepG2 cells as the MMP-9 concentration increased (Fig. 2J), confirming that MMP-9 has metalloproteinase activity. A mutant MMP-9 protein (MMP-9-mut) lacking metalloproteinase activity was generated, in which 3 histidines (H) in the catalytic domain of wild-type MMP-9 (MMP-9-wt) were replaced by lysines (K). HepG2 cells were then transfected with pCMV-Tag2B, pCMV-Tag2B-MMP-9-wt, and pCMV-Tag2B-MMP-9-mut. The results confirmed that MMP-9-wt had enzyme activity and that MMP-9-mut lacked enzyme activity in HepG2 cells (Fig. 2K). Our results indicated that HBeAg, HBsAg, and HBV RNAs were upregulated by both MMP-9-wt and MMP-9-mut (Fig. 2L to N). These results indicated that the enzyme activity of MMP-9 is not required for the function of MMP-9 in the activation of HBV replication. These results demonstrated that MMP-9 plays a stimulatory role in the activation of HBV replication in hepatocytes.
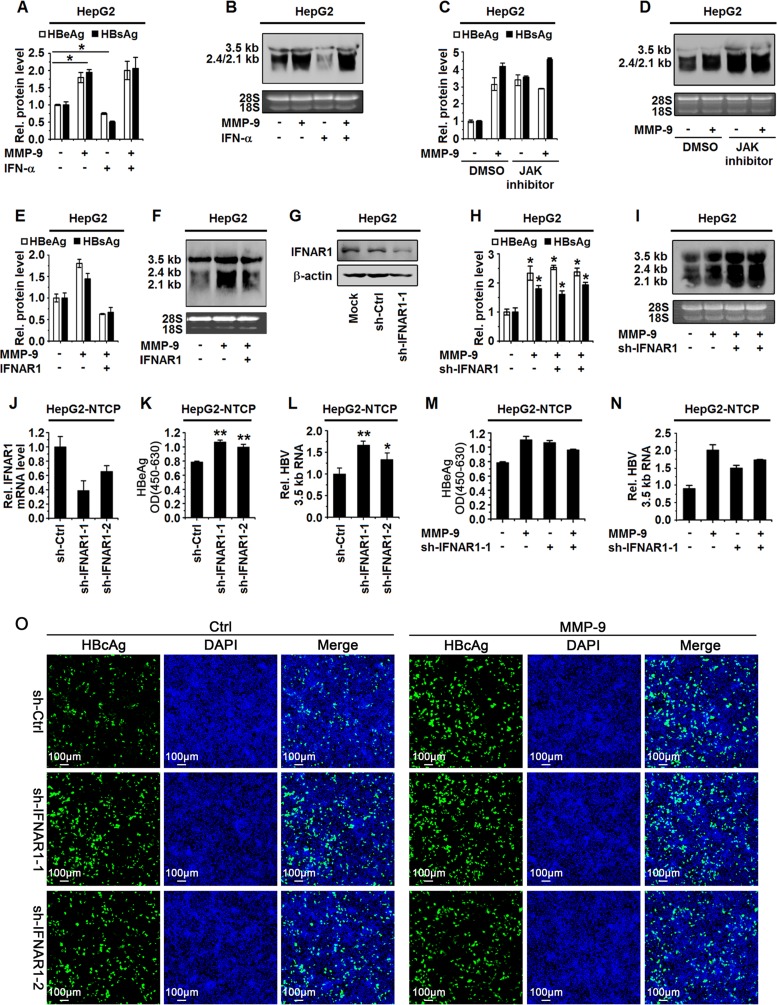
MMP-9 facilitates HBV replication in the HepG2-NTCP infection system.
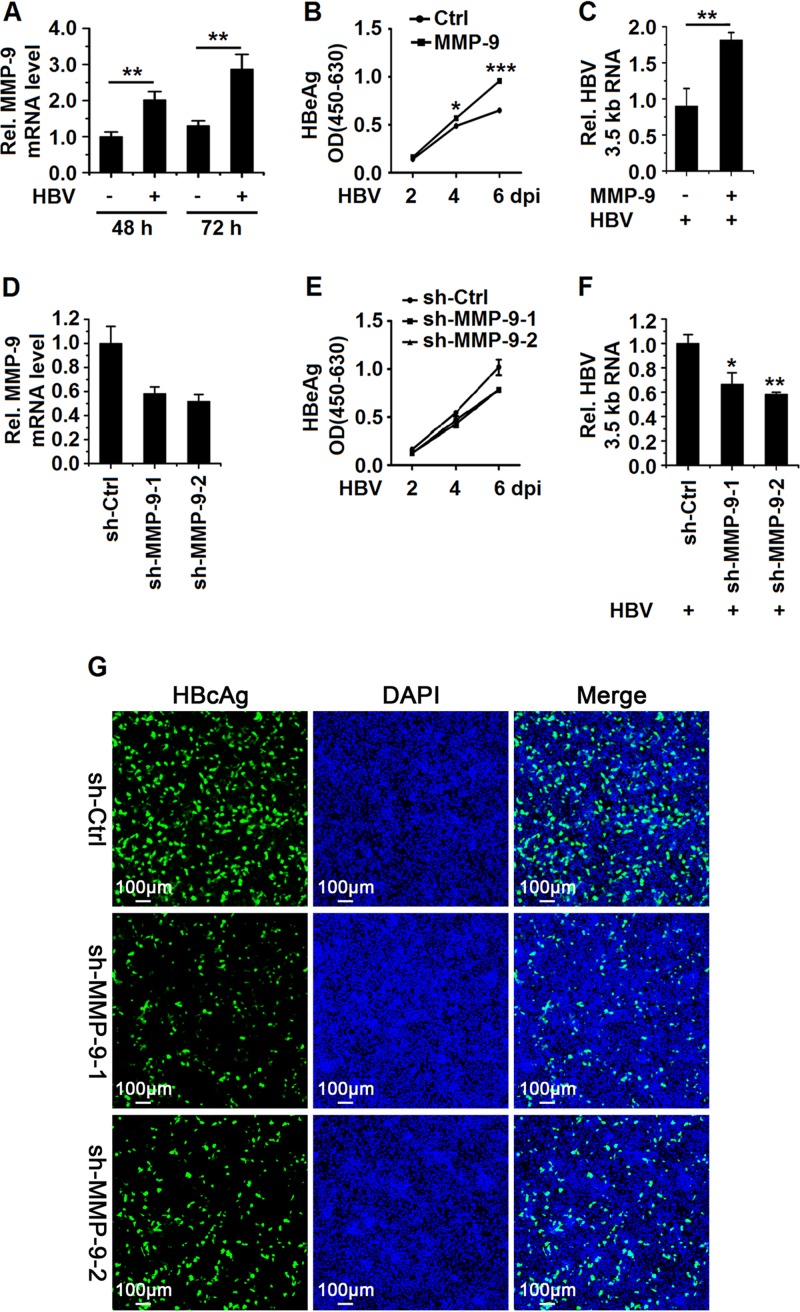
To investigate the effect of MMP-9 on the HBV replication cycle, HepG2-NTCP cells were infected with or without HBV by inoculation with or without the supernatants of HepaAD38 cells (19, 20). MMP-9 mRNA expression was induced in HBV-infected HepG2-NTCP cells (Fig. 3A). HepG2-NTCP cells were transfected with pCMV-Tag2B-MMP-9 and then infected with HBV. The secretion of HBeAg (Fig. 3B) and the expression of HBV 3.5-kb RNA (Fig. 3C) were activated by MMP-9. In addition, HepG2-NTCP cells were transfected with sh-MMP-9-1 and sh-MMP-9-2 and then infected with HBV. The levels of MMP-9 were downregulated by sh-MMP-9-1 and sh-MMP-9-2, indicating that these shRNAs are effective (Fig. 3D). The secretion of HBeAg (Fig. 3E) and the expression of HBV 3.5-kb RNA (Fig. 3F) were attenuated in the presence of sh-MMP-9-1 and sh-MMP-9-2. Similarly, intracellular HBcAg levels were reduced in the presence of sh-MMP-9-1 and sh-MMP-9-2 (Fig. 3G). Taken together, we demonstrated that MMP-9 facilitates HBV replication in the HepG2-NTCP infection system.
FIG 3.
MMP-9 facilitates HBV replication in a HepG2-NTCP infection system. (A) HepG2-NTCP cells were mock infected or infected with HBV at 1,000 GEq for the indicated times. MMP-9 mRNA levels were measured by qPCR. (B and C) HepG2-NTCP cells were transfected with the vector or pCMV-Tag2B-MMP-9. Twenty-four hours later, cells were infected with HBV at 1,000 GEq. (B) HBeAg levels in the supernatants of infected cells at the indicated times were determined by an ELISA. OD, optical density. (C) The levels of HBV 3.5-kb RNA at 6 days postinfection (dpi) were measured by qPCR. (D to G) HepG2-NTCP cells were transfected with sh-Ctrl, sh-MMP-9-1, or sh-MMP-9-2. Twenty-four hours later, cells were infected with HBV at 1,000 GEq. (E) HBeAg levels in the supernatants of infected cells at the indicated times were determined by an ELISA. (D and F) The mRNA levels of MMP-9 (D) and HBV 3.5-kb RNA (F) at 6 days postinfection were measured by qPCR. (G) The expression of HBcAg in cells was analyzed by immunostaining at 8 days postinfection. Graphs show means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Type I IFN signaling is upregulated by HBV infection and downregulated by MMP-9.
The mechanism by which MMP-9 regulates HBV replication was evaluated. Previous studies showed that during HBV infection, the activation of innate responses is predominantly weak or transient (21–23). However, other studies reported that HBV elicits a strong and specific innate antiviral response that results in the noncytopathic clearance of HBV DNA in HepaRG cells, and IFN-β and ISGs were induced in HepG2 cells by HBV baculovirus (Bac-HBV) (24). In this study, we determined the effects of HBV infection and replication on the innate immune response in HepG2-NTCP cells.
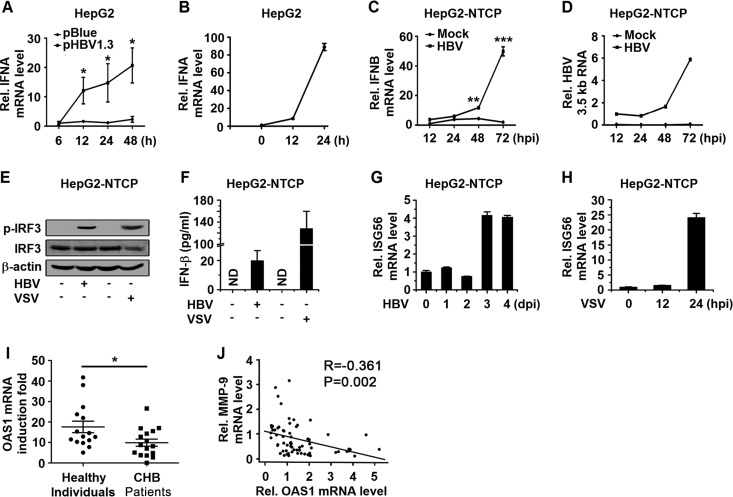
Initially, we showed that the gene encoding IFN-α (IFNA) mRNA was facilitated by HBV in pHBV1.3-transfected HepG2 cells (Fig. 4A). For comparison, IFNA mRNA was induced significantly by infection with vesicular stomatitis virus (VSV) in HepG2 cells (Fig. 4B). In addition, IFNB mRNA was enhanced by HBV infection in HepG2-NTCP cells in a time-dependent manner (Fig. 4C), and this induction was correlated with the expression of HBV 3.5-kb RNA (Fig. 4D). Moreover, the phosphorylated IRF3 (p-IRF3) protein (Fig. 4E) and the IFN-β protein (Fig. 4F) were induced by HBV and significantly activated by VSV in HepG2-NTCP cells. Finally, the expression of an interferon-stimulated gene (ISG56) was facilitated by HBV (Fig. 4G) and significantly induced by VSV (Fig. 4H). Taken together, these results suggested that HBV infection and replication induce a type I IFN response in HepG2-NTCP cells and HepG2 cells.
