ABSTRACT
To maximize the coding potential of viral genomes, internal ribosome entry sites (IRES) can be used to bypass the traditional requirement of a 5′ cap and some/all of the associated translation initiation factors. Although viral IRES typically contain higher-order RNA structure, an unstructured sequence of about 84 nucleotides (nt) immediately upstream of the Turnip crinkle virus (TCV) coat protein (CP) open reading frame (ORF) has been found to promote internal expression of the CP from the genomic RNA (gRNA) both in vitro and in vivo. An absence of extensive RNA structure was predicted using RNA folding algorithms and confirmed by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) RNA structure probing. Analysis of the IRES region in vitro by use of both the TCV gRNA and reporter constructs did not reveal any sequence-specific elements but rather suggested that an overall lack of structure was an important feature for IRES activity. The CP IRES is A-rich, independent of orientation, and strongly conserved among viruses in the same genus. The IRES was dependent on eIF4G, but not eIF4E, for activity. Low levels of CP accumulated in vivo in the absence of detectable TCV subgenomic RNAs, strongly suggesting that the IRES was active in the gRNA in vivo. Since the TCV CP also serves as the viral silencing suppressor, early translation of the CP from the viral gRNA is likely important for countering host defenses. Cellular mRNA IRES also lack extensive RNA structures or sequence conservation, suggesting that this viral IRES and cellular IRES may have similar strategies for internal translation initiation.
IMPORTANCE Cap-independent translation is a common strategy among positive-sense, single-stranded RNA viruses for bypassing the host cell requirement of a 5′ cap structure. Viral IRES, in general, contain extensive secondary structure that is critical for activity. In contrast, we demonstrate that a region of viral RNA devoid of extensive secondary structure has IRES activity and produces low levels of viral coat protein in vitro and in vivo. Our findings may be applicable to cellular mRNA IRES that also have little or no sequences/structures in common.
KEYWORDS: IRES, translation, carmovirus, cap-independent translation, Turnip crinkle virus, unstructured RNA, internal ribosome entry site, translational control
INTRODUCTION
Eukaryotic mRNAs utilize a cap-dependent translation where the 40S ribosomal subunit is recruited to a 5′ 7-methylguanosine (m7G) cap with the assistance of eukaryotic initiation factors (eIFs). Translation initiation is largely governed by eIF4F, which consists of eIF4E and eIF4G in plants. eIF4E is responsible for binding to the 5′ cap structure, while eIF4G is a scaffolding protein that interacts with eIF4E and poly(A) binding protein (PABP) bound to the 3′ poly(A) tail, thus circularizing the mRNA (1). The 43S preinitiation complex, composed of the ribosomal 40S subunit bound to eIF3/eIF5 and eIF2-Met-tRNAiMet, is attracted to the 5′ cap via binding of eIF3 to eIF4G and then scans in the 5′-to-3′ direction until an initiation codon in good context is reached (2, 3). The 60S ribosome subunit then joins the 40S subunit with the assistance of eIF5B to form the 80S initiation complex, which is followed by polypeptide elongation and eventual termination.
Dependence on the 5′ end for translation initiation in eukaryotes generally leads to monocistronic translation, with a single polypeptide translated from each mRNA. To increase the number of genes that can be expressed from a genome of limited size, many eukaryotic RNA viruses have evolved alternative translation initiation strategies, including leaky scanning, “stop/go” reinitiation, ribosome shunting, and internal initiation (4). Such noncanonical translation allows for polycistronic translation as well as for either effective competition with host translation or continued translation after virus-induced shutdown of host cap-dependent translation.
Noncanonical translation initiation can be facilitated by the presence of internal ribosome entry sites (IRES), which are segments of RNA that recruit ribosome subunits and can be used for initiation of 5′-proximal translation in the absence of a 5′ cap or for translation of otherwise inaccessible internal open reading frames (ORFs) (4, 5). Animal virus IRES, first discovered in picornaviruses, are often >450 nucleotides (nt) long and are generally located immediately upstream of the translated ORF (6, 7). Although viral IRES often lack conservation of the primary nucleotide sequence, related IRES maintain strong conservation of secondary structure (8). An interesting trend is that IRES with the most stable structures tend to require the fewest initiation factors (9). For example, the dicistrovirus Cricket paralysis virus IRES is highly structured and can directly bind ribosomes in the absence of additional factors, while the less structured picornavirus IRES require some canonical eIF proteins (e.g., eIF4G, eIF4A, and eIF4B) (10). To date, it is not possible to predict which initiation factors are required for a particular IRES based on sequence or structure alone, so this must be determined experimentally.
