ABSTRACT
The impact of mosquito-borne flavivirus infections worldwide is significant, and many critical aspects of these viruses' biology, including virus-host interactions, host cell requirements for replication, and how virus-host interactions impact pathology, remain to be fully understood. The recent reemergence and spread of flaviviruses, including dengue virus (DENV), West Nile virus (WNV), and Zika virus (ZIKV), highlight the importance of performing basic research on this important group of pathogens. MicroRNAs (miRNAs) are small, noncoding RNAs that modulate gene expression posttranscriptionally and have been demonstrated to regulate a broad range of cellular processes. Our research is focused on identifying pro- and antiflaviviral miRNAs as a means of characterizing cellular pathways that support or limit viral replication. We have screened a library of known human miRNA mimics for their effect on the replication of three flaviviruses, DENV, WNV, and Japanese encephalitis virus (JEV), using a high-content immunofluorescence screen. Several families of miRNAs were identified as inhibiting multiple flaviviruses, including the miRNA miR-34, miR-15, and miR-517 families. Members of the miR-34 family, which have been extensively characterized for their ability to repress Wnt/β-catenin signaling, demonstrated strong antiflaviviral effects, and this inhibitory activity extended to other viruses, including ZIKV, alphaviruses, and herpesviruses. Previous research suggested a possible link between the Wnt and type I interferon (IFN) signaling pathways. Therefore, we investigated the role of type I IFN induction in the antiviral effects of the miR-34 family and confirmed that these miRNAs potentiate interferon regulatory factor 3 (IRF3) phosphorylation and translocation to the nucleus, the induction of IFN-responsive genes, and the release of type I IFN from transfected cells. We further demonstrate that the intersection between the Wnt and IFN signaling pathways occurs at the point of glycogen synthase kinase 3β (GSK3β)–TANK-binding kinase 1 (TBK1) binding, inducing TBK1 to phosphorylate IRF3 and initiate downstream IFN signaling. In this way, we have identified a novel cellular signaling network with a critical role in regulating the replication of multiple virus families. These findings highlight the opportunities for using miRNAs as tools to discover and characterize unique cellular factors involved in supporting or limiting virus replication, opening up new avenues for antiviral research.
IMPORTANCE MicroRNAs are a class of small regulatory RNAs that modulate cellular processes through the posttranscriptional repression of multiple transcripts. We hypothesized that individual miRNAs may be capable of inhibiting viral replication through their effects on host proteins or pathways. To test this, we performed a high-content screen for miRNAs that inhibit the replication of three medically relevant members of the flavivirus family: West Nile virus, Japanese encephalitis virus, and dengue virus 2. The results of this screen identify multiple miRNAs that inhibit one or more of these viruses. Extensive follow-up on members of the miR-34 family of miRNAs, which are active against all three viruses as well as the closely related Zika virus, demonstrated that miR-34 functions through increasing the infected cell's ability to respond to infection through the interferon-based innate immune pathway. Our results not only add to the knowledge of how viruses interact with cellular pathways but also provide a basis for more extensive data mining by providing a comprehensive list of miRNAs capable of inhibiting flavivirus replication. Finally, the miRNAs themselves or cellular pathways identified as modulating virus infection may prove to be novel candidates for the development of therapeutic interventions.
KEYWORDS: Wnt signaling, flavivirus, interferons, microRNA
INTRODUCTION
Flaviviruses are small, positive-sense, enveloped viruses that are transmitted by arthropods such as mosquitoes or ticks. Viruses of this family that infect humans are a significant cause of morbidity and mortality worldwide and include dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), and the recently emerging pathogen Zika virus (ZIKV) (1–4). Due their relatively small genome (10 to 11 kb), flaviviruses are highly dependent on the host cell to complete their life cycle, and many aspects of the host-pathogen interface still remain to be elucidated (5–7). Research into the host functions supporting and limiting flaviviral replication both informs our understanding of the host-pathogen interface as well as provides promising avenues toward the discovery of antiviral therapeutics.
MicroRNAs (miRNAs) are small (20- to 23-nucleotide [nt]), noncoding RNAs that play major roles in regulating a wide range of cellular processes. Through complementarity between the “seed” sequence (nt 2 to 8) of the miRNA and sequences in the 3′ untranslated regions (UTRs) of target transcripts, cellular mRNAs are targeted to the RNA-induced silencing complex (RISC), whereby deadenylation, destabilization, and sometimes direct cleavage result in the repression of the expression of the transcript. Due to the small region of complementarity dictating miRNA targeting, it is estimated that a single miRNA can cause the repression of hundreds of transcripts, and many examples in which a single miRNA acts on multiple factors within a specific cellular process have been found (8, 9).
The role of miRNAs during viral infection has been the subject of considerable study and some controversy. Several studies demonstrated that viral infection results in significant changes to the expression profiles of cellular miRNAs (10–17). These observed changes in miRNA expression may represent an arm of the host response to viral infection or manipulation of endogenous host cell pathways by the virus in order to promote a cellular environment more conducive to viral replication. In agreement with this possibility, multiple studies have identified miRNAs that inhibit the replication of various viruses (15, 18–21). In contrast, cells lacking miRNAs due to the genetic ablation of Dicer have been shown to support the replication of several RNA viruses at levels equivalent to those in control cells with an intact miRNA population. Furthermore, the degradation of host miRNAs via the expression of the vaccinia virus gene product VP55 has been shown to have very little effect on the cellular response to double-stranded RNA (dsRNA) or type I interferon (IFN) (22, 23), although the loss of miRNAs results in increased cytokine production. Thus, the role of miRNAs in regulating viral replication may still require further study. Nonetheless, because cellular miRNAs are predicted to target multiple components of host cell pathways, we hypothesize that miRNAs may also be used as tools to elucidate host cell functions required for viral replication, even if the endogenous miRNA does not normally play a role in the replication of the virus of interest.
In this study, we use a high-content screen to identify miRNAs that inhibit the replication of three medically important flaviviruses, DENV, WNV, and Japanese encephalitis virus (JEV). We find that some of the most potent inhibitors of these viruses are members of the miRNA miR-34 family and that this antiviral activity extends to other virus families as well. Members of the miR-34 family (miR-34a/c and -449a/c) have been studied extensively for their role in development and cancer and have been demonstrated to act as potent tumor suppressors. miR-34a is often deleted or downregulated in tumors, and it is induced by p53 (24). This miRNA has been demonstrated to be a potent antioncogenic treatment in multiple animal models, indicating its potential for acting against a broad range of cancer types, including prostate and lung tumors (25–27). miR-34-regulated targets have been identified in multiple growth-related pathways, including cell proliferation, apoptosis, and the cell cycle. miR-34a has also been shown to regulate multiple components of Wnt/β-catenin signal transduction (24, 28), a critical pathway mediating cellular growth, development, and tumorigenesis. Recently, several groups demonstrated that cross talk between the Wnt pathway and the cellular innate immune pathways can occur (29–34). Due to previously reported observations that miR-34 family members regulate Wnt signaling and that interfacing occurs between IFN and Wnt signaling, we investigate the role of this cross talk in the antiviral activity of miR-34a and demonstrate that the repression of Wnt signaling by the miR-34 family potentiates type I IFN signaling in response to viral infection.
RESULTS
High-content screen of human miRNAs to identify antiflaviviral miRNAs.