FIG 4.
HBV infection activates type I IFN signaling in HepG2-NTCP cells. (A) HepG2 cells were transfected with pBlue or pHBV1.3 for the indicated times, and the mRNA levels of IFNA were determined by qPCR. (B) HepG2 cells were infected with VSV (MOI = 1) for the indicated times, and the mRNA levels of IFNA were determined by qPCR. (C and D) HepG2-NTCP cells were mock infected or infected with HBV at 1,000 GEq per cell. IFNB mRNA levels (C) and HBV 3.5-kb RNA levels (D) were analyzed by qPCR at the indicated times postinfection. hpi, hours postinfection. (E and F) HepG2-NTCP cells were mock infected or infected with HBV at 1,000 GEq for 6 days or infected with or without VSV at an MOI of 1 for 24 h. (E) Protein levels of p-IRF3, IRF3, and β-actin were determined by Western blotting. (F) Protein levels of IFN-β in the culture supernatants were determined by an ELISA. (G and H) HepG2-NTCP cells were infected with HBV (1,000 GEq) or VSV (MOI = 1) for the indicated times, and ISG56 mRNA levels were measured by qPCR. (I) PBMCs freshly isolated from CHB patients (n = 16) and healthy individuals (n = 15) were stimulated with or without IFN-α (300 U/ml) for 12 h. OAS1 mRNA levels were measured by qPCR. Points represent the fold induction of OAS1 mRNA for each sample. (J) MMP-9 and OAS1 mRNA levels in PBMCs of CHB patients (n = 69) were measured by qPCR, and the correlation of mRNA levels of OAS1 and MMP-9 was analyzed by using SPSS software. Graphs show means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The effects of IFN stimulation on PBMCs from CHB patients and healthy donors were then determined. PBMCs freshly isolated from CHB patients and healthy individuals were treated with IFN-α. IFN-α-induced oligoadenylate synthetase 1 (OAS1) mRNA levels were significantly higher in PBMCs freshly isolated from healthy individuals than in PBMCs freshly isolated from CHB patients (Fig. 4I). In addition, the mRNA level of OAS1 was inverse correlated with the mRNA level of MMP-9 in PBMCs of CHB patients (Fig. 4J). These results suggested that type I IFN signaling is downregulated in CHB patients and that MMP-9 may be involved in this regulation. Taken together, our results indicated that HBV infection initially induces type I IFN signaling, but such activation is downregulated by MMP-9.
MMP-9 facilitates HBV replication through attenuating IFNAR1 function and IFN/JAK/STAT signaling.
The mechanism by which MMP-9 activates HBV replication was evaluated. Since IFN/JAK/STAT signaling accomplishes antiviral activities by activating ISG production (25), we determined whether MMP-9 activates HBV replication through regulating the IFN/JAK/STAT pathway. It was suggested previously that HBV may evade the innate immune response (23), and HBV is detected by immune cells and hepatocytes to activate the innate immune response (17, 24). Initially, we evaluated the effect of HBV on the production of IFN-α in HepG2 cells. Type I IFN is facilitated by HBV in HepG2 and HepG2-NTCP cells (Fig. 4A and C), suggesting that HBV enhances type I IFN in HepG2 cells. Thus, we further clarified whether MMP-9 activates HBV replication through regulating IFN action. HepG2 cells were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 and treated with recombinant human IFN-α (rhIFN-α). HBeAg, HBsAg, and HBV RNAs were activated by MMP-9 and repressed by rhIFN-α, whereas rhIFN-α-mediated repression was attenuated by MMP-9 (Fig. 5A). In addition, HBV RNAs were also enhanced by MMP-9 and repressed by rhIFN-α, whereas rhIFN-α-mediated repression was reduced by MMP-9 (Fig. 5B). These results demonstrated that MMP-9 downregulates IFN-α action in the repression of HBV replication.
FIG 5.
MMP-9 facilitates HBV replication by repressing IFN-α action. (A and B) HepG2 cells were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 for 12 h and then treated with rhIFN-α for 36 h. (A) HBeAg and HBsAg in the supernatants were assayed by an ELISA. (B) HBV RNAs were detected by Northern blotting, and 18S and 28S rRNAs were used as loading controls. (C and D) HepG2 cells were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 for 12 h and then treated with JAK inhibitor I for 36 h. (C) HBeAg and HBsAg in the supernatants were assayed by an ELISA. (D) HBV RNAs were detected by Northern blot analyses. (E and F) HepG2 cells were cotransfected with pHBV1.3, pCMV-Tag2B-MMP-9, and pCAGGS-IFNAR1 for 48 h. (E) HBeAg and HBsAg in the supernatants were assayed by an ELISA. (F) HBV RNA levels were determined by Northern blotting. (G) HepG2 cells were transfected with sh-IFNAR1-1, and the protein level of IFNAR1 was determined by Western blotting. (H and I) HepG2 cells were cotransfected with sh-IFNAR1, pHBV1.3, and pCMV-Tag2B-MMP-9 for 48 h. (H) HBeAg and HBsAg in the supernatants were assayed by an ELISA. (I) HBV RNAs were detected by Northern blot analyses. (J to L) HepG2-NTCP cells were transfected with the vector, sh-IFNAR1-1, or sh-IFNAR1-2. Twenty-four hours later, cells were infected with HBV at 1,000 GEq. At 6 days postinfection, the mRNA level of IFNAR1 (J) was measured by qPCR, the HBeAg level in the supernatants of infected cells was determined by an ELISA (K), and the mRNA level of HBV 3.5-kb RNA was measured by qPCR (L). (M and N) HepG2-NTCP cells were transfected with the vector or pCMV-Tag2B-MMP-9 and sh-Ctrl or sh-IFNAR1-1. Twenty-four hours later, cells were infected with HBV at 1,000 GEq. At 6 days postinfection, HBeAg levels in the supernatants of infected cells were determined by an ELISA (M), and the levels of HBV 3.5-kb RNA were measured by qPCR (N). (O) HepG2-NTCP cells were transfected with the vector or pCMV-Tag2B-MMP-9 and sh-Ctrl, sh-IFNAR1-1, or sh-IFNAR1-2. Twenty-four hours later, cells were infected with HBV at 1,000 GEq. The expression of HBcAg in the cells was analyzed by immunostaining at 8 days postinfection. Results show means ± standard deviations (n = 3). *, P < 0.05.
IFN-α acts through IFN-α/β receptors (IFNAR1 and -2), which are associated with TYK2 and JAK1, respectively (6). The effect of MMP-9 on IFN action was further determined by using JAK inhibitor I, which blocks IFN/JAK/STAT signaling. HepG2 cells were cotransfected with pHBV1.3 and pCMV-Tag2B-MMP-9 and then treated with JAK inhibitor I. The results revealed that HBeAg and HBsAg were facilitated in the presence of MMP-9, JAK inhibitor I, and MMP-9 plus JAK inhibitor I (Fig. 5C), and HBV RNAs were upregulated by MMP-9, JAK inhibitor I, and MMP-9/JAK inhibitor I (Fig. 5D), indicating that MMP-9 facilitates HBV replication in a manner similar to that of JAK inhibitor I and suggesting that MMP-9 mediates HBV replication through repressing JAK1 function.
Moreover, the role of IFNAR1 in the MMP-9-regulated activation of HBV replication was determined in HepG2 cells cotransfected with pHBV1.3, pCMV-Tag2B-MMP-9, and pCAGGS-IFNAR1. The results showed that HBeAg and HBsAg were upregulated by MMP-9, but MMP-9-mediated activation was downregulated by IFNAR1 (Fig. 5E), and HBV RNAs were enhanced by MMP-9, but MMP-9-mediated activation was reduced by IFNAR1 (Fig. 5F), indicating that IFNAR1 overcomes MMP-9 function in the activation of HBV replication. Furthermore, the effect of MMP-9 on IFNAR1 in the MMP-9-regulated activation of HBV replication was determined by using short hairpin RNA (shRNA) specific for IFNAR1 (sh-IFNAR1-1), which effectively downregulated IFNAR1 protein expression (Fig. 5G). HepG2 cells were cotransfected with sh-IFNAR1-1, pHBV1.3, and pCMV-Tag2B-MMP-9. The results revealed that HBeAg and HBsAg were enhanced by MMP-9, sh-IFNAR1-1, and MMP-9 plus sh-IFNAR1-1 (Fig. 5H), and HBV RNAs were upregulated by MMP-9, sh-IFNAR1-1, and MMP-9/sh-IFNAR1 (Fig. 5I). In addition, HepG2-NTCP cells were transfected with sh-IFNAR1-1 or sh-IFNAR1-2, which effectively downregulated IFNAR1 mRNA expression (Fig. 5J), and then infected with HBV. The secretion of HBeAg (Fig. 5K) and the expression of HBV 3.5-kb RNA (Fig. 5L) were upregulated by sh-IFNAR1-1 or sh-IFNAR1-2 in the HepG2-NTCP infection system. Moreover, HepG2-NTCP cells were cotransfected with sh-IFNAR1-1 and pCMV-Tag2B-MMP-9 and then infected with HBV. The secretion of HBeAg (Fig. 5M), the expression of HBV 3.5-kb RNA (Fig. 5N), and the intracellular level of HBcAg (Fig. 5O) were induced by MMP-9, but MMP-9-mediated induction was attenuated in the presence of sh-IFNAR1 (Fig. 5O). These results indicated that MMP-9 activates HBV replication in a manner similar to that of sh-IFNAR1 and suggested that MMP-9 may mediate HBV replication through repressing IFNAR1. Taken together, we demonstrated that MMP-9 facilitates HBV replication by attenuating IFNAR1 function and IFN/JAK/STAT signaling.
MMP-9 represses the actions of type I IFNs and the productions of antiviral proteins in response to HBV infection.
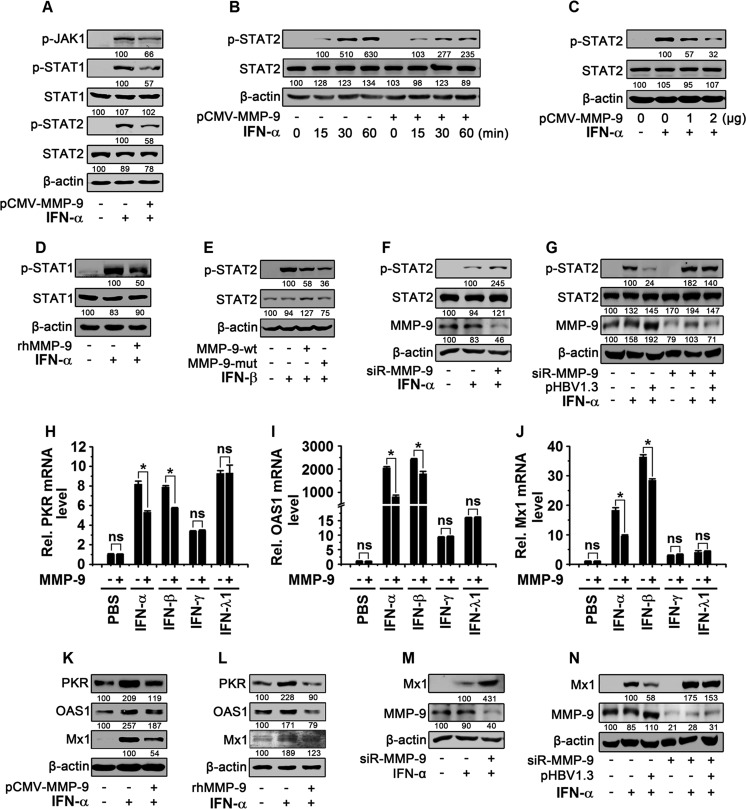
The mechanism by which MMP-9 regulates the IFN/JAK/STAT pathway was evaluated. The effect of the overexpression of MMP-9 on IFN/JAK/STAT signaling was evaluated in HepG2 cells transfected with pCMV-Tag2B-MMP-9 and treated with rhIFN-α. We showed that p-JAK1, p-STAT1, and p-STAT2 were upregulated by rhIFN-α and downregulated by MMP-9 (Fig. 6A); p-STAT2 was enhanced by rhIFN-α in a time-dependent manner, but such activation was attenuated by MMP-9 (Fig. 6B), and p-STAT2 was activated by rhIFN-α, but such activation was downregulated by MMP-9 in a dose-dependent manner (Fig. 6C). In addition, the effect of recombinant human MMP-9 (rhMMP-9) on the regulation of IFN/JAK/STAT was evaluated in HepG2 cells treated with rhIFN-α and rhMMP-9. The results showed that p-STAT1 was activated by rhIFN-α and repressed by rhMMP-9 (Fig. 6D). It was reported previously that MMP-9 cleaves the IFN-β protein to abolish its activity (26). We evaluated the effect of MMP-9 on the activity of IFN-β in HepG2 cells transfected with pCMV-Tag2B-MMP-9-wt or pCMV-Tag2B-MMP-9-mut and treated with rhIFN-β. p-STAT2 was activated by rhIFN-β, but such activation was repressed by both MMP-9-wt and MMP-9-mut (Fig. 6E), indicating that MMP-9 attenuates IFN-β activity and that metalloproteinase activity is not required for MMP-9 in the repression of IFN-β.