The largest family of RNA plant viruses, the Potyviridae, share synteny with the Picornaviridae but differ in terms of IRES characteristics. Potyvirus IRES are much shorter than picornavirus IRES (60 to 200 nt), and the %GC is much lower (<30% compared to 60%) (11). Using folding algorithms, potyvirus IRES are predicted to lack extensive secondary structure, while the high GC content of picornavirus IRES results in very stable RNA structures (11). Despite the low %GC in the Tobacco etch virus (TEV) 5′ untranslated region (UTR), three pseudoknots have been identified that play an important role in translational enhancement (12). The 5′ UTR of the potyvirus Turnip mosaic virus (TuMV) contains an IRES that maintains activity even when replaced by the reverse complement sequence (13). This indicates that neither the RNA sequence nor RNA structure is critical for the TuMV IRES. Although “true IRES” appear to function independently in bicistronic reporter constructs and thus do not require sequences elsewhere for activity, an interplay between a 5′ UTR IRES and the 3′ UTR has been demonstrated for TEV (14). Similar 5′ IRES are absent in tombusviruses and luteoviruses, which instead contain elements in or near their 3′ UTRs, known as 3′ cap-independent translation enhancers (3′ CITEs), which enhance translation from their genomic RNA (gRNA) 5′ ends, usually through long-distance RNA-RNA interactions with 5′-proximal hairpins (15).
Internal ORFs in plant viruses can also be translated from an IRES. The coat proteins (CPs) of many monopartite plant viruses are encoded by the 3′-terminal gene, which is mainly translated from a subgenomic RNA (sgRNA). Some of these plant viruses contain an IRES immediately upstream of the CP ORF that can direct translation from the gRNA. The crucifer-infecting Tobacco mosaic virus (crTMV) contains a 148-nt IRES upstream of the CP ORF that has activity across kingdoms in plant, yeast, and human cell-based assays (16, 17). An IRES upstream of the CP in carmoviruses (Tombusviridae family) is thought to exist because the CP also functions as the silencing suppressor (18, 19), which is likely needed early in infection, prior to synthesis of high levels of sgRNA (20, 21). Based on studies with the carmovirus Turnip crinkle virus (TCV), the CP sequesters small interfering RNAs (siRNAs) and longer double-stranded RNAs (22) to prevent formation of active RNA-induced silencing complexes (RISC) that consist of at least one Argonaute protein (AGO). In addition, the CP binds to and inhibits Ago1 and/or Ago2 (23, 24), the main effector proteins that use siRNAs generated during an infection as guides for targeting the viral gRNA (25, 26).
In the carmovirus Hibiscus chlorotic ringspot virus (HCRSV), an IRES was defined within a 100-nt region upstream of the CP ORF that contains a critical 18-nt primary RNA sequence (20). The secondary structure of the HCRSV IRES was not determined, but IRES activity was enhanced by an unidentified interaction with the 3′ UTR (20). The IRES upstream of the carmovirus Pelargonium flower break virus (PFBV) CP ORF is 80 nt long and, when disrupted, caused a defect in virus accumulation in vivo (21). The structure of the PFBV IRES was predicted in silico to lack extensive secondary structure, similar to the TuMV IRES.
Studies of carmoviral IRES have mainly been performed using full-length gRNA or reporter constructs and wheat germ extracts (WGE). IRES secondary structures, if any, have not been defined experimentally, and evidence is lacking that CP IRES produce low levels of CP directly from the gRNA in vivo. In addition, it is not known if expression from the carmoviral IRES requires any eukaryotic translation initiation factors. Since TCV is the most extensively studied carmovirus, we used this system to clarify the requirements for internal translation of CP. The TCV gRNA is 4,053 nt long and lacks a 5′ cap and 3′ poly(A) tail. Cap-independent translation of the gRNA utilizes a 3′ CITE that assumes a tRNA-shaped structure and enhances translation initiation at the 5′ end through 60S ribosome recruitment in the absence of any long-distance interactions (27, 28). The five TCV gRNA overlapping ORFs encode the 5′ replicase-associated protein p28 and its ribosomal readthrough product, p88, which is the RNA-dependent RNA polymerase (RdRp); two movement proteins, p8 and p9, which are expressed from sgRNA1; and the 38-kDa CP, which is expressed from sgRNA2.
In this study, we identified a region immediately upstream of the TCV CP ORF that functions as an IRES both in vitro and in vivo. Deletion analyses indicated that no specific sequences within this region were required for IRES activity but rather that a stretch of about 84 A-rich unstructured bases was important. Despite a lack of sequence conservation among carmoviruses, bioinformatic analyses suggest conservation of A-rich regions upstream of their CP ORFs. eIF4G, but not eIF4E, was required for optimal CP IRES activity. In the absence of detectable sgRNA1 and sgRNA2, low levels of CP were apparently expressed from the gRNA that were sufficient for wild-type (WT) levels of gRNA accumulation in vivo. These results open the possibility that low-level IRES activity mediated by unstructured regions is more widespread throughout viral and eukaryotic genomes than previously thought.