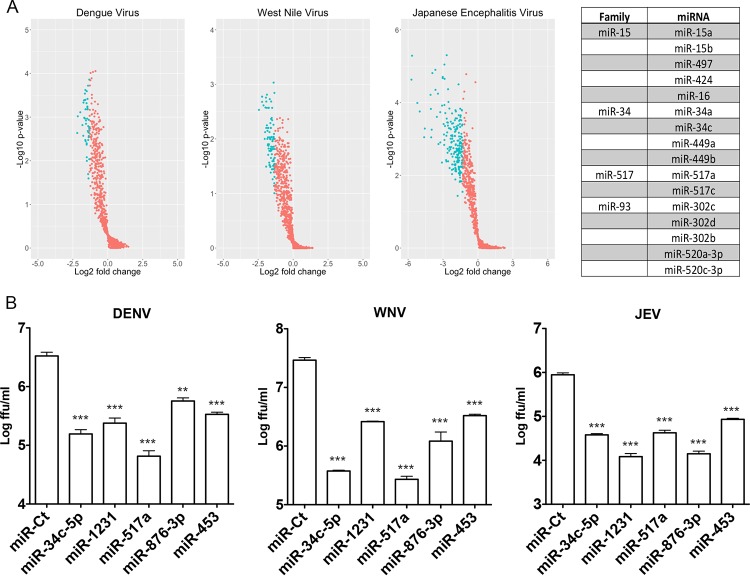
Given the requirement of viruses for host physiological processes to complete their life cycle and the wide range of cellular functions that are regulated by host miRNAs, the identification of miRNAs that support/limit viral replication has the potential to enhance our understanding of cellular pathways that are important for viral infection. Therefore, we used a high-content immunofluorescence assay to screen known human miRNAs for their effects on the replication of three different flaviviruses, DENV, WNV, and JEV. HeLa cells transfected with individual miRNAs were infected with each virus and fixed at various times postinfection (p.i.), and the viral envelope was detected by using an antiflaviviral antibody. Of the ∼1,200 miRNAs screened, 45, 93, and 204 were found to significantly (P < 0.05) inhibit DENV, WNV, and JEV infection, respectively, by more than 60% (Fig. 1A, blue). miRNAs that displayed >20% toxicity, as quantified by the loss of nuclear staining, were eliminated from the analysis. The complete results from the miRNA library against DENV, WNV, and JEV are provided in Table S1 in the supplemental material. Although only five miRNAs met the criteria for inhibiting all three viruses (miR-34c-5p, miR-1231, miR-517a, miR-876-3p, and miR-453), several miRNA families (i.e., miRNAs with the same seed sequence that are predicted to repress some or all of the same targets) were overrepresented among the lists of miRNAs that significantly inhibited each of the viruses, including the miR-34, miR-517, miR-15, miR-148, and miR-573 families (Fig. 1A, right). Confirmation of the activity of the panflaviviral inhibitors identified was performed by using focus-forming assays to determine the level of inhibition of the production of infectious virus (Fig. 1B).
FIG 1.
High-content microRNA screen against three flaviviruses, DENV, WNV, and JEV. (A) HeLa cells were transfected with miRNAs from the Dharmacon/Thermo Scientific v16.0 library and infected with DENV, WNV, or JEV (MOI = 0.5 FFU/cell) at 48 h posttransfection in 96-well plates in triplicate. At 24 h (WNV and JEV) or 48 h (DENV) posttransfection, cells were fixed and immunostained with antiflavivirus envelope antibody/AF488-anti-mouse IgG and DAPI to visualize nuclei. Total AF488 and DAPI signals were quantitated and normalized to those for miRNA control-transfected wells. miRNAs causing >20% toxicity were eliminated from the analysis. Volcano plots indicate the performance of miRNAs against DENV, WNV, and JEV infections. Active miRNAs (>60% inhibition; P value of <0.05) are denoted in blue. Highly represented miRNA families with activity against all three flaviviruses are shown at the right. (B) HeLa cells were reverse transfected with the indicated miRNAs and infected with DENV, WNV, and JEV at an MOI of 0.5 FFU/cell at 48 h posttransfection. Supernatants were collected at 48 h, and infectious virus was quantitated by a focus-forming assay.
miR-34 family members are potent inhibitors of flavivirus replication.
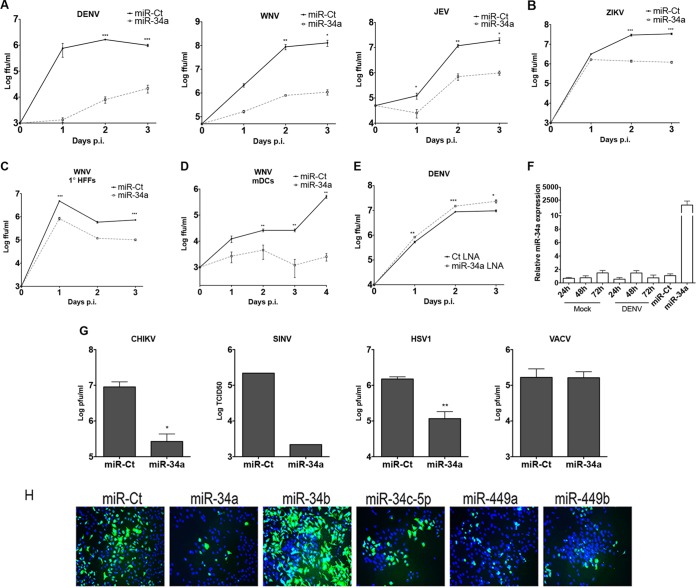
Further confirmation of the inhibitory effect of miR-34a against DENV, WNV, and JEV was carried out by examining the effect of miR-34a transfection on flaviviral replication in multistep growth curves. miR-34a transfection potently inhibited infection by all three flaviviruses compared to control miRNA (miR-Ct) transfection (Fig. 2A). Inhibition of WNV infection by miR-34a was also examined in primary human foreskin fibroblasts (HFFs) (Fig. 2C) and murine bone marrow-derived dendritic cells (BM-DCs) (Fig. 2D) to confirm antiviral activity in primary cell types with relevance to flavivirus infection. To determine the breadth of antiviral activity, miR-34a transfections were carried out, followed by infection by multiple virus types, including ZIKV (strain PRVABC059), chikungunya virus (CHIKV), Sindbis virus (SINV), herpes simplex virus 1 (HSV-1), and vaccinia virus (VACV). As shown in Fig. 2B and G, miR-34a exhibited significant inhibitory activity against ZIKV, SINV, CHIKV, and HSV-1, while VACV replication was unaffected. The effect of the inhibition of miRNA activity was investigated by using miR-34a-targeting locked nucleic acid (LNA) as well. Transfection of a miR-34a-targeting LNA resulted in a modest but significant enhancement of DENV replication (Fig. 2E), suggesting that endogenous miR-34a plays a role in suppressing flaviviral replication. Interestingly, miR-34a levels were unaffected by DENV infection (Fig. 2F). Other miRNAs with the same seed sequence as those of miR-34a, miR34c, and miR-449a/b (GCAGUG) were also examined and found to repress DENV infection as well (Fig. 2H). Interestingly, miR-34b, whose seed sequence is offset by a single nucleotide, did not inhibit DENV infection, demonstrating the specificity of this antiviral activity for miR-34 family-targeted transcripts. These results suggest a relatively broad range of antiviral activity, indicating a possible involvement of the cellular innate response pathways in the antiviral activity of the miR-34 family.