FIG 6.
MMP-9 inhibits IFN-α/β downstream effectors. (A) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9 for 48 h and treated with rhIFN-α (300 IU/ml) for 30 min. (B) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9 for 48 h and treated with rhIFN-α (300 IU/ml) for different times. (C) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9 at different concentrations for 48 h and treated with rhIFN-α for 30 min. (D) HepG2 cells (2 × 106) were treated with rhMMP-9 (50 ng/ml) for 3 h and treated with rhIFN-α for 30 min. (E) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9-wt or pCMV-Tag2B-MMP-9-mut for 48 h and treated with rhIFN-β (10 ng/ml) for 30 min. (F) HepG2 cells (2 × 106) were transfected with siR-MMP-9 (100 nM) for 48 h and treated with rhIFN-α for 30 min. (G) HepG2 cells (2 × 106) were transfected with pHBV1.3 (1 μg) and siR-MMP-9 (100 nM) for 48 h and treated with rhIFN-α for 30 min. For panels A to G, levels of proteins expressed in treated cells were determined by Western blot analyses using the corresponding antibodies, as indicated. (H to J) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9 for 36 h and treated with rhIFN-α (300 IU/ml), rhIFN-β (10 ng/ml), rhIFN-γ (50 ng/ml), and rhIFN-λ1 (100 ng/ml) for 12 h. PKR mRNA (H), OAS1 mRNA (I), and Mx1 mRNA (J) levels were measured by qPCR. (K) HepG2 cells (2 × 106) were transfected with pCMV-Tag2B-MMP-9 for 36 h and treated with rhIFN-α for 12 h. (L) HepG2 cells (2 × 106) were treated with rhMMP-9 (50 ng/ml) for 3 h and treated with rhIFN-α for 12 h. (M) HepG2 cells (2 × 106) were transfected with siR-MMP-9 (100 nM) for 36 h and treated with rhIFN-α for 12 h. (N) HepG2 cells (2 × 106) were cotransfected with pHBV1.3 (1 μg) and siR-MMP-9 (100 nM) for 36 h and treated with rhIFN-α for 12 h. In panels K to N, levels of proteins expressed in treated cells were determined by Western blot analyses using the corresponding antibodies, as indicated. Results show means ± standard deviations (n = 3). ns, nonsignificant; *, P < 0.05.
Moreover, the effect of the knockdown of MMP-9 on the regulation of IFN/JAK/STAT was evaluated in HepG2 cells transfected with siR-MMP-9 and treated with rhIFN-α. The results revealed that p-STAT2 was induced by IFN-α and further enhanced by siR-MMP-9 (Fig. 6F). It was reported previously that HBV suppresses IFN activity in vivo and in vitro and activates MMP-9 in hepatocytes and hepatoma cells (9, 11, 27). In this study, we demonstrated that MMP-9 is upregulated in PBMCs of CHB patients and activated by HBV in PBMCs, macrophages, and HepG2-NTCP cells in vitro. Thus, we further determined whether MMP-9 plays a role in the HBV-mediated regulation of IFN in HepG2 cells transfected with pHBV1.3 and siR-MMP-9 and treated with IFN-α. The results showed that p-STAT2 was induced by IFN-α and repressed by HBV, but the HBV-mediated reduction of p-STAT2 levels was blocked by siR-MMP-9 (Fig. 6G), indicating that MMP-9 plays an inhibitory role in the HBV-mediated activity of IFN-α. Therefore, we demonstrated that the overexpression of MMP-9 downregulates IFN/JAK/STAT signaling, whereas the knockdown of MMP-9 upregulates IFN/JAK/STAT signaling, and thus, we revealed that MMP-9 plays an inhibitory role in the HBV-mediated activation of IFN/JAK/STAT signaling.
Since IFN activates the JAK/STAT pathway to induce ISGs (28), we investigated the effect of MMP-9 on the productions of ISGs, double-stranded RNA-activated protein kinase (PKR), OAS1, and interferon-induced GTP-binding protein 1 (Mx1). HepG2 cells were transfected with pCMV-Tag2B-MMP-9 and treated with rhIFN-α (type I IFN), rhIFN-β (type I IFN), rhIFN-γ (type II IFN), and rhIFN-λ1 (type III IFN). The results revealed that PKR mRNA (Fig. 6H), OAS1 mRNA (Fig. 6I), and Mx1 mRNA (Fig. 6J) were activated by rhIFN-α, rhIFN-β, rhIFN-γ, and rhIFN-λ1, but the activations mediated by rhIFN-α and rhIFN-β were attenuated by MMP-9, whereas the activations induced by IFN-γ and IFN-λ1 were not affected by MMP-9 (Fig. 6H to J). These results suggested that MMP-9 attenuates the activities of type I IFNs but not the actions of type II and III IFNs. The effect of MMP-9 on the regulation of type I IFN was further determined in HepG2 cells transfected with pCMV-Tag2B-MMP-9 or incubated with rhMMP-9 and treated with rhIFN-α. The results showed that the PKR, OAS1, and Mx1 proteins were induced by rhIFN-α, but such activations were repressed by MMP-9 (Fig. 6K) and rhMMP-9 (Fig. 6L). In addition, we determined the role of siR-MMP-9 in the regulation of type I IFN in HepG2 cells transfected with siR-MMP-9 and treated with rhIFN-α. The results showed that Mx1 was induced by rhIFN-α and further enhanced by siR-MMP-9 (Fig. 6M). Finally, HepG2 cells were cotransfected with pHBV1.3 and siR-MMP-9 and treated with rhIFN-α. The results revealed that the Mx1 protein was stimulated by rhIFN-α and repressed by HBV, but HBV-mediated repression was attenuated by siR-MMP-9 (Fig. 6N). Taken together, our results demonstrated that MMP-9 attenuates the activity of IFN-α/β and represses the production of antiviral proteins in response to HBV stimulation.
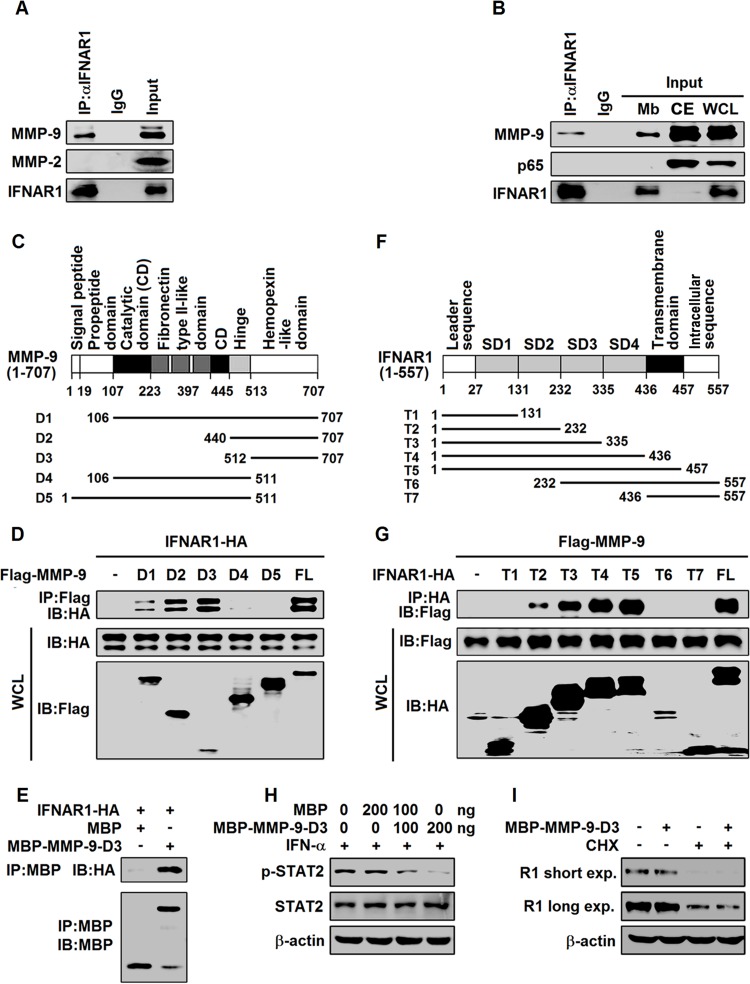
The hemopexin-like domain of MMP-9 interacts with the extracellular domain of IFNAR1 to block binding with IFN-α.
The expression of type I IFN receptors (IFNAR1/2) is modulated under physiological and pathological conditions (29–31), and IFN activates JAK/STAT signaling by binding to IFNAR1/2 (6). Since MMP-9 interacts with the membrane form of Ku80 (32), we determined whether MMP-9 can interact with IFNAR1. Coimmunoprecipitation (co-IP) results showed that MMP-9, but not MMP-2, was coimmunoprecipitated with IFNAR1 in differentiated THP-1 cells (Fig. 7A). In addition, membrane extracts (Mbs), cytosolic extracts (CEs), and whole-cell lysates (WCLs) of differentiated THP-1 cells were separated and prepared, and the membrane extracts were immunoprecipitated with anti-IFNAR1. The results showed that IFNAR1 was detected in the immunoprecipitates, Mbs, and WCLs but not in CEs, and MMP-9 was detected in the immunoprecipitates, Mbs, WCLs, and CEs, whereas p65 was detected in WCLs and CEs but not in immunoprecipitates or Mbs, as expected (Fig. 7B), suggesting that IFNAR1 interacts with MMP-9 in the membrane fractions of differentiated THP-1 cells.
FIG 7.
MMP-9 interacts with IFNAR1 at the cell surface. (A) Differentiated THP-1 cells (5 × 106) were lysed, and cell lysates were immunoprecipitated with anti-IFNAR1 or IgG. The immunoprecipitates and WCLs were analyzed by Western blotting with anti-MMP-9, anti-MMP-2, and anti-IFNAR1. (B) Mbs of differentiated THP-1 cells (5 × 106) were immunoprecipitated with anti-IFNAR1 or IgG. The immunoprecipitates, Mbs, CEs, and WCLs were analyzed by Western blotting with anti-p65, anti-MMP-9, and anti-IFNAR1. (C) Schematic diagram of wild-type MMP-9 (MMP-9) and truncated mutants (MMP-9-D1 to -D5). (D) HEK293T cells were cotransfected with plasmid pCAGGS-IFNAR1 expressing HA-tagged IFNAR1 and plasmid p3×FLAG-CMV-14-MMP-9 expressing Flag-tagged MMP-9 (FL) or plasmids p3×FLAG-CMV-14-MMP-9-D1 to -D5 expressing Flag-tagged MMP-9-D1 to -D5. Cell lysates were immunoprecipitated with anti-Flag. The immunoprecipitates and WCLs were analyzed by Western blotting with the indicated antibodies. IB, immunoblotting. (E) MBP pulldown assay with lysates from IFNAR1-HA-expressing HEK293T cells and MBP or MBP-MMP-9-D3 purified from E. coli cells. After pulldown, the precipitates were analyzed by Western blotting with anti-HA and anti-MBP. (F) Schematic diagram of wild-type IFNAR1 (IFNAR1) and truncated mutants (IFNAR1-T1 to -T7). (G) HEK293T cells were cotransfected with plasmid p3×FLAG-CMV-14-MMP-9 and plasmid pCAGGS-IFNAR1 expressing full-length HA-tagged IFNAR1 or plasmids pCAGGS-IFNAR1-T1 to -T7 expressing HA-tagged truncated IFNAR1 (IFNAR1-T1 to -T7). Cells were lysed, and cell lysates were immunoprecipitated with anti-HA antibody. The immunoprecipitates and WCLs were analyzed by Western blotting with anti-Flag and anti-HA. (H) HepG2 cells were treated with MBP or MBP–MMP-9-D3 for 3 h and treated with rhIFN-α for 30 min. p-STAT2, STAT2, and β-actin levels were determined by Western blotting. (I) HepG2 cells were treated with MBP or MBP–MMP-9-D3 for 12 h and treated with CHX (50 μg/ml) for 4 h. IFNAR1 and β-actin levels were determined by Western blotting.
The domains of MMP-9 that interact with IFNAR1 were analyzed by generating five plasmids expressing truncated MMP-9 (MMP-9-D1 to -D5), in which MMP-9 was fused to a Flag tag (Fig. 7C). HEK293T cells were then cotransfected with pCAGGS-IFNAR1 expressing hemagglutinin (HA)-tagged IFNAR1 along with plasmid p3×FLAG-CMV-14-MMP-9 expressing Flag-tagged MMP-9 (full length [FL]) or plasmids (p3×FLAG-CMV-14-MMP-9-D1 to -D5) expressing Flag-tagged MMP-9-D1 to -D5, respectively. Coimmunoprecipitation revealed that MMP-9, MMP-9-D1, MMP-9-D2, and MMP-9-D3 were precipitated with IFNAR1 in HEK293T cells, but MMP-9-D4 and MMP-9-D5 failed to do so (Fig. 7D), suggesting that the hemopexin-like domain is required for the binding of MMP-9 to IFNAR1. A maltose-binding protein (MBP) pulldown assay with lysates from IFNAR1-HA-expressing HEK293T cells and MBP–MMP-9-D3 purified from Escherichia coli further revealed that MBP–MMP-9-D3 interacted with IFNAR1-HA in HEK293T cell lysates (Fig. 7E), confirming that the hemopexin-like domain of MMP-9 interacts with IFNAR1.
The domains of IFNAR1 that interact with MMP-9 were determined by generating seven plasmids expressing truncated IFNAR1 (IFNAR1-T1 to -T7), in which the truncated IFNAR1 proteins were fused to an HA tag (Fig. 7F). HEK293T cells were then cotransfected with plasmid p3×FLAG-CMV-14-MMP-9 and plasmid pCAGGS-IFNAR1 expressing full-length HA-tagged IFNAR1 or plasmids pCAGGS-IFNAR1-T1 to -T7 expressing HA-tagged truncated IFNAR1 (IFNAR1-T1 to -T7). Co-IP revealed that IFNAR1 and IFNAR1-T2 to -T5 were precipitated with MMP-9, but IFNAR1-T1, IFNAR1-T6, and IFNAR1-T7 failed to interact (Fig. 7G), suggesting that the extracellular domain is required for the binding of IFNAR1 to MMP-9.
The MMP-9 hemopexin domain determines substrate specificity and interacts with cell surface receptors and Ku (33). IFNAR1 N-terminal domains (subdomain 1 [SD1], SD2, and SD3) are required for IFN-α recognition, and the membrane-proximal domain (SD4) is critical for the formation of a functional signaling complex. Since we showed that IFNAR1 SD2 was required for the interaction with MMP-9, and SD2 to SD4 retained full binding affinity, we speculated that MMP-9 may interfere with the binding of IFN-α to IFANR1. To confirm this speculation, HepG2 cells were incubated with MBP–MMP-9-D3 and treated with IFN-α. p-STAT2 was upregulated by rhIFN-α and downregulated by MBP–MMP-9-D3 (Fig. 7H), suggesting that MMP-9-D3 attenuates the action of IFN-α. Moreover, HepG2 cells were incubated with MBP–MMP-9-D3 and treated with an inhibitor of protein biosynthesis (cycloheximide [CHX]). The results showed that the IFNAR1 protein was not affected by MBP–MMP-9-D3 even in the presence of CHX (Fig. 7I), indicating that MMP-9-D3 has no effect on the production of the IFNAR1 protein, and thus, MMP-9 may inhibit IFNAR1 activity in binding to IFN-α. Taken together, we revealed that the hemopexin-like domain of MMP-9 interacts with the extracellular domain of IFNAR1 to repress the activity IFNAR1 in binding to IFN-α.
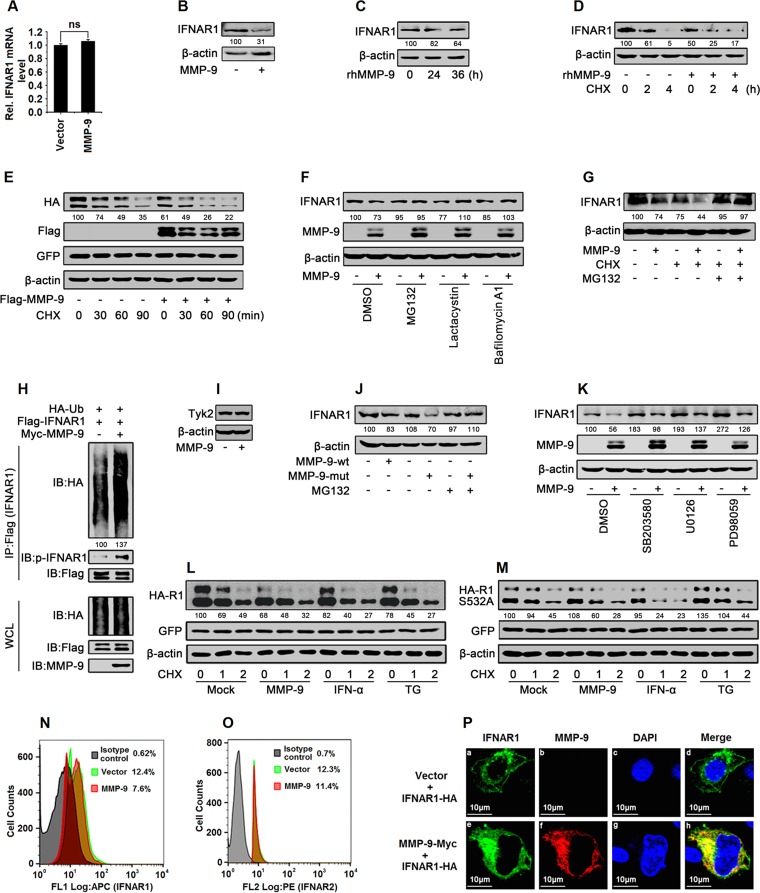
MMP-9 facilitates the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1.
IFNAR1 mRNA was not affected by MMP-9 in HepG2 cells (Fig. 8A), but the IFNAR1 protein was downregulated by MMP-9 and rhMMP-9 in HepG2 cells (Fig. 8B and C), suggesting that MMP-9 may regulate IFNAR1 at the posttranslational level. We then determined whether MMP-9 plays a role in the regulation of IFNAR1 stability. A CHX chase assay showed that endogenous IFNAR1 was downregulated in the presence of CHX, and such a reduction was further facilitated by rhMMP-9 (Fig. 8D); similarly, exogenously expressed IFNAR1 was attenuated by CHX, and such a reduction was further enhanced by MMP-9 (Fig. 8E), suggesting that MMP-9 promotes IFNAR1 degradation.
FIG 8.
MMP-9 promotes the phosphorylation, ubiquitination, degradation, and subcellular distribution of IFNAR1. (A and B) HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 48 h. Cells were harvested. IFNAR1 mRNAs were analyzed by qPCR (A), and IFNAR1 and β-actin proteins were detected by Western blotting (B). (C) HepG2 cells were treated with rhMMP-9 (50 ng/ml) for different times. Cells were harvested, and IFNAR1 and β-actin were detected by Western blotting. (D) HepG2 cells were treated with rhMMP-9 (50 ng/ml) for 12 h and treated with CHX (50 μg/ml) for the indicated times. Cells were harvested, and IFNAR1 and β-actin were detected by Western blotting. (E) HEK293T cells were cotransfected with pCAGGS-IFNAR1 and pCMV-Tag2B-MMP-9 for 48 h and treated with CHX (50 μg/ml) for the indicated times. The proteins were analyzed by Western blotting using the corresponding antibodies. (F) HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 42 h and treated with proteasome inhibitors (MG132 and lactacystin) and a lysosome inhibitor (bafilomycin A1) for 6 h. Cells were harvested, and IFNAR1, MMP-9, and β-actin were detected by Western blot analyses. (G) HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 42 h and treated with MG132 (20 μM) for 2 h and with CHX (50 μg/ml) for 4 h. Cells were harvested, and IFNAR1 and β-actin were detected by Western blot analyses. (H) HEK293T cells were cotransfected with pCDNA3.1-HA-Ub, pCDNA3.1-3×Flag-IFNAR1, and pcDNA3.1-Myc or pcDNA3.1-Myc-MMP-9 and treated with MG132 (20 μM) for 9 h. Cell lysates were denatured and subjected to IP with anti-Flag. The immunoprecipitates and WCLs were analyzed by Western blotting with the indicated antibodies. (I) HepG2 cells were transfected with pCMV-Tag2B or pCMV-Tag2B-MMP-9 for 48 h. TYK2 and β-actin protein levels expressed in the treated cells were determined by Western blot analyses using the corresponding antibodies. (J) HepG2 cells were transfected with pCMV-Tag2B-MMP-9-wt or pCMV-Tag2B-MMP-9-mut for 42 h and treated with MG132 (20 μM) for 6 h. Cells were harvested, and IFNAR1 and β-actin were detected by Western blot analyses. (K) HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 42 h and treated with the signaling component inhibitors SB203580 (p38 MAPK inhibitor), U0126 (ERK1/2 inhibitor), and PD98059 (MEK inhibitor) for 6 h. Cells were harvested, and levels of IFNAR1, MMP-9, and β-actin proteins were detected by Western blot analyses. (L and M) HEK293T cells were cotransfected with pCAGGS-IFNAR1 (L) or pCAGGS-IFNAR1-S532A (M) and pCMV-Tag2B-MMP-9 for 33 h and treated with rhIFN-α or TG (1 μM) for 1 h and with CHX (50 μg/ml) for the indicated times. Levels of IFNAR1-HA, IFNAR1-S532A–HA, GFP, and β-actin proteins were determined by Western blotting. (N and O) HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 48 h and then harvested. IFNAR1 protein (N) and IFNAR2 protein (O) levels were analyzed by flow cytometry. APC, allophycocyanin; PE, phycoerythrin. (P) HepG2 cells were cotransfected with pCAGGS-IFNAR1 and pcDNA3.1-Myc-MMP-9 for 24 h. Cells were immunostained with anti-Myc and anti-HA antibodies, and the nucleus was stained by DAPI and analyzed by confocal microscopy. Results show means ± standard deviations. ns, nonsignificant.