FIG 2.
miR-34 family members are broadly acting antiviral miRNAs. (A) HeLa cells were reverse transfected with a control miRNA (miR-Ct) or miR-34a and infected with DENV, WNV, and JEV at an MOI of 0.1 FFU/cell at 48 h posttransfection. Supernatants were collected at the indicated times postinfection and assayed for viral titers by a focus-forming assay. (B) ZIKV (strain PRVABC59) infections were carried out as described above for panel A at an MOI of 5 FFU/cell. (C) HFFs were transfected with miR-Ct or miR-34a and infected with WNV at an MOI of 3 FFU/cell at 48 h posttransfection. (D) Primary BM-DCs were electroporated with miR-Ct or miR-34a as described in Materials and Methods. At 48 h postelectroporation, cells were infected with WNV at an MOI of 0.01 FFU/cell. (E) HeLa cells were transfected with control or miR-34a LNAs and then infected with DENV at an MOI of 0.5 particles/cell. Supernatants were collected at the indicated times postinfection and assayed by a focus-forming assay. (F) HeLa cells were mock treated, infected with DENV at an MOI of 5 FFU/cell, or transfected with miR-Ct/miR-34a. At the indicated times postinfection or at 48 h posttransfection, total RNA was isolated, and the miR-34a expression level was quantitated by RT-qPCR (normalized to the U6 snRNP small RNA). (G) HeLa cells transfected with miR-Ct or miR-34a were infected with CHIKV, SINV, HSV-1, or VACV at an MOI of 0.5 particles/cell. Supernatants were collected at 24 to 48 h postinfection, and infectious units were quantitated by a plaque assay (CHIKV, HSV-1, and VACV) or a TCID50 assay (SINV) (results are representative of data from >3 independent experiments) (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.005). (H) HeLa cells were transfected with miR-34 family members (miR-34a/c and miR-449a/b), miR-Ct, or miR-34b and infected with DENV at an MOI of 5 FFU/cell at 48 h posttransfection. Cells were fixed at 48 h postinfection and immunostained with antienvelope/DAPI.
Repression of Wnt signaling controls miR-34a antiviral activity.
Extensive research on the role of miR-34 family members in cancer has characterized these miRNAs as tumor suppressors and identified miR-34-repressed targets in multiple oncogenic pathways, including Wnt/β-catenin signaling, the cell cycle, and apoptosis (reviewed in reference 24). Initiation of Wnt signaling occurs through the binding of soluble Wnt factors to the low-density lipoprotein receptor-related protein (LRP)/Frizzled receptor at the cell surface. The activation of this receptor complex leads to a downstream cascade that culminates in the repression of glycogen synthase kinase 3β (GSK3β) activity. GSK3β exists at steady state in a complex that includes Axin, adenomatous polyposis coli protein (APC), and β-catenin. GSK3β constitutively phosphorylates β-catenin, targeting it for degradation through the ubiquitin-proteasome system. The activation of the Wnt pathway and the subsequent inhibition of GSK3β activity release β-catenin from degradation, allowing it to translocate to the nucleus and initiate the transcription of Wnt-responsive transcripts (e.g., c-jun and c-myc) (reviewed in references 35 and 36).
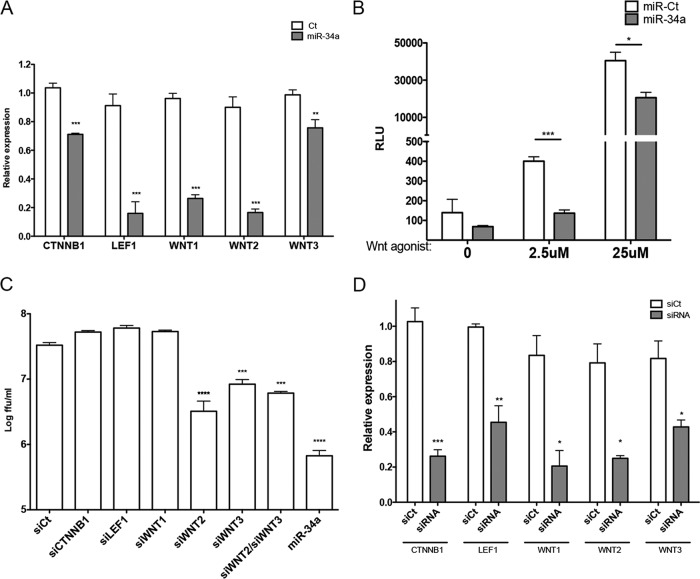
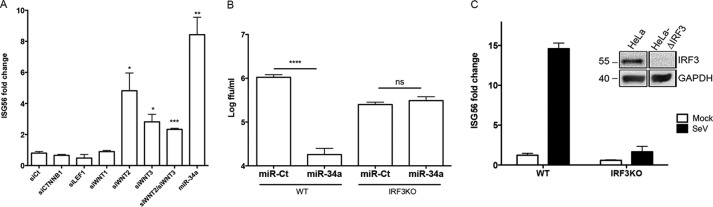
To confirm the repression of Wnt signaling by miR-34a in our system, the effect of transfection on transcript levels of characterized factors of the Wnt pathway was examined by reverse transcriptase quantitative PCR (RT-qPCR). Consistent with previously reported results (28), miR-34a transfection significantly inhibited the production of CTTNB1, LEF1, WNT1, WNT2, and WNT3 (Fig. 3A). Additionally, the effect on Wnt signaling was quantified by using a previously characterized reporter system (pTOP-FLASH) (37). As expected, miR-34a transfection repressed Wnt signaling in both the absence and the presence of the activation of this pathway by a Wnt agonist (Fig. 3B). These results confirm the suppressive effects of miR-34a on Wnt activation in our model system. To determine whether these effects are responsible for the antiflaviviral effects of the miR-34 family, small interfering RNA (siRNA) knockdown of CTNNB1, LEF1, WNT1, WNT2, and WNT3 was performed, followed by DENV infection. Only the knockdown of WNT2 or WNT3 displayed significant inhibition of DENV infection, although this inhibition did not reach the level of that with miR-34a transfection (Fig. 3C), indicating a role for both WNT2 and WNT3 in miR-34-mediated inhibition of viral infection. The cotransfection of siRNAs targeting WNT2 and WNT3 displayed no additive effect on replication. Significant levels of knockdown by each siRNA were confirmed by RT-qPCR (Fig. 3D). These results implicate WNT2 and WNT3 in the antiviral effects of miR-34a but suggest that other targets are contributing to its activity.
FIG 3.
Repression of Wnt pathway factors by miR-34a dictates antiviral activity. (A) Total RNA isolated from miR-Ct- and miR-34a-transfected cells was assayed for transcript levels of the indicated Wnt pathway factors by RT-qPCR (*, P value of <0.05; **, P value of <0.01; ***, P value of <0.005). (B) HeLa cells were transfected with miR-Ct or miR-34a, followed by transfection 24 h later with pTOP-FLASH, a plasmid that expresses luciferase under the control of a Wnt-responsive promoter. Cells were treated at 24 h post-plasmid transfection with 2.5 μM and 25 μM Wnt agonist II to induce the Wnt/β-catenin pathway for 24 h. The luciferase level was quantitated by using the OneGlo luciferase system. (C) HeLa cells were transfected with siRNAs targeting the indicated transcripts or miR-Ct and -34a for 48 h, followed by infection with DENV at an MOI of 0.1 FFU/cell. Quantitation of infectious virus in supernatants collected at 3 days postinfection was performed by a focus-forming assay. (D) HeLa cells were transfected with the indicated siRNAs for 48 h. Total RNA was assayed for transcript levels of the indicated Wnt pathway factors by RT-qPCR.
miR-34a inhibits flavivirus infection through activation of type I interferon signaling.