IFNAR1 is hydrolyzed through the lysosomal pathway, and the ubiquitination of IFNAR1 is required for endocytosis (29). Here, the pathways involved in the MMP-9-mediated degradation of IFNAR1 were validated in HepG2 cells treated with proteasome inhibitors (MG132 and lactacystin) and a lysosome inhibitor (bafilomycin A1). IFNAR1 degradation was promoted by MMP-9 but attenuated by MG132, lactacystin, and bafilomycin A1 (Fig. 8F), indicating that proteasome and lysosome pathways were involved in the MMP-9-mediated degradation of IFNAR1. In addition, IFNAR1 degradation was facilitated by MMP-9 in the presence of CHX, whereas MG132 protected IFNAR1 from MMP-9-mediated degradation (Fig. 8G), confirming that MMP-9 promotes IFNAR1 degradation depending on ubiquitination. Moreover, HEK293T cells were cotransfected with pCDNA3.1-HA-Ub (ubiquitin), pCDNA3.1-3×Flag-IFNAR1, or pcDNA3.1-Myc-MMP-9 and treated with MG132. Co-IP results showed that MMP-9 enhanced both the ubiquitination and phosphorylation of IFNAR1 (Fig. 8H). The N terminus of TYK2 sustains the level of IFNAR1 by inhibiting its endocytosis and degradation (34). We showed that TYK2 production was not affected by MMP-9 (Fig. 8I), suggesting that TYK2 is not involved in the MMP-9-mediated regulation of IFNAR1. The effect of MMP-9 enzyme activity on the degradation of IFNAR1 was also evaluated. Like MMP-9-wt, MMP-9-mut facilitated IFNAR1 degradation (Fig. 8J), indicating that the enzyme activity of MMP-9 is not required for IFNAR1 degradation. Taken together, we revealed that MMP-9 facilitates the phosphorylation, ubiquitination, and degradation of IFNAR1.
The ligand-dependent and ligand-independent pathways are involved in the regulation of IFNAR1 degradation (31). In the ligand-dependent pathway, IFN-α/ββ induces IFNAR1 phosphorylation on a specific Ser residue (Ser535), which leads to the recruitment of E3 ubiquitin ligase followed by IFNAR1 ubiquitination and lysosomal degradation (29, 30). In the ligand-independent pathway, the unfolded-protein response (UPR) activates p38 kinase, which subsequently phosphorylates IFNAR1 at Ser532 (35–38). We previously reported that extracellular signal-regulated kinase (ERK) is involved in the degradation of IFNAR1 (39, 40). Here, the signaling pathways that participated in the MMP-9-mediated degradation of IFNAR1 were evaluated in HepG2 cells transfected with pCMV-Tag2B-MMP-9 and treated with the signaling component inhibitors SB203580 (p38 mitogen-activated protein kinase [MAPK] inhibitor), U0126 (ERK1/2 inhibitor), and PD98059 (MEK inhibitor). The results showed that IFNAR1 degradation was facilitated by MMP-9 in the presence of SB203580, U0126, and PD98059 (Fig. 8K), suggesting that p38 and ERK are not required for the degradation of IFNAR1 mediated by MMP-9. The effect of Ser532 phosphorylation on the MMP-9-mediated degradation of IFNAR1 was also evaluated. HEK293T cells were cotransfected with pCMV-Tag2B-MMP-9 and pCAGGS-IFNAR1 or pCAGGS-IFNAR1-S532A (expressing the IFNAR1-S532A mutant) and treated with CHX. The cells were further treated with IFN-α to induce the ligand-dependent and Ser532-independent degradation of IFNAR1 or treated with an inducer of the UPR (thapsigargin [TG]) to stimulate the ligand-independent and Ser532-dependent degradation of IFNAR1. The results showed that IFNAR1 degradation was facilitated by MMP-9, IFN-α, and TG (Fig. 8L), whereas IFNAR1-S532A degradation was promoted by MMP-9 and IFN-α but not by TG (Fig. 8M). These results demonstrated that MMP-9 facilitates IFNAR1 degradation in a p38-, ERK-, and Ser532 phosphorylation-independent manner.
Upon phosphorylation and ubiquitination, IFNAR1 was redistributed from the cell surface to the perinuclear compartments. Flow cytometry analyses revealed that the level of IFNAR1 (Fig. 8N), but not IFNAR2 (Fig. 8O), on the cell surface was reduced by MMP-9. Confocal microscopy showed that IFNAR1 was partially distributed on the cell surface in the absence of MMP-9 (Fig. 8Pa to d), whereas most of the IFNAR1 protein was internalized in the presence of MMP-9 (Fig. 8Pe to h). These results suggested that MMP-9 regulates the subcellular distribution of IFNAR1. Taken together, our results demonstrated that MMP-9 promotes the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1 and revealed that MMP-9 facilitates IFNAR1 degradation in a p38-, ERK-, S532 phosphorylation-, TYK2-, and enzyme activity-independent manner.
MMP-9 enhances VSV replication by attenuating IFN antiviral action.
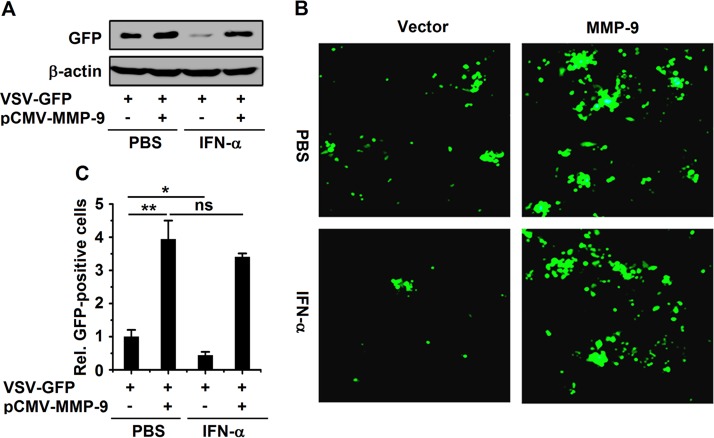
Since MMP-9 enhances HBV replication by attenuating IFN antiviral action, MMP-9 may play a broad role in the regulation of other viruses. To prove this hypothesis, we investigated the role of MMP-9 in the replication of VSV by using an recombinant enhanced green fluorescent protein (EGFP)-expressing vesicular stomatitis virus strain (VSV-GFP) as described previously (41). HepG2 cells were transfected with pCMV-Tag2B-MMP-9, treated with rhIFN-α, and infected with VSV-GFP. The production of the VSV protein (as indicated by GFP) was upregulated by MMP-9 and downregulated by rhIFN-α, but IFN-α-mediated repression was recovered by MMP-9 (Fig. 9A). Similarly, fluorescence microscopy images showed that VSV replication (as indicated by intensity) was stimulated by MMP-9 and repressed by rhIFN-α, but IFN-α-mediated repression was suppressed by MMP-9 (Fig. 9B and C). Moreover, the numbers of GFP-positive cells were enhanced by MMP-9 and reduced by rhIFN-α, whereas the IFN-α-mediated reduction was attenuated by MMP-9 (Fig. 9C). Taken together, our results demonstrated that MMP-9 facilitates the replication of VSV by attenuating the antiviral action of IFN.
FIG 9.
MMP-9 facilitates the replication of VSV by attenuating IFN-α action. HepG2 cells were transfected with pCMV-Tag2B-MMP-9 for 24 h and infected with VSV-GFP (MOI = 0.01) for 24 h. GFP levels were measured by Western blotting (A) and fluorescence microscopy (B), and relative levels of GFP-positive cells are shown in graph (C). Results show means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01; ns, nonsignificant.
DISCUSSION
Chronic HBV infection is a major cause of CHB, liver cirrhosis, and HCC, but the mechanism underlying the development of chronic infection is largely unknown. Previous studies showed that the MMP-9 level is elevated in hepatocytes and in the sera of patients with CHB and HCC (10, 42). This study revealed that MMP-9 is activated in PBMCs of CHB patients and in leukocytes. MMP-9 has multiple functions involved in development, angiogenesis, apoptosis, inflammation, and cancer growth. MMP-9 is regulated by HBV (9), human immunodeficiency virus type 1 (HIV-1) (43), and hepatitis C virus (HCV) (44, 45). The effect of MMP-9 on the replication of RSV was controversial. Different reports showed that MMP-9 exerts proviral activity (12, 13) or antiviral activity (14) on RSV, but the mechanism by which MMP-9 modulates HBV replication is unknown. Here, we demonstrated that MMP-9 facilitates the replications of HBV and VSV by suppressing IFN activity and revealed a novel mechanism by which HBV represses host immunity to maintain replication through the activation of MMP-9.
HBV is known as a “stealth virus,” as the activation of innate responses is predominantly weak or absent during HBV infection (21–23). A stealth virus is not detected by pattern recognition receptors (PRRs) or is able to inhibit nascent innate responses. A chimpanzee study showed that HBV infection does not induce a strong modulation of gene expression in the liver compared to that induced by HCV infection (23). A weak and transient innate response is observed upon HBV infection in human liver progenitor HepaRG cells and primary human hepatocytes (46). However, HBV elicits a strong and specific innate antiviral response resulting in the noncytopathic clearance of HBV DNA in HepaRG cells, and IFN-β and ISGs are induced in HepG2 cells based on a baculovirus vector transduction system (Bac-HBV) (24). In addition, acute infection with woodchuck hepatitis virus (WHV) induces immune genes immediately after infection (47). A transient but slight activation of the IFN-α gene was also detected in human hepatocytes infected by HBV in chimeric mice (48). In this study, we demonstrated that HBV infection initially induces an innate immune response, but such activation is subsequently repressed by MMP-9. Thus, we suggest that MMP-9 plays an important role in the suppression of IFN signaling in response to HBV infection.
Initially, HBV stimulates MMP-9 expression in leukocytes and hepatocytes. Subsequently, MMP-9 counteracts host innate immunity to facilitate HBV replication through attenuating type I IFN action, IFN/JAK/STAT signaling, and ISG production. Host innate immunity is the first line of defense against invading pathogens (49). The activation of innate immunity by Toll-like receptor (TLR) agonists results in the suppression of HBV in mice and chimpanzees (50, 51). However, the innate immune response is repressed by HBV with various mechanisms. It was reported previously that HBV inhibits STAT1 nuclear translocation (52) and activates phosphatase 2A (PP2A) expression to inhibit IFN-α signaling in hepatocytes (53). We demonstrated previously that collagen triple-helix repeat-containing 1 (CTHRC1) facilitates HBV replication by suppressing IFN action and disturbing the JAK/STAT cascade (39). Here, we discovered a new role of MMP-9 in the regulation of viral replication through a novel mechanism. Interestingly, we demonstrated that MMP-9 also represses IFNAR1 activity through promoting the phosphorylation, ubiquitination, subcellular distribution, and degradation of the IFNAR1 protein. It was reported previously that a lower level of IFNAR1 is associated with a higher risk of liver diseases (54), and polymorphisms of the IFNAR1 promoter are associated with CHB and HCC susceptibility (55, 56). IFNAR1 is downregulated by HBx in hepatocytes and in woodchucks infected with WHV (57, 58), indicating that HBV/WHV may establish chronic infection by repressing IFNAR1. Our results demonstrated that HBV attenuates IFNAR1 through the activation of MMP-9, which provides a new explanation as to how HBV establishes chronic infection.
Ligand binding or cellular signaling induces IFNAR1 phosphorylation and ubiquitination-dependent degradation. Ser532 phosphorylation and p38 kinase activity are required for IFNAR1 degradation in a ligand-independent fashion (31). Here, we revealed that MMP-9 promotes IFNAR1 degradation through lysosome pathways, whereas Ser532 phosphorylation and p38 activity are not required for the MMP-9-mediated degradation of IFNAR1 in response to HBV stimulation and demonstrated that MMP-9 facilitates IFNAR1 degradation in an ERK- and TYK2-independent manner. Therefore, we provided a new mechanism underlying the regulation of IFNAR1 degradation mediated by MMP-9. As a multidomain protein, MMP-9 not only cleaves the extracellular matrix, cell surface substrates, and intracellular substrates (59, 60) but also exerts noncatalytic signaling effects. The MMP-9 hemopexin domain induces intracellular signaling to prevent B cell apoptosis independent of the catalytic function (61) and interacts with the cell surface protein Ku to regulate cellular invasion (32). Here, we showed that the MMP-9 hemopexin-like domain interacts with the IFNAR1 extracellular domain on the cell surface, and such an interaction blocks the IFN-induced phosphorylation of STAT2, suggesting that MMP-9 may promote IFNAR1 degradation by interaction. However, the metalloproteinase activity is dispensable for MMP-9 function in the regulation of HBV replication and IFNAR1 degradation. Since the hemopexin-like domain of MMP-9 (MMP-9-D3) has no effect on IFNAR1 expression, it is possible that MMP-9 interacts with IFNAR1 by the hemopexin-like domain and modulates the stability of IFNAR1 by another domain(s). Matrix metalloproteinases are regulators of the tumor microenvironment (62). We discovered new roles of MMP-9 in the regulation of host innate immunity and viral replication. The excess expression of MMP-9 induced by HBV in the liver of patients with CHB may serve as a regulator of the liver microenvironment to facilitate chronic and persistent infection that may lead to the development of liver cirrhosis and HCC.
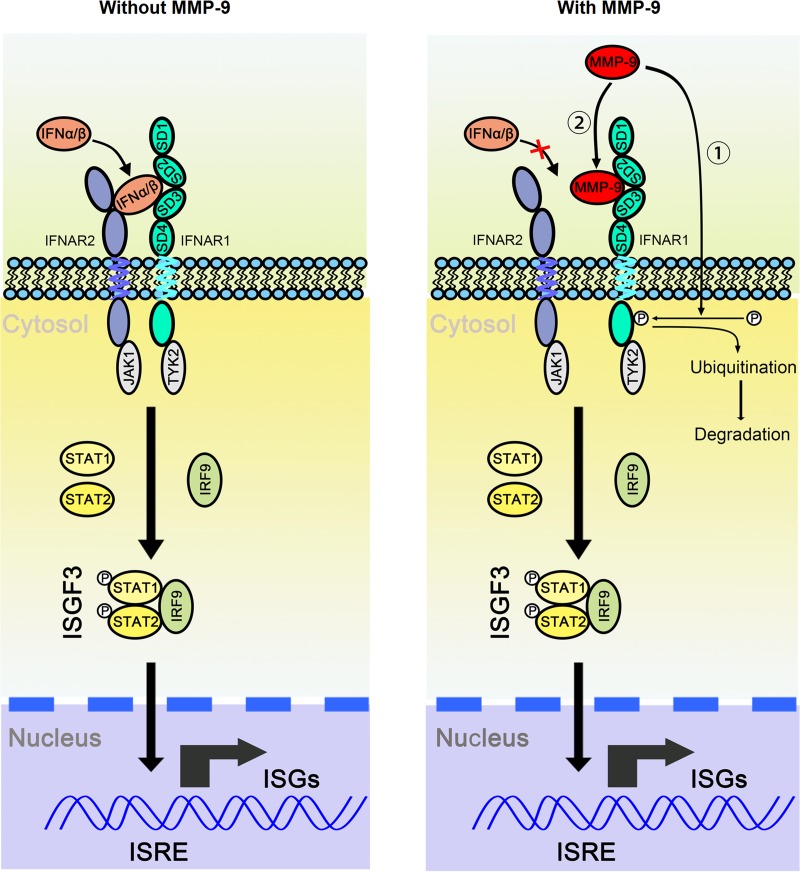
In conclusion, we identified a novel positive-feedback regulation loop between HBV replication and MMP-9 production (Fig. 10). On one hand, HBV activates MMP-9 in HBV-infected patients, HBV-stimulated leukocytes, and HBV-infected HepG2-NTCP cells. On the other hand, MMP-9 facilitates HBV replication through repressing IFN/JAK/STAT signaling, interacting with IFNAR1 to block its binding with IFN-α, and promoting the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1. Therefore, HBV may take advantage of MMP-9 function to establish or maintain chronic infection by repressing innate immunity.
FIG 10.
MMP-9 facilitates HBV replication by repressing IFN/JAK/STAT signaling and IFNAR1 function. We propose a novel positive-feedback regulation loop between HBV replication and MMP-9 production. Initially, HBV activates MMP-9 in infected patients and in HBV-stimulated leukocytes. Subsequently, MMP-9 facilitates HBV replication through repressing IFN/JAK/STAT signaling, interacting with IFNAR1 to block its binding with IFN-α, and promoting the phosphorylation, ubiquitination, subcellular distribution, and degradation of IFNAR1. ISRE, interferon-stimulated response element.
MATERIALS AND METHODS
Clinical samples.
Blood samples were obtained from 85 patients with HBV infection admitted to RenMin Hospital of Wuhan University, China. All patients were confirmed to be HBV positive but negative for hepatitis A, C, D, and E viruses and HIV. PBMCs from 55 healthy individuals with no history of liver disease were randomly selected from the Wuhan blood donation center.
Written informed consent was obtained from each patient. This study was conducted according to the principles of the Declaration of Helsinki and approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, in accordance with its guidelines for the protection of human subjects. The Institutional Review Board of the College of Life Sciences, Wuhan University, approved the collection of blood samples for this research, in accordance with guidelines for the protection of human subjects. Written informed consent was obtained from each participant.
PBMC isolation.
PBMCs were isolated by density centrifugation of fresh peripheral venous blood samples diluted 1:1 in pyrogen-free phosphate-buffered saline (PBS) over Histopaque (Haoyang Biotech). Cells were washed twice in PBS and resuspended in culture medium (RPMI 1640) supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). Freshly isolated PBMCs were cultured at 1 × 106 cells/ml.
Viruses and infection.
For the stimulation of PBMCs and macrophages, HBV inoculums were culture supernatants from HepG2.2.15 cells, the HBV stock titer (genome equivalents [GEq] per milliliter) was assessed by using qPCR, and the multiplicity of infection (MOI) was defined as the number of GEq per cell. For the preparation of UV-inactivated HBV, the virus was dispersed in a tissue culture dish, and a compact UV lamp was placed directly above the dish for 30 min. The recombinant EGFP-expressing vesicular stomatitis virus strain (VSV-GFP) (41) was amplified in Vero cells, and the titer was determined on Vero cells by fluorescence microscopy.
For infection of HepG2-NTCP cells (provided by Ying Zhu, Wuhan University, China), HBV inoculums were concentrated 100-fold from the supernatants of HepaAD38 cells (provided by Ying Zhu of Wuhan University, China) by ultracentrifugation. For infection, HepG2-NTCP cells were seeded onto collagen I-coated plates in Dulbecco's modified Eagle medium (DMEM) for 6 h, and the medium was then changed to primary hepatocyte maintenance medium (PMM) with 2% fetal bovine serum (FBS). PMM is Williams' E medium supplemented with insulin-transferrin-selenium solution (catalog no. I3146; Sigma), 2 mM l-glutamine, 10 ng/ml of human epidermal growth factor (EGF), 18 μg/ml of hydrocortisone, 40 ng/ml of dexamethasone, 2% dimethyl sulfoxide (DMSO), 100 U/ml of penicillin, and 100 μg/ml of streptomycin for 12 h. Cells were then infected with 1,000 GEq per cell of HBV in PMM containing 4% (wt/vol) polyethylene glycol 8000 (PEG 8000) for 16 h. The virus-containing medium was removed, and cells were washed four times and further incubated in PMM. The medium was changed every other day (19, 20).
Cell culture and transfection.
Human hepatocellular HepG2 cells, HepG2.2.15 cells, and Vero cells, obtained from the American Type Culture Collection (ATCC); human hepatocarcinoma Huh7 cells and human embryonic kidney HEK293T cells, obtained from the China Center for Type Culture Collection (CCTCC) (Wuhan, China); HepaAD38 cells; and HepG2-NTCP cells were grown in DMEM supplemented with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate at 37°C in 5% CO2. Human monocytic leukemia THP-1 cells, obtained from the CCTCC, were grown in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate at 37°C in 5% CO2. For differentiation, THP-1 cells were incubated with phorbol-12-myristate-13-acetate (PMA) (100 nM), the supernatants were removed 12 h later, and cells were grown in normal medium without PMA for another 24 h. HepG2, HepG2-NTCP, Huh7, and HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Plasmids, small interfering RNAs, and reagents.
The coding regions of MMP-9 (GenBank accession number NM_004994) and IFNAR1 (NM_000629) were generated by PCR amplification. For MMP-9, the PCR product was inserted into the EcoRI and XhoI sites of pCMV-Tag2B. For the full-length and truncated forms of MMP-9, the PCR products were inserted into the EcoRI and XbaI sites of p3×Flag-CMV-14. In addition, the PCR product of full-length MMP-9 was inserted into the XhoI and EcoRI sites of pcDNA3.1/Myc-His(−)B. For the full-length and truncated forms of IFNAR1, the PCR products were inserted into the EcoRI and XhoI sites of pCAGGS, followed by an HA tag at its carboxy terminus. In addition, the PCR product of full-length IFNAR1 was inserted into the EcoRI and XhoI sites of pcDNA3.1, proceeded by a 3×Flag tag. To generate mutational MMP-9, three histidines (H) (indicated by underlining) contained in the sequence 401HEFGHALGLDH411 were replaced by lysines (K). HBV-1.3 was generated from HepG2.2.15 cells (genotype D, subtype ayw; GenBank accession no. U95551), digested with EcoRI and SalI, and inserted into pBluescript II (Invitrogen).
Specific siRNAs were purchased from Guangzhou RiboBio, and the sequence of the siRNA against MMP-9 was reported previously (63). Oligonucleotides targeting MMP-9 and IFNAR1 were cloned into pLKO.1. Antibodies against p-JAK1 (catalog no. sc-101716), OAS1 (catalog no. sc-98424), PKR (catalog no. sc-100378), STAT2 (catalog no. sc-476), MMP-2 (catalog no. sc-13594), and p65 (catalog no. sc-109) were purchased from Santa Cruz Technology Company. Antibodies against MMP-9 (catalog no. 3852), p-STAT1 (catalog no. 9167), and STAT1 (catalog no. 9172) were purchased from Cell Signaling Technology. Anti-p-STAT2 (catalog no. 07-224) was purchased from Millipore. Anti-IFNAR1 (catalog no. ab45172) was purchased from Abcam. Anti-p-IFNAR1 (catalog no. P4623-1V), anti-Flag (catalog no. F1804), and anti-HA (catalog no. H6908) were purchased from Sigma-Aldrich. Anti-HBcAg (catalog no. B0586) was purchased from Dako. Antibodies against β-actin (catalog no. 60008-1-Ig), GFP (catalog no. 66002-1-Ig), Mx1 (catalog no. 13750-1-AP), and MBP (catalog no. 66003-1) were purchased from Proteintech Group. Flow cytometry Abs specific for IFNAR1 and IFNAR2 were purchased from R&D Systems. Human hepatitis B Ig served as an HBV-neutralizing Ab and was purchased from Hualan Bio (Hualan Biological Engineering, Xinxiang, China). rhMMP-9 was purchased from Calbiochem.
The inhibitors LY-294002, SP600125, PD98059, and SB203580 were purchased from Tocris Bioscience. U0126, MG132, and PMA were purchased from Sigma-Aldrich. Bafilomycin A1, lactacystin, and CHX were purchased from Cayman Chemical Company. JAK inhibitor I was purchased from Merck.
Quantitative RT-PCR analysis.