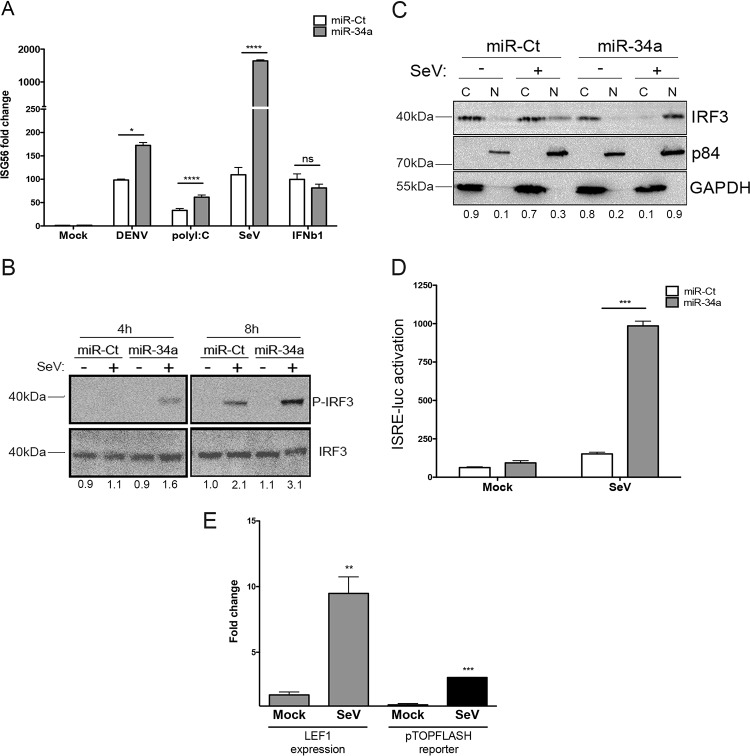
Although the Wnt/β-catenin signaling pathway has been extensively characterized as being critical for the regulation of development, cell-cell interactions, cancer progression, and stem cell control, recent research has implicated this signaling pathway in the modulation of the type I IFN response as well (29–31, 33, 34, 38). To determine whether miR-34a is affecting the IFN pathway in our system, miR-34a-transfected HeLa cells were treated with various stimuli, and the induction of IFN signaling was quantitated by RT-qPCR of interferon-stimulated gene 56 (ISG56) (also known as IFIT1). miR-34a transfection was found to enhance the induction of the IFN response when cells were treated with DENV, poly(I:C), or Sendai virus (SeV) but not IFN-β (Fig. 4A). Notably, ISG56 induction was not observed in miR-34a-transfected cells in the absence of a stimulus, demonstrating that it does not act as a direct inducer of ISG56 gene expression but rather potentiates the response to exogenous stimuli. The lack of potentiation of IFN-β-induced transcription, downstream of the IFN-α/β receptor, suggests that the effects observed are being carried out during the initial phase of IFN signaling, upstream of IRF activation and prior to IFN-α/β release. Given that the miR-34a-associated ISG induction differential was highest with SeV infection, we used this stimulus for subsequent experiments.
FIG 4.
miR-34a transfection potentiates type I interferon signaling. (A) miR-Ct/-34a-transfected HeLa cells were treated with the indicated stimuli [DENV at an MOI of 5 FFU/cell for 30 h, poly(I:C) transfected at 5 μg/ml for 18 h, and SeV at a 1:1,000 dilution for 6 h], and total RNA was isolated. Relative ISG56 levels were quantified by RT-qPCR and normalized to β-actin levels (*, P value of <0.01; **, P value of <0.005; ***, P value of <0.001; ns, not significant). (B) miR-Ct/-34a-transfected cells were treated with SeV for the indicated times, and total protein was analyzed by Western blotting for phosphorylated and total IRF3 levels. Results are representative of data from at least three independent experiments. Fold changes were calculated by measuring the integrated density by using ImageJ software and normalized to values for the control. (C) miR-Ct/-34a-transfected cells were treated with SeV for 6 h. Cytoplasmic (C) and nuclear (N) fractions were collected and analyzed by Western blotting for total IRF3, p84 (nuclear marker), and GAPDH (cytoplasmic marker). Results are representative of data from at least three independent experiments. (D) Supernatants from miR-Ct/-34a-transfected, SeV-treated cells were collected at 6 h and placed onto ISRE-luciferase-expressing telomerized human fibroblasts. At 18 h posttreatment, cells were lysed, and luciferase levels were quantified by using the OneGlo assay reagent. (E) HeLa cells were treated with SeV for 18 h. Total RNA was isolated and assayed for the Wnt-responsive transcript LEF1 or β-actin by TaqMan RT-qPCR. (Left) Fold changes were calculated by using the ΔΔCT method. HeLa cells were transfected with the pTOP-FLASH reporter plasmid and then treated at 48 h posttransfection with Wnt agonist II or SeV. (Right) Cells were lysed and assayed for luciferase activity at 18 h posttreatment.
IRF3 is a key player in the IFN induction process. IRF3 activation is triggered by various pattern recognition receptors, resulting in its phosphorylation and nuclear localization and the subsequent expression of IFN-β. In the presence of miR-34a, SeV infection was found to induce early and potent activation of IRF3 phosphorylation (Fig. 4B). Accordingly, nuclear translocation of IRF3 also occurred to a high degree in miR-34a-transfected HeLa cells following SeV treatment (Fig. 4C). To quantitate the secretion of type I IFNs, supernatants from miRNA-transfected, SeV-treated cells were collected and used to treat IFN-stimulated responsive element (ISRE)–luciferase human reporter cells (39). In agreement with the observed effects of miR-34a on IRF3 activation, high levels of type I IFNs were detected in the supernatants of miR-34a-transfected cells (Fig. 4D). These results indicate that miR-34a is a potent enhancer of IFN induction and that this activation may be responsible for the observed antiviral activity of this miRNA family.
To further dissect the role of the Wnt pathway in the cellular response to a viral stimulus, the effect of SeV treatment on Wnt signaling was examined by the quantification of the level of a Wnt-responsive transcript (Fig. 4E, left) or activation of the pTOP-FLASH reporter vector (Fig. 4E, right) following treatment with SeV for 18 h. In both readouts, activation of Wnt signaling by SeV was observed. These data suggest that the initiation of the innate immune response triggers a corresponding response through Wnt signaling to act as a damper to prevent the overstimulation of interferon signaling.
To confirm the role of the specific miR-34a targets previously identified to inhibit DENV infection (Fig. 3C), siRNA knockdown of CTNNB1, LEF1, WNT1, WNT2, and WNT3, or the simultaneous knockdown of both WNT2 and WNT3, was performed, followed by SeV treatment. As expected, the knockdown of either of WNT2 or WNT3 had an enhancing effect on ISG56 transcription (Fig. 5A), demonstrating that the repressive function of miR-34a on WNT2 and WNT3 is directly related to its observed induction of IFN signaling.
FIG 5.
Repression of flavivirus infection occurs through IRF3-mediated interferon signaling. (A) miR-Ct/-34a- or siRNA-transfected cells were treated with SeV for 6 h. Total RNA was analyzed for ISG56 levels as described in the text. (B) Parental and IRF3KO HeLa cells were transfected with miR-Ct/-34a, followed by infection with DENV at an MOI of 5 FFU/cell. Supernatants were collected at 48 h, and infectious virus quantified by a focus-forming assay. WT, wild type. (C) Confirmation of IRF3 knockout by Western blotting (inset) and reactivity to SeV treatment by ISG56 RT-qPCR.