Quantitative reverse transcription-PCR (RT-PCR) analysis was performed to determine the relative mRNA levels. Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA was reverse transcribed with oligo(dT). Real-time PCR was performed with a Light Cycler 480 instrument (Roche), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The following primers were used: MMP-9 forward primer 5′-CCTCTGGAGGTTCGACGTG-3′, MMP-9 reverse primer 5′-AACTCACGCGCCAGTAGAAG-3′, IFNAR1 forward primer 5′-GCGCGAACATGTAACTGGTG-3′, IFNAR1 reverse primer 5′-ATTCCCGACAGACTCATCGC-3′, IFNA forward primer 5′-TTTCTCCTGCCTGAAGGACAG-3′, IFNA reverse primer 5′-GCTCATGATTTCTGCTCTGACA-3′, IFNB forward primer 5′-GCCGCATTGACCATGTATGAGA-3′, IFNB reverse primer 5′-GAGATCTTCAGTTTCGGAGGTAAC-3′, ISG56 forward primer 5′-AGCCAGCTGTCCTCACAGAC-3′, ISG56 reverse primer 5′-CTTCTACCACTGTTTCATGC-3′, PKR forward primer 5′-AAAGCGAACAAGGAGTAAG-3′, PKR reverse primer 5′-GATGATGCCATCCCGTAG-3′, OAS1 forward primer 5′-TTCCGTCCATAGGAGCCAC-3′, OAS1 reverse primer 5′-AAGCCCTACGAAGAATGTC-3′, Mx1 forward primer 5′-TTCAGCACCTGATGGCCTATC-3′, Mx1 reverse primer 5′-TGGATGATCAAAGGGATGTGG-3′, HBV 3.5-kb RNA forward primer 5′-GAGTGTGGATTCGCACTCC, HBV 3.5-kb RNA reverse primer 5′-GAGGCGAGGGAGTTCTTCT, GAPDH forward primer 5′-GGAAGGTGAAGGTCGGAGTCAACGG-3′, and GAPDH reverse primer 5′-CTCGCTCCTGGAAGATGGTGATGGG-3′. Data were normalized to the GAPDH expression level in each sample.
Western blot analysis and coimmunoprecipitation.
For Western blot analysis, cells were lysed in PBS (pH 7.4) that contained 1% Triton X-100, 1 mM EDTA, and a protease inhibitor mixture (Roche). Cell lysates (50 μg) were separated by 12% SDS-PAGE and then transferred onto a nitrocellulose membrane (Millipore). The membranes were blocked in 5% nonfat dried milk before incubation with specific antibodies. Blots were detected with the Clarity Western ECL substrate (Bio-Rad). The separation of cytosolic and membrane extracts was performed as described previously (32). For coimmunoprecipitation, cells were lysed in IP lysis buffer (0.025 M Tris-HCl, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol [pH 7.4]). Cell lysates were mixed with specific antibodies or IgG and protein G-agarose beads (GE Healthcare) and rocked overnight at 4°C. Beads were washed five times with lysis buffer, denatured with SDS-PAGE loading buffer, and separated by SDS-PAGE, with subsequent immunoblot analysis. The gray density of Western blots was measured by using ImageJ software (National Institutes of Health, Bethesda, MD).
Zymography assay.
MMP-9 proteinase activity was detected by a gelatin zymography assay as described previously (44). The cell lysates or supernatants were separated in SDS-PAGE gels containing 1 mg/ml gelatin (Sigma-Aldrich). After electrophoresis, the gel was washed twice (45 min each wash) with 2.5% Triton X-100 (Sigma-Aldrich), followed by a 30-min wash with 50 mM Tris-HCl (pH 7.6) containing 5 mM CaCl2, 1 μM ZnCl2, and 0.02% sodium azide. The gel was then incubated overnight at 37°C in the same buffer, after which it was stained with 0.25% Coomassie brilliant blue R-250 (Sigma-Aldrich) for 2 h and then destained.
Enzyme-linked immunosorbent assay.
Cell culture supernatants were collected to detect the levels of HBeAg and HBsAg with an enzyme-linked immunosorbent assay (ELISA) kit (Ke Hua Bio Engineering, Shanghai, China) according to the manufacturer's instructions. Cell culture supernatants were collected to detect IFN-β levels with an ELISA kit (Elabscience, Wuhan, China) according to the manufacturer's instructions.
Northern blot analysis and HBV DNA analysis.
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Ten micrograms of total RNA was separated in a 1% agarose gel containing 2.2 M formaldehyde and transferred onto a nylon membrane (GE Healthcare). To detect HBV RNAs, the membrane was probed with a digoxigenin (DIG)-labeled RNA probe and immunoblotted with the DIG Northern Starter kit (Roche). The 28S and 18S rRNAs were used as loading controls.
At 96 h posttransfection, the cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.0], 0.5% NP-40) on ice. Nuclei were pelleted by centrifugation for 1 min at 10,000 × g. The supernatant was adjusted with 10 mM MgCl2 and treated with DNase I for 1 h at 37°C. The enzymes were inactivated by incubation at 75°C for 15 min in the presence of 10 mM EDTA. Protein was digested with proteinase K in the presence of 1% SDS. Nucleic acids were purified by phenol-chloroform extraction and ethanol precipitation. Extracellular encapsidated HBV DNA was extracted as described previously, with modifications (64). Ten microliters of the cell culture supernatant was treated with DNase I for 1 h at 37°C, and the enzymes were then inactivated by incubation at 75°C for 15 min in the presence of 10 mM EDTA. After this, the mixture was added to 100 μl lysis buffer (20 mM Tris-HCl, 20 mM EDTA, 50 mM NaCl, and 0.5% SDS) containing proteinase K. After incubation at 50°C overnight, viral DNA was isolated by phenol-chloroform extraction and ethanol precipitation. HBV DNA was subjected to TaqMan real-time PCR in a Light Cycler instrument (Roche) by using PCR primers (5′-AGAAACAACACATAGCGCCTCAT-3′ and 5′-TGCCCCATGCTGTAGATCTTG-3′) and probe (5′-TGTGGGTCACCATATTCTTGGG-3′).
Flow cytometry.
HepG2 cells were Fc blocked with human IgG for 15 min at 4°C prior to staining. Cells in cold PBS (containing 2% bovine serum albumin [BSA]) were incubated with specific Abs for 1 h at 4°C. Cells were washed with cold PBS (containing 2% BSA) twice and then analyzed by using a FACSCalibur instrument (Beckman Coulter).
Denatured immunoprecipitation and ubiquitination assays.
A ubiquitination assay was performed as described previously (65). The cells were lysed in IP lysis buffer containing 1% SDS and heated for 5 min. The samples were diluted 1:9 with IP lysis buffer and centrifuged at 12,000 × g for 10 min at 4°C. The supernatants were subjected to immunoprecipitation with anti-Flag, and the immunoprecipitates and WCLs were analyzed by 8% SDS-PAGE and immunoblotting.
MBP pulldown assay.
MBP and MBP–MMP-9-D3 were expressed by E. coli BL21 cells and purified. The cell extracts were prepared by ultrasonication and incubated with amylose resins (New England BioLabs). The MBP and MBP–MMP-9-D3 immobilized resins were washed 5 times with cold PBS. The resulting immobilized amylose resins were incubated with HEK293T lysates, which overexpressed IFNAR1-HA. The resins were washed 5 times with cold PBS, denatured with SDS-PAGE loading buffer, and analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence.
HepG2 cells were grown in glass-bottom dishes. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde for 15 min, washed three times with PBS, permeabilized with PBS containing 0.2% Triton X-100 for 5 min, washed three times with PBS, and blocked with PBS containing 5% bovine serum albumin for 30 min at room temperature. The cells were then incubated with anti-HA and anti-Myc overnight at 4°C, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG and cy3-conjugated goat anti-mouse IgG for 45 min and staining with 4′,6-diamidino-2-phenylindole (DAPI). The cells were viewed by confocal laser microscopy (FluoView FV1000; Olympus).
Statistical analysis.
The results are presented as means ± standard deviations. Student's t test for paired samples was used to determine statistical significance. Differences were considered statistically significant at a P value of ≤0.05.
ACKNOWLEDGMENTS
We thank Mingzhou Chen of Wuhan University, China, for kindly providing VSV-GFP and Ying Zhu of Wuhan University, China, for kindly providing the HepG2-NTCP cells and HepaAD38 cells.
This work was supported by research grants from the Major State Basic Research Development Program (973 Program) (2012CB518900), the National Natural Science Foundation of China (31230005, 81471942, 81171525, 31270206, and 31200134), and the National Mega Project on Major Infectious Disease Prevention (2012ZX10002006-003 and 2012ZX10004-207).
We disclose no conflicts.
REFERENCES
- 1.Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. 2006. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol Rev 28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 2.Liaw YF, Chu CM. 2009. Hepatitis B virus infection. Lancet 373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu R, Fan R, Hou J. 2014. Chronic hepatitis B virus infection: epidemiology, prevention, and treatment in China. Front Med 8:135–144. doi: 10.1007/s11684-014-0331-5. [DOI] [PubMed] [Google Scholar]
- 6.Uzé G, Schreiber G, Piehler J, Pellegrini S. 2007. The receptor of the type I interferon family. Curr Top Microbiol Immunol 316:71–95. [DOI] [PubMed] [Google Scholar]
- 7.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Björklund M, Koivunen E. 2005. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta 1755:37–69. [DOI] [PubMed] [Google Scholar]
- 9.Chung TW, Kim JR, Suh JI, Lee YC, Chang YC, Chung TH, Kim CH. 2004. Correlation between plasma levels of matrix metalloproteinase (MMP)-9/MMP-2 ratio and alpha-fetoproteins in chronic hepatitis carrying hepatitis B virus. J Gastroenterol Hepatol 19:565–571. doi: 10.1111/j.1440-1746.2004.03344.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayasaka A, Suzuki N, Fujimoto N, Iwama S, Fukuyama E, Kanda Y, Saisho H. 1996. Elevated plasma levels of matrix metalloproteinase-9 (92-kd type IV collagenase gelatinase B) in hepatocellular carcinoma. Hepatology 24:1058–1062. doi: 10.1002/hep.510240513. [DOI] [PubMed] [Google Scholar]
- 11.Chung TW, Lee YC, Kim CH. 2004. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J 18:1123–1125. [DOI] [PubMed] [Google Scholar]
- 12.Kong MY, Whitley RJ, Peng N, Oster R, Schoeb TR, Sullender W, Ambalavanan N, Clancy JP, Gaggar A, Blalock JE. 2015. Matrix metalloproteinase-9 mediates RSV infection in vitro and in vivo. Viruses 7:4230–4253. doi: 10.3390/v7082817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo SJ, Yun YJ, Lyu MA, Woo SY, Woo ER, Kim SJ, Lee HJ, Park HK, Kook YH. 2002. Respiratory syncytial virus infection induces matrix metalloproteinase-9 expression in epithelial cells. Arch Virol 147:229–242. doi: 10.1007/s705-002-8316-1. [DOI] [PubMed] [Google Scholar]
- 14.Dabo AJ, Cummins N, Eden E, Geraghty P. 2015. Matrix metalloproteinase 9 exerts antiviral activity against respiratory syncytial virus. PLoS One 10:e0135970. doi: 10.1371/journal.pone.0135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vartio T, Hovi T, Vaheri A. 1982. Human macrophages synthesize and secrete a major 95,000-dalton gelatin-binding protein distinct from fibronectin. J Biol Chem 257:8862–8866. [PubMed] [Google Scholar]
- 16.Chung TW, Moon SK, Lee YC, Kim JG, Ko JH, Kim CH. 2002. Enhanced expression of matrix metalloproteinase-9 by hepatitis B virus infection in liver cells. Arch Biochem Biophys 408:147–154. doi: 10.1016/S0003-9861(02)00522-2. [DOI] [PubMed] [Google Scholar]
- 17.Hosel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, Langenkamp A, Falk C, Buning H, Rose-John S, Protzer U. 2009. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology 50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 18.Nagase H, Woessner JF Jr. 1999. Matrix metalloproteinases. J Biol Chem 274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 19.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Faelth M, Stindt J, Koeniger C, Nassal M, Kubitz R, Sueltmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher SP, Chin DJ, Ji Y, Iniguez AL, Taillon B, Swinney DC, Ravindran P, Cheng DT, Bitter H, Lopatin U, Ma H, Klumpp K, Menne S. 2012. Transcriptomic analysis of the woodchuck model of chronic hepatitis B. Hepatology 56:820–830. doi: 10.1002/hep.25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn C, Peppa D, Khanna P, Nebbia G, Jones M, Brendish N, Lascar RM, Brown D, Gilson RJ, Tedder RJ, Dusheiko GM, Jacobs M, Klenerman P, Maini MK. 2009. Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterology 137:1289–1300. doi: 10.1053/j.gastro.2009.06.054. [DOI] [PubMed] [Google Scholar]