To examine whether IRF3-dependent induction of the IFN response is required for the antiviral role of miR-34a, IRF3 knockout (IRF3KO) HeLa cells were produced by using clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 genome editing (39). IRF3 protein levels (Fig. 5C, inset) and the responsiveness of the cells to SeV stimulation were tested to ensure complete knockdown (Fig. 5C). The requirement of IRF3 for the miR-34a-mediated inhibition of DENV replication was then tested by miRNA transfection of either parental cells or IRF3KO cells, followed by DENV infection. Surprisingly, IRF3KO cells did not support a higher level of viral replication, which is likely due to the various mechanisms of innate signaling antagonism employed by DENV (40–42). Nonetheless, in contrast to the potent inhibition observed in miR-34a-transfected wild-type cells, replication in IRF3KO cells was unaffected by the presence of miR-34a (Fig. 5B), demonstrating a direct role for IRF3 in the antiviral activity associated with miR-34a.
Cross talk between the Wnt and IFN pathways occurs through direct binding of TBK1 and GSK3β.
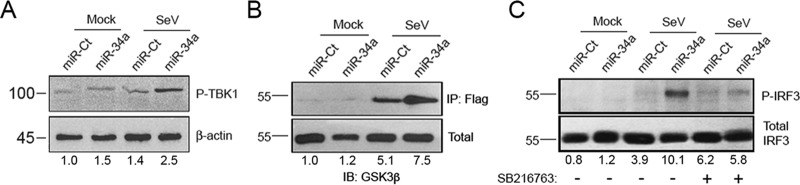
Previous research implicated several different mechanisms through which the Wnt and IFN pathways interact. Examples include the β-catenin-mediated activation of IFN-responsive transcription, GSK3α/β nuclear translocation and repression of IRFs, and direct interactions between GSK3 and IRF3 or TANK-binding kinase 1 (TBK1) (the IRF3-directed kinase) (29, 31–34, 43, 44). Notably, this cross talk between pathways was suggested to have both positive and negative impacts on the induction of the IFN response. To investigate the specific mechanism in this system through which we observe an inverse relationship between Wnt activation and IFN signaling, the phosphorylation status of TBK1 was determined, and we observed increased TBK1 phosphorylation at Ser172 following miR-34a/SeV treatment (Fig. 6A), suggesting that the activation of GSK3β induced by Wnt pathway inhibition indeed promotes the GSK3β/TBK1 interaction, as was previously reported (44). Additionally, using immunoprecipitation of Flag-tagged TBK1 and subsequent immunoblotting for GSK3β (Fig. 6B), we found that miR-34a transfection followed by SeV infection promotes direct interactions between these two factors. Finally, the requirement of GSK3β activity for miR-34a-mediated IRF3 phosphorylation was determined by using a GSK3β inhibitor (SB216763). We found that GSK3β kinase activity is required for the observed effects on the IRF3 phosphorylation of miR-34a (Fig. 6C). These results collectively demonstrate that the cross talk between the Wnt and IFN pathways occurs at the point of GSK3β-TBK1 binding and that this interaction promotes the proinflammatory, antiviral activity of miR-34a.
FIG 6.
Interferon and Wnt pathways intersect through direct GSK3β/TBK1 interaction. (A) miR-Ct/-34a-transfected HeLa cells were mock or SeV treated for 6 h. Total protein was assessed for phosphorylated TBK1 and β-actin levels by Western blotting. Results are representative of data from at least three independent experiments. (B) miR-Ct/-34a-transfected cells were transfected with a plasmid encoding Flag-tagged TBK1 at 24 h post-miRNA transfection. At 24 h post-plasmid transfection, cells were treated with SeV for 6 h, lysed, and subjected to immunoprecipitation with Flag-Sepharose beads. Immunoprecipitated and total fractions were analyzed by Western blotting for GSK3β levels. IB, immunoblotting. (C) miR-Ct/-34a-transfected cells were treated with SeV as described above, with or without the GSK3β inhibitor SB216763 (5 μM). Phosphorylated and total IRF3 levels were assessed by Western blotting. Results are representative of data from at least three independent experiments. Fold changes were calculated by measuring the integrated density using ImageJ software and normalized to the values for the control.
DISCUSSION
In this study, we use a high-content screen of a human miRNA library to identify miRNAs that have an inhibitory effect on the replication of three flaviviruses, DENV, WNV, and JEV. The identification of the miR-34 family as potent antiflaviviral miRNAs with some inhibitory effects that extend beyond the flavivirus family led us to investigate this well-characterized miRNA family for its role in influencing the host-pathogen interface.
Viruses are highly dependent on cellular processes to complete their life cycles. Although a great deal has been discovered about host-virus interactions, much remains to be learned regarding host pathways that support and limit virus replication. Because of the low level of complementarity between a miRNA seed sequence and a transcript 3′ UTR required to mediate repression, a single miRNA has the capacity to regulate a large number of factors and can culminate in the repression of an entire signaling pathway. This study complements previous siRNA-based screens in which the knockdown of individual proteins has been used to identify host factors that influence virus replication (45–48). Thus, we believe that this approach has great potential for facilitating the discovery of novel host pathways that modulate virus replication, which could lead to the description of novel therapeutic targets.
We have focused on the miR-34 family, which has been shown to target more than a dozen transcripts within the Wnt/β-catenin signaling pathway, and the transfection of these miRNAs results in the potent repression of Wnt signaling (24). We found that this network intersects the IRF3 terminal signaling pathway and that its repression results in the induction of the IRF3-mediated antiviral response when cells are exposed to pathogen-associated molecular patterns (PAMPs) (e.g., viral double-stranded RNA). These results suggest that under constitutive conditions, factors in the Wnt pathway are functioning to control inflammatory responses and that the release of this repression through increased miR-34 expression is the mechanism by which these miRNAs exert their antiviral properties.
Previous studies also implicated the Wnt pathway as being important for viral infection. A recent genome-wide siRNA screen identified multiple components of the Wnt pathway as positively acting upon Rift Valley fever virus as well as other members of the bunyavirus family (49). The activation of aspects of Wnt signaling and stabilization of β-catenin have also been observed in cells latently infected with the gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus (50). The role of the Wnt pathway during the replication of these viruses has been hypothesized to involve cell replication or differentiation (49, 51), but the role of the modulation of innate immunity may also be considered in light of our results. Multiple studies (29–34, 38, 52) described a relationship between Wnt signaling and the innate response, with differing hypotheses about the specific relationship of these pathways. Purported intersections include β-catenin-mediated activation of IFN-responsive transcription, repression of IRFs by GSK3α/β, and direct interactions between GSK3 and IRF3 or TBK1, resulting in the promotion of IRF- and NF-κB-responsive transcription. Our results indicate that the binding of TBK1 to GSK3β and the activation of its kinase activity are the primary aspects of the cross talk between the pathways that dictates the antiviral effects of miR-34a within this model system. The work presented here suggests that the Wnt pathway can function to modulate the host inflammatory response, perhaps as a means of dampening innate signaling and preventing toxic effects on the cell. These data provide a clearer understanding of the intersection between these two critical host pathways, Wnt signaling and interferon induction.
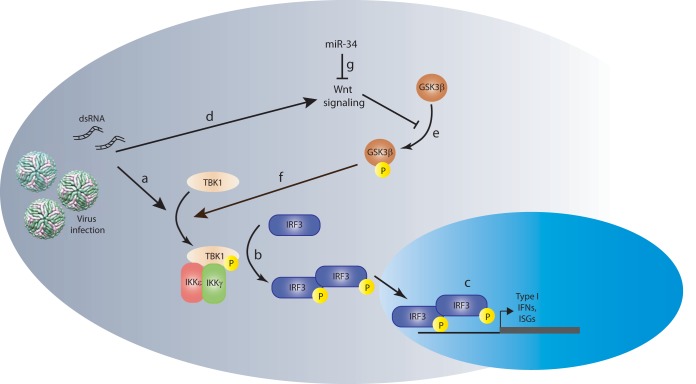
We also show that the inhibition of endogenous miR-34a results in a modest increase in DENV replication (Fig. 2E), suggesting that miR-34a normally serves to dampen Wnt signaling, allowing TBK1-mediated phosphorylation of IRF3 in response to PAMP detection. High levels of exogenously introduced miR-34a, as used in the screen and experiments described above, make the cells exquisitely sensitive to PAMP recognition, resulting in a temporal and quantitative increase in IFN signaling and rendering the cells more resistant to virus infection. Our model, in which miR-34 and Wnt act antagonistically to fine-tune the IFN response, is shown in Fig. 7. In this model, the induction of the innate response by virus infection or other stimuli through pattern recognition/RIG-I-like receptors occurs in concert with the activation of the Wnt pathway (Fig. 7a and d). Innate signaling culminates in the phosphorylation of TBK1, which acts on IRF3 to promote the transcription of IFN-β and ISGs (Fig. 7b). Our data and those reported previously by others (38, 44) suggest a role for phosphorylated GSK3β in regulating the activation of TBK1 (Fig. 7f). miR-34a repression of Wnt signaling (Fig. 7g), which suppresses the phosphorylation of GSK3β (e), results in a GSK3β/TBK1 interaction that promotes IRF3 phosphorylation and the subsequent transcriptional activation of IFN-β and other ISGs, promoting an antiviral response (c). We note that this model is consistent with previously suggested roles for miRNAs as regulators of inflammatory responses. Several studies have shown that cellular stress, including viral infection, can result in the poly-ADP-ribosylation of proteins of the RISC, relieving the miRNA-mediated inhibition of mRNA expression (53, 54), leading to the hypothesis that miRNAs provide a block to the IFN response, which is released upon infection and RISC inactivation. Additionally, miR-146a has been implicated in a negative-feedback loop regulating NF-κB-mediated transcription, preventing hyperinflammation in response to PAMPs (55, 56), while the depletion of the total cellular miRNA population by VACV VP55 results in increased cytokine expression (23).
FIG 7.
Model of the Wnt-IFN signaling intersection. Innate activation by viral infection or dsRNA treatment (via Toll-like or RIG-I-like receptors) induces TBK1 phosphorylation (a), subsequent IRF3 phosphorylation/homodimerization (b), and translocation into the nucleus, where it induces the transcription of type I IFNs and interferon-stimulated genes (c). Simultaneous activation of the Wnt signaling pathway by viral infection (d) culminates in the repression of GSK3b phosphorylation (e), which feeds back positively into the IFN signaling pathway through interactions with TBK1 (f). The intersection between these two pathways suggests that the Wnt pathway can function to modulate the host inflammatory response as a way of controlling innate signaling. Inhibition of Wnt signaling by miR-34 (g) results in enhanced signaling through IRF3, promoting an antiviral state in the cell. IKK, IκB kinase.
In addition to defining novel virus-host interfaces, miRNAs also have therapeutic potential. The capacity for miRNAs for use in vivo is currently being investigated in the context of cancer therapies. A number of tumor-suppressive miRNAs are currently in various stages of development as replacement therapies for a range of human cancers. miR-34a has demonstrated great promise in animal models of myelomas and hepatocellular, lung, and breast cancers and is now being tested in human trials for efficacy against hepatic and hematologic cancers (reviewed in reference 57). Although challenges remain in terms of the delivery and targeting of miRNAs as therapeutics, these studies illustrate the great potential for the in vivo use of these molecules. Furthermore, our results suggest that small-molecule antagonists of Wnt signaling, which have been suggested for use as anticancer therapeutics (58), may also be capable of functioning as antiviral molecules.
In summary, this study not only demonstrates a novel strategy for antiviral discovery but also reveals the potential for discovering novel host networks involved in viral replication and antiviral responses.
MATERIALS AND METHODS
Cell culture and reagents.
HEK293 cells (Microbix), HeLa cells (ATCC), telomerized human foreskin fibroblasts (tHFs) (gift from V. DeFilippis, Oregon Health & Science University [OHSU]), and HFFs (Clonetics) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin G-sodium, and 100 μg/ml streptomycin sulfate (Invitrogen). Primary murine BM-DCs were prepared as previously described (59). Briefly, bone marrow cells were aspirated from the femurs and tibias of C57BL/6 mice and cultured in 20 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 20 ng/ml interleukin-4 (IL-4) (Peprotech) for 5 days prior to electroporation. SB216763 was obtained from Sigma (catalog number S3442), and Wnt agonist II was obtained from EMD Biosciences (catalog number SKL2001).
Antibodies.
In this study, antibodies recognizing the following proteins were obtained from the indicated sources: IRF3 (Santa Cruz Biotechnology), phospho-IRF3 (Ser386; Epitomics), p84 (Genetex), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam), GSK3β (Cell Signaling Technology), phospho-TBK1 (Ser172; Cell Signaling Technology), and a Flag epitope (Sigma). The hybridoma producing panflavivirus anti-E antibody 4G2 (60) was obtained from the ATCC. The hybridoma was maintained and antibody was purified from the culture supernatant by the Vaccine and Gene Therapy Institute monoclonal antibody core facility (OHSU).
Virus strains.
DENV2 (New Guinea C) and Sindbis virus (AR339 strain) were obtained from the ATCC. JEV (SA-14-2-8) was obtained from Robert Tesh, World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch. West Nile virus (385-99) was described previously (61), and DENV, WNV, and JEV strains were passaged twice on C6/36 cells and purified by centrifugation as previously described (62). Virus titers were determined by a focus-forming assay (63). Herpes simplex virus 1 (F1 strain) was a kind gift from A. Hill (Oregon Health & Science University). Vaccinia virus (Western Reserve strain) was a kind gift from M. Slifka (Oregon Health & Science University). CHIKV was a kind gift from D. Streblow (Oregon Health & Science University). Zika virus (strain PRVABC59) was obtained from the Centers for Disease Control and Prevention.
High-content screen.
miRNA mimics representing all miRNAs annotated with miRBase v16.0 were obtained from Dharmacon/Thermo Scientific. Reverse transfection of miRNAs was performed by the distribution of 2.5 pmol miRNA, 0.2 μl RNAiMAX (Invitrogen), and 20 μl Opti-MEM (Gibco) per well into 96-well plates, followed by an overlay with 1.2 × 104 HeLa cells. At 48 h posttransfection (p.t.), cells were infected with DENV2 (NGC), WNV (NY99), or JEV (SA-14-2-8) at a multiplicity of infection (MOI) of 0.5 focus-forming units (FFU) per cell. Cells were fixed at 24 h p.i. (WNV and JEV) or 48 h p.i. (DENV) and stained for the viral envelope (E) protein as previously described (63). Nine fields per well were imaged for E positivity (Alexa Fluor 488 [AF488]) and nuclear staining (4′,6-diamidino-2-phenylindole [DAPI]) by using an Opera LX high-content imaging system (PerkinElmer) and analyzed with Acapella software (PerkinElmer) by determining the pixel intensity for each image after background (area of no cells) subtraction. Screens were performed in triplicate. Following the reading of all replicate plates, the results were quantile normalized across all plates, and ratios of viral protein expression and cell nuclei were calculated for each well compared to the control values. Significant effects on viral replication were determined by fitting a linear model per miRNA, with P values calculated by using a one-sided t test with null values equaling 1. Wells with <80% DAPI staining were determined to be cytotoxic and eliminated from the analysis. Significant inhibitors of viral replication were assessed as those that inhibited viral replication by >60% with raw P values of <0.05.