- 23.Wieland S, Thimme R, Purcell RH, Chisari FV. 2004. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A 101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucifora J, Durantel D, Testoni B, Hantz O, Levrero M, Zoulim F. 2010. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology 51:63–72. doi: 10.1002/hep.23230. [DOI] [PubMed] [Google Scholar]
- 25.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelissen I, Martens E, Van Den Steen PE, Proost P, Ronsse I, Opdenakker G. 2003. Gelatinase B/matrix metalloproteinase-9 cleaves interferon-beta and is a target for immunotherapy. Brain 126:1371–1381. doi: 10.1093/brain/awg129. [DOI] [PubMed] [Google Scholar]
- 27.Bertoletti A, Ferrari C. 2012. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut 61:1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 28.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar KG, Tang W, Ravindranath AK, Clark WA, Croze E, Fuchs SY. 2003. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J 22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. 2009. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian J, Zheng H, Huangfu WC, Liu J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. 2011. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog 7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monferran S, Paupert J, Dauvillier S, Salles B, Muller C. 2004. The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. EMBO J 23:3758–3768. doi: 10.1038/sj.emboj.7600403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piccard H, Van den Steen PE, Opdenakker G. 2007. Hemopexin domains as multifunctional liganding modules in matrix metalloproteinases and other proteins. J Leukoc Biol 81:870–892. doi: 10.1189/jlb.1006629. [DOI] [PubMed] [Google Scholar]
- 34.Gauzzi MC, Barbieri G, Richter MF, Uze G, Ling L, Fellous M, Pellegrini S. 1997. The amino-terminal region of Tyk2 sustains the level of interferon alpha receptor 1, a component of the interferon alpha/beta receptor. Proc Natl Acad Sci U S A 94:11839–11844. doi: 10.1073/pnas.94.22.11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya S, HuangFu WC, Liu J, Veeranki S, Baker DP, Koumenis C, Diehl JA, Fuchs SY. 2010. Inducible priming phosphorylation promotes ligand-independent degradation of the IFNAR1 chain of type I interferon receptor. J Biol Chem 285:2318–2325. doi: 10.1074/jbc.M109.071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya S, Qian J, Tzimas C, Baker DP, Koumenis C, Diehl JA, Fuchs SY. 2011. Role of p38 protein kinase in the ligand-independent ubiquitination and down-regulation of the IFNAR1 chain of type I interferon receptor. J Biol Chem 286:22069–22076. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Carvalho LP, Bhattacharya S, Carbone CJ, Kumar KG, Leu NA, Yau PM, Donald RG, Weiss MJ, Baker DP, McLaughlin KJ, Scott P, Fuchs SY. 2009. Mammalian casein kinase 1alpha and its leishmanial ortholog regulate stability of IFNAR1 and type I interferon signaling. Mol Cell Biol 29:6401–6412. doi: 10.1128/MCB.00478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Plotnikov A, Banerjee A, Suresh Kumar KG, Ragimbeau J, Marijanovic Z, Baker DP, Pellegrini S, Fuchs SY. 2008. Ligand-independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun 367:388–393. doi: 10.1016/j.bbrc.2007.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai L, Zhang W, Tan L, Yang H, Ge M, Zhu C, Zhang R, Cao Y, Chen J, Luo Z, Ho W, Liu F, Wu K, Wu J. 2015. Hepatitis B virus hijacks CTHRC1 to evade host immunity and maintain replication. J Mol Cell Biol 7:543–556. doi: 10.1093/jmcb/mjv048. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q, Gong R, Qu J, Zhou Y, Liu W, Chen M, Liu Y, Zhu Y, Wu J. 2012. Activation of the Ras/Raf/MEK pathway facilitates hepatitis C virus replication via attenuation of the interferon-JAK-STAT pathway. J Virol 86:1544–1554. doi: 10.1128/JVI.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boritz E, Gerlach J, Johnson JE, Rose JK. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J Virol 73:6937–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arii S, Mise M, Harada M, Furutani M, Ishigami S, Niwano M, Mizumoto M, Fukumoto M, Imamura M. 1996. Overexpression of matrix metalloproteinase 9 gene in hepatocellular carcinoma with invasive potential. Hepatology 24:316–322. doi: 10.1002/hep.510240206. [DOI] [PubMed] [Google Scholar]
- 43.Misse D, Esteve PO, Renneboog B, Vidal M, Cerutti M, St Pierre Y, Yssel H, Parmentier M, Veas F. 2001. HIV-1 glycoprotein 120 induces the MMP-9 cytopathogenic factor production that is abolished by inhibition of the p38 mitogen-activated protein kinase signaling pathway. Blood 98:541–547. doi: 10.1182/blood.V98.3.541. [DOI] [PubMed] [Google Scholar]
- 44.Lu L, Zhang Q, Wu K, Chen X, Zheng Y, Zhu C, Wu J. 2015. Hepatitis C virus NS3 protein enhances cancer cell invasion by activating matrix metalloproteinase-9 and cyclooxygenase-2 through ERK/p38/NF-kappaB signal cascade. Cancer Lett 356:470–478. doi: 10.1016/j.canlet.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 45.Nunez O, Fernandez-Martinez A, Majano PL, Apolinario A, Gomez-Gonzalo M, Benedicto I, Lopez-Cabrera M, Bosca L, Clemente G, Garcia-Monzon C, Martin-Sanz P. 2004. Increased intrahepatic cyclooxygenase 2, matrix metalloproteinase 2, and matrix metalloproteinase 9 expression is associated with progressive liver disease in chronic hepatitis C virus infection: role of viral core and NS5A proteins. Gut 53:1665–1672. doi: 10.1136/gut.2003.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luangsay S, Gruffaz M, Isorce N, Testoni B, Michelet M, Faure-Dupuy S, Maadadi S, Ait-Goughoulte M, Parent R, Rivoire M, Javanbakht H, Lucifora J, Durantel D, Zoulim F. 2015. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol 63:1314–1322. doi: 10.1016/j.jhep.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Guy CS, Mulrooney-Cousins PM, Churchill ND, Michalak TI. 2008. Intrahepatic expression of genes affiliated with innate and adaptive immune responses immediately after invasion and during acute infection with woodchuck hepadnavirus. J Virol 82:8579–8591. doi: 10.1128/JVI.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lutgehetmann M, Bornscheuer T, Volz T, Allweiss L, Bockmann JH, Pollok JM, Lohse AW, Petersen J, Dandri M. 2011. Hepatitis B virus limits response of human hepatocytes to interferon-alpha in chimeric mice. Gastroenterology 140:2074–2083. doi: 10.1053/j.gastro.2011.02.057. [DOI] [PubMed] [Google Scholar]
- 49.Seth RB, Sun LJ, Chen ZJJ. 2006. Antiviral innate immunity pathways. Cell Res 16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 50.Wu J, Huang S, Zhao X, Chen M, Lin Y, Xia Y, Sun C, Yang X, Wang J, Guo Y, Song J, Zhang E, Wang B, Zheng X, Schlaak JF, Lu M, Yang D. 2014. Poly(I:C) treatment leads to interferon-dependent clearance of hepatitis B virus in a hydrodynamic injection mouse model. J Virol 88:10421–10431. doi: 10.1128/JVI.00996-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanford RE, Guerra B, Chavez D, Giavedoni L, Hodara VL, Brasky KM, Fosdick A, Frey CR, Zheng J, Wolfgang G, Halcomb RL, Tumas DB. 2013. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology 144:1508–1517. doi: 10.1053/j.gastro.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JL, Wu M, Zhang XN, Zhang W, Zhang ZQ, Chen LX, He J, Zheng Y, Chen CC, Wang F, Hu YW, Zhou XH, Wang C, Xu Y, Lu MJ, Yuan ZH. 2013. Hepatitis B virus polymerase impairs interferon-alpha-induced STAT activation through inhibition of importin-alpha 5 and protein kinase C-delta. Hepatology 57:470–482. doi: 10.1002/hep.26064. [DOI] [PubMed] [Google Scholar]
- 53.Christen V, Treves S, Duong FHT, Heim MH. 2007. Activation of endoplasmic reticulum stress response by hepatitis viruses up-regulates protein phosphatase 2A. Hepatology 46:558–565. doi: 10.1002/hep.21611. [DOI] [PubMed] [Google Scholar]
- 54.He XX, Chang Y, Jiang HJ, Tang F, Meng FY, Xie QH, Li PY, Song YH, Lin JS. 2010. Persistent effect of IFNAR-1 genetic polymorphism on the long-term pathogenesis of chronic HBV infection. Viral Immunol 23:251–257. doi: 10.1089/vim.2009.0102. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J, Huang JD, Poon VK, Chen DQ, Chan CC, Ng F, Guan XY, Watt RM, Lu L, Yuen KY, Zheng BJ. 2009. Functional dissection of an IFN-alpha/beta receptor 1 promoter variant that confers higher risk to chronic hepatitis B virus infection. J Hepatol 51:322–332. doi: 10.1016/j.jhep.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 56.Zhou J, Lu L, Yuen MF, Lam TW, Chung CP, Lam CL, Zhang B, Wang S, Chen Y, Wu SH, Poon VK, Ng F, Chan CC, Jiang S, Yuen KY, Zheng BJ. 2007. Polymorphisms of type I interferon receptor 1 promoter and their effects on chronic hepatitis B virus infection. J Hepatol 46:198–205. doi: 10.1016/j.jhep.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 57.Fan H, Zhu Z, Wang Y, Zhang X, Lu Y, Tao Y, Fan W, Wang Z, Wang H, Roggendorf M, Lu M, Wang B, Yang D. 2012. Molecular characterization of the type I IFN receptor in two woodchuck species and detection of its expression in liver samples from woodchucks infected with woodchuck hepatitis virus (WHV). Cytokine 60:179–185. doi: 10.1016/j.cyto.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 58.Cho IR, Oh M, Koh SS, Malilas W, Srisuttee R, Jhun BH, Pellegrini S, Fuchs SY, Chung YH. 2012. Hepatitis B virus X protein inhibits extracellular IFN-alpha-mediated signal transduction by downregulation of type I IFN receptor. Int J Mol Med 29:581–586. doi: 10.3892/ijmm.2012.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cauwe B, Opdenakker G. 2010. Intracellular substrate cleavage: a novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit Rev Biochem Mol Biol 45:351–423. doi: 10.3109/10409238.2010.501783. [DOI] [PubMed] [Google Scholar]
- 60.Cauwe B, Van den Steen PE, Opdenakker G. 2007. The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit Rev Biochem Mol Biol 42:113–185. doi: 10.1080/10409230701340019. [DOI] [PubMed] [Google Scholar]
- 61.Redondo-Munoz J, Ugarte-Berzal E, Terol MJ, Van den Steen PE, del Cerro MH, Roderfeld M, Roeb E, Opdenakker G, Garcia-Marco JA, Garcia-Pardo A. 2010. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia B cell survival through its hemopexin domain. Cancer Cell 17:160–172. doi: 10.1016/j.ccr.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 62.Kessenbrock K, Plaks V, Werb Z. 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanceau J, Truchet S, Bauvois B. 2003. Matrix metalloproteinase-9 silencing by RNA interference triggers the migratory-adhesive switch in Ewing's sarcoma cells. J Biol Chem 278:36537–36546. doi: 10.1074/jbc.M304300200. [DOI] [PubMed] [Google Scholar]
- 64.Tian YJ, Chen WL, Ou JHJ. 2011. Effects of interferon-alpha/beta on HBV replication determined by viral load. PLoS Pathog 7:e1002159. doi: 10.1371/journal.ppat.1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, Zhong B, Shu HB. 2014. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]