Viral growth curves.
HeLa cells were transfected with 10 pmol miR-34a (Dharmacon), miR-Ct (RISC-free negative control; Dharmacon), or the miRcury LNA inhibitor (miR-34a) (catalog number 4100982; Exiqon) in 24-well plates. At 48 h p.t., cells were infected with DENV, WNV, JEV, or ZIKV at an MOI of 0.1 FFU/cell. At the indicated times p.i., supernatants were collected, and the amount of virus was quantitated by a focus-forming assay on Vero cells. CHIKV, SINV, HSV-1, and VACV infections were performed at an MOI of 0.5 FFU/cell. Supernatants were collected at 48 h postinfection and assayed for infectious particles by a 50% tissue culture infective dose (TCID50) assay (SINV) or a plaque assay (CHIKV, HSV-1, and VACV).
RT-qPCR.
Total RNA was isolated by using TRIzol. RT-qPCR was performed with 100 ng RNA by using the RNA-to-Ct (catalog number 4392653; Life Technologies) assay system with TaqMan assays for LEF1 (catalog number Hs01547250_m1), CTNNB1 (catalog number Hs00355049_m1), WNT1 (catalog number Hs01011247_m1), WNT2 (catalog number Hs00608224_m1), WNT3 (catalog number Hs00902257_m1), ISG56 (catalog number Hs03027069_s1), ACTB1 (catalog number Hs99999903_m1), hsa-miR-34a (catalog number 000426), and U6 snRNP (catalog number 001973). The relative expression level was quantitated by using the ΔΔCT method relative to values for β-actin.
siRNA transfections.
Control, CTNNB1 (catalog number s437), LEF1 (catalog number s27618), WNT1 (catalog number s14863), WNT2 (catalog number s14867), and WNT3 (s14869) siRNAs were obtained from Life Technologies. Transfections were performed with 10 pmol siRNA and 0.8 μl Lipofectamine RNAiMAX in 24-well plates. Infections were carried out at 48 h p.t. at 0.1 FFU/cell, and supernatants were quantitated by a focus-forming assay. For BM-DCs, 500,000 cells were electroporated with 30 pmol miR-34a or a control miRNA mimic by using the Amaxa mouse dendritic cell Nucleofector kit (Lonza) according to the manufacturer's protocol. At 48 h posttransfection, cells were infected with WNV at 0.01 PFU/cell. Supernatants were collected and assayed for infectious virus as described above.
Luciferase assays.
Twenty-four hours after transfection with miR-Ct/miR-34a, cells were transfected with TOP-FLASH (catalog number 12456; Addgene) and then treated with a Wnt agonist (catalog number 681667; Calbiochem) for 24 h before luciferase quantitation using the OneGlo luciferase reagent (catalog number E6110; Promega) according to the manufacturer's instructions. Supernatants from miRNA-transfected, SeV-treated cells were collected at 6 h posttreatment and placed onto tHFs expressing an ISRE-luciferase reporter plasmid (64). Luciferase levels were quantitated at 24 h posttreatment as described above.
IRF3 knockout cell production.
IRF3 knockout HeLa cells were produced by using CRISPR-Cas9 technology as previously described (39, 65). HeLa cells were transduced with lentiviruses expressing IRF3-targeting guide RNA and Cas9 endonuclease, and positive transductants were selected with 3 μg/ml puromycin. Monoclonal populations were obtained by limited dilution and assayed for IRF3 knockout by Western blotting and innate reactivity to SeV treatment.
Immunoprecipitations.
At the indicated times posttransfection/posttreatment, cells were harvested in 1% Triton X-100-containing lysis buffer, and immunoprecipitations were performed by using anti-Flag-Sepharose beads (catalog number A2220; Sigma) for 1 to 2 h. Samples were eluted from beads with SDS-PAGE loading buffer and analyzed by Western blotting.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yih-tai Chen of the Oregon Translational Research Development Institute for performing image analysis of the high-content assay, Victor DeFilippis and Meaghan Hancock for critical readings of the manuscript, and Christopher Parkins for technical assistance.
This work was supported by NIH grants and contracts R21 AI101282 and HHSN272201100017C.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02388-16.
REFERENCES
- 1.Special Programme for Research & Training in Tropical Diseases (TDR). 2007. Report of Dengue Scientific Working Group. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Hayes EB, Komar N, Nasci RS, Montgomery SP, O'Leary DR, Campbell GL. 2005. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis 11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Flohic G, Porphyre V, Barbazan P, Gonzalez JP. 2013. Review of climate, landscape, and viral genetics as drivers of the Japanese encephalitis virus ecology. PLoS Negl Trop Dis 7:e2208. doi: 10.1371/journal.pntd.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischl W, Bartenschlager R. 2011. Exploitation of cellular pathways by dengue virus. Curr Opin Microbiol 14:470–475. doi: 10.1016/j.mib.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Acosta EG, Kumar A, Bartenschlager R. 2014. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Adv Virus Res 88:1–109. doi: 10.1016/B978-0-12-800098-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan MN, Garcia-Blanco MA. 2014. Targeting host factors to treat West Nile and dengue viral infections. Viruses 6:683–708. doi: 10.3390/v6020683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JL, Grey FE, Uhrlaub JL, Nikolich-Zugich J, Hirsch AJ. 2012. Induction of the cellular microRNA, Hs_154, by West Nile virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors. J Virol 86:5278–5287. doi: 10.1128/JVI.06883-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. 2008. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol 82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, Wu Z, Peng Y, Liu X, Lu J, Wang L, Pan Q, He ML, Li XP. 2010. MicroRNA-10b induced by Epstein-Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer Lett 299:29–36. doi: 10.1016/j.canlet.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, Doukas T, Paranjape S, Polacek C, dos Santos FB, Jalili R, Babrzadeh F, Gharizadeh B, Grimm D, Kay M, Koike S, Sarnow P, Ronaghi M, Ding SW, Harris E, Chow M, Diamond MS, Kirkegaard K, Glenn JS, Fire AZ. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng X, Li Y, Walters KA, Rosenzweig ER, Lederer SL, Aicher LD, Proll S, Katze MG. 2009. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics 10:373. doi: 10.1186/1471-2164-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slonchak A, Shannon RP, Pali G, Khromykh AA. 2016. Human microRNA miR-532-5p exhibits antiviral activity against West Nile virus via suppression of host genes SESTD1 and TAB3 required for virus replication. J Virol 90:2388–2402. doi: 10.1128/JVI.02608-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 17.Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol 82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santhakumar D, Forster T, Laqtom NN, Fragkoudis R, Dickinson P, Abreu-Goodger C, Manakov SA, Choudhury NR, Griffiths SJ, Vermeulen A, Enright AJ, Dutia B, Kohl A, Ghazal P, Buck AH. 2010. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci U S A 107:13830–13835. doi: 10.1073/pnas.1008861107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashraf U, Zhu B, Ye J, Wan S, Nie Y, Chen Z, Cui M, Wang C, Duan X, Zhang H, Chen H, Cao S. 2016. MicroRNA-19b-3p modulates Japanese encephalitis virus-mediated inflammation via targeting RNF11. J Virol 90:4780–4795. doi: 10.1128/JVI.02586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Ye J, Ashraf U, Li Y, Wei S, Wan S, Zohaib A, Song Y, Chen H, Cao S. 2016. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol 90:3722–3734. doi: 10.1128/JVI.03242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, Li KC, Cheng CL, Cheng HW, Lee JY, Lee CN, Yang PC. 2011. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9:58–69. doi: 10.1016/j.chom.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Bogerd HP, Skalsky RL, Kennedy EM, Furuse Y, Whisnant AW, Flores O, Schultz KL, Putnam N, Barrows NJ, Sherry B, Scholle F, Garcia-Blanco MA, Griffin DE, Cullen BR. 2014. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol 88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguado LC, Schmid S, Sachs D, Shim JV, Lim JK, ten Oever BR. 2015. MicroRNA function is limited to cytokine control in the acute response to virus infection. Cell Host Microbe 18:714–722. doi: 10.1016/j.chom.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha YH, Kim NH, Park C, Lee I, Kim HS, Yook JI. 2012. miRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle 11:1273–1281. doi: 10.4161/cc.19618. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. 2010. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. 2011. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim NH, Kim HS, Kim N-G, Lee I, Choi H-S, Li X-Y, Kang SE, Cha SY, Ryu JK, Na JM, Park C, Kim K, Lee S, Gumbiner BM, Yook JI, Weiss SJ. 2011. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal 4:ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baril M, Es-Saad S, Chatel-Chaix L, Fink K, Pham T, Raymond VA, Audette K, Guenier AS, Duchaine J, Servant M, Bilodeau M, Cohen E, Grandvaux N, Lamarre D. 2013. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signaling pathway as negative regulator of virus-induced innate immune responses. PLoS Pathog 9:e1003416. doi: 10.1371/journal.ppat.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hack K, Reilly L, Proby C, Fleming C, Leigh I, Foerster J. 2012. Wnt5a inhibits the CpG oligodeoxynucleotide-triggered activation of human plasmacytoid dendritic cells. Clin Exp Dermatol 37:557–561. doi: 10.1111/j.1365-2230.2012.04362.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. 2010. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol 11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 32.Khan KA, Do F, Marineau A, Doyon P, Clement JF, Woodgett JR, Doble BW, Servant MJ. 2015. Fine-tuning of the RIG-I-like receptor/interferon regulatory factor 3-dependent antiviral innate immune response by the glycogen synthase kinase 3/beta-catenin pathway. Mol Cell Biol 35:3029–3043. doi: 10.1128/MCB.00344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, Coyne CB, Sarkar SN. 2011. PKC alpha regulates Sendai virus-mediated interferon induction through HDAC6 and beta-catenin. EMBO J 30:4838–4849. doi: 10.1038/emboj.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillesheim A, Nordhoff C, Boergeling Y, Ludwig S, Wixler V. 2014. β-Catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-κB signaling. Cell Commun Signal 12:29. doi: 10.1186/1478-811X-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaus A, Birchmeier W. 2008. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 36.Giles RH, van Es JH, Clevers H. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta 1653:1–24. [DOI] [PubMed] [Google Scholar]
- 37.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13:680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 38.Qin Y, Liu Q, Tian S, Xie W, Cui J, Wang RF. 2016. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3beta to TBK1. Cell Res 26:613–628. doi: 10.1038/cr.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sali TM, Pryke KM, Abraham J, Liu A, Archer I, Broeckel R, Staverosky JA, Smith JL, Al-Shammari A, Amsler L, Sheridan K, Nilsen A, Streblow DN, DeFilippis VR. 2015. Characterization of a novel human-specific STING agonist that elicits antiviral activity against emerging alphaviruses. PLoS Pathog 11:e1005324. doi: 10.1371/journal.ppat.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalrymple NA, Cimica V, Mackow ER. 2015. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio 6:e00553-15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muñoz-Jordán JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A 100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J-T, Chang L-S, Chen C-J, Doong S-L, Chang C-W, Chen M-R. 2014. Glycogen synthase kinase 3 negatively regulates IFN regulatory factor 3 transactivation through phosphorylation at its linker region. Innate Immun 20:78–87. doi: 10.1177/1753425913485307. [DOI] [PubMed] [Google Scholar]
- 44.Lei C-Q, Zhong B, Zhang Y, Zhang J, Wang S, Shu H-B. 2010. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 33:878–889. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 45.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. 2014. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog 10:e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. 2005. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev 19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harmon B, Bird SW, Schudel BR, Hatch AV, Rasley A, Negrete OA. 15 August 2016. A genome-wide RNAi screen identifies a role for Wnt/beta-catenin signaling during Rift Valley fever virus infection. J Virol doi: 10.1128/JVI.00543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat Med 9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 51.Hayward SD, Liu J, Fujimuro M. 2006. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci STKE 2006:re4. [DOI] [PubMed] [Google Scholar]
- 52.Shi M, Cho H, Inn KS, Yang A, Zhao Z, Liang Q, Versteeg GA, Amini-Bavil-Olyaee S, Wong LY, Zlokovic BV, Park HS, Garcia-Sastre A, Jung JU. 2014. Negative regulation of NF-kappaB activity by brain-specific tripartite motif protein 9. Nat Commun 5:4820. doi: 10.1038/ncomms5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seo GJ, Kincaid RP, Phanaksri T, Burke JM, Pare JM, Cox JE, Hsiang TY, Krug RM, Sullivan CS. 2013. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. 2011. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med 208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gulla A, Tagliaferri P, Tassone P, Caraglia M. 2014. mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 3:e194. doi: 10.1038/mtna.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dihlmann S, von Knebel Doeberitz M. 2005. Wnt/beta-catenin-pathway as a molecular target for future anti-cancer therapeutics. Int J Cancer 113:515–524. doi: 10.1002/ijc.20609. [DOI] [PubMed] [Google Scholar]
- 59.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BR, Silverman RH, Gale M Jr, Diamond MS. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol 80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henchal EA, Gentry MK, McCown JM, Brandt WE. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg 31:830–836. [DOI] [PubMed] [Google Scholar]
- 61.Xiao SY, Guzman H, Zhang H, Travassos da Rosa AP, Tesh RB. 2001. West Nile virus infection in the golden hamster (Mesocricetus auratus): a model for West Nile encephalitis. Emerg Infect Dis 7:714–721. doi: 10.3201/eid0704.017420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medigeshi GR, Hirsch AJ, Brien JD, Uhrlaub JL, Mason PW, Wiley C, Nikolich-Zugich J, Nelson JA. 2009. West Nile virus capsid degradation of claudin proteins disrupts epithelial barrier function. J Virol 83:6125–6134. doi: 10.1128/JVI.02617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shum D, Smith JL, Hirsch AJ, Bhinder B, Radu C, Stein DA, Nelson JA, Fruh K, Djaballah H. 2010. High-content assay to identify inhibitors of dengue virus infection. Assay Drug Dev Technol 8:553–570. doi: 10.1089/adt.2010.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Fruh K. 2010. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J Virol 84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.