RESULTS
Large deletions upstream of the CP ORF reduce CP synthesis in vitro.
Previous reports on HCRSV and PFBV suggested that an ∼150-nt region immediately upstream of the CP initiation codon and within the ORF for the movement protein (i.e., p9 for TCV) had IRES activity (20, 21). These studies were unable to identify sequences/structures important for internal translation in WGE assays, and what precisely comprised a carmovirus CP IRES remained unclear. The goals of the current study were to (i) determine the location of the sequence within the TCV gRNA that permitted internal initiation of CP translation, (ii) determine if this sequence had IRES activity defined as functioning out of context by use of reporter constructs, (iii) identify any critical structure or motifs within this sequence, (iv) determine whether components of eIF4F were necessary for activity, and (v) determine if CP was expressed in vivo in the absence of sgRNA1 and sgRNA2.
Typical WGE translation assays using TCV gRNA as the template yielded high levels of p28 and trace amounts of the ribosomal readthrough product, p88 (Fig. 1B, lane 1). Two additional bands of moderate intensity were observed between 35 and 40 kDa. The larger product, migrating at 38 kDa, corresponded to the CP (29), and the product at 35 kDa corresponded to translation initiating within the p88 ORF, at position 1393 (determined by altering individual AUG start codons [data not shown]). The identity of CP was confirmed by introducing an in-frame UGA stop codon after 5 amino acids within the CP ORF that abolished CP synthesis (Fig. 1B, lane CP-UGA). The level of CP expression from the gRNA in vitro was 8-fold lower than the level of expression from equimolar amounts of sgRNA2 (Fig. 1D). To rule out the possibility of RNA degradation resulting in CP expression from cleaved RNA, Northern blotting was used to demonstrate that the input RNA remained intact throughout the incubation period (Fig. 1E, compare lanes for 0 min versus 90 min).
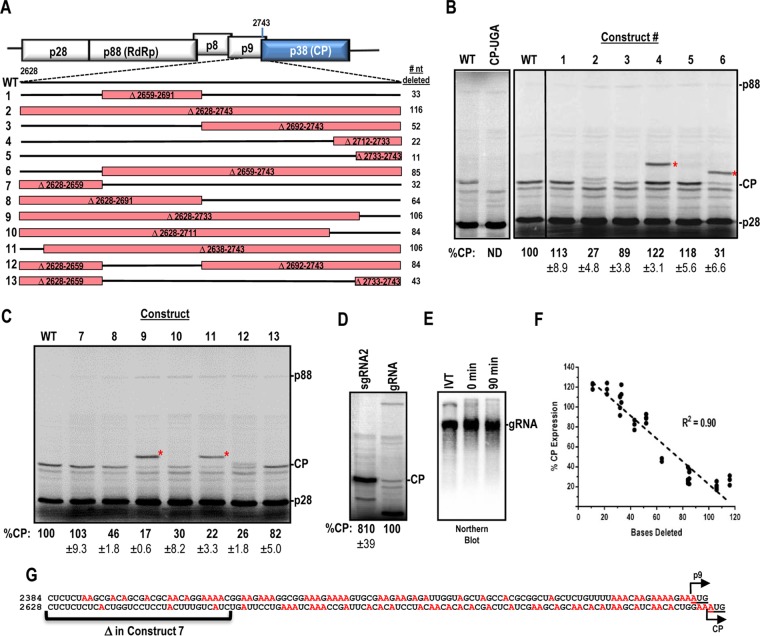
FIG 1.
Deletion analysis of a region required for internal expression of the TCV CP. (A) Deletions generated in a 116-nt region immediately upstream of the CP ORF. Deleted regions are boxed in red. Construct designations are shown to the left, and numbers of nucleotides deleted are shown to the right. (B and C) In vitro translation of constructs 1 to 13 by use of WGE. CP-UGA contains an in-frame stop codon at position 2759 in the TCV genome. Asterisks denote locations of p9-CP fusion proteins that result from internal translation initiation at the p9 ORF for some constructs. Data were normalized to p28 levels, and means and standard deviations are shown. Data are from at least three independent experiments. (D) CP expression in WGE, using equimolar amounts of sgRNA2 and gRNA. (E) Analysis of RNA integrity during the WGE assay. After 90 min of incubation in WGE, the mixture was subjected to Northern blotting using a gRNA probe. Purified RNA transcripts (IVT) and the 0-min time point were used as controls. (F) Inverse correlation between CP expression and deletion size. Normalized CP expression was plotted against the number of nucleotides deleted for each construct in panel A. Linear regression was used to determine the goodness of fit as shown by the R2 value. (G) A-rich region upstream of the p9 and CP ORFs. Adenine residues are shown in red. Initiation codons are underlined.
Deletion of a 116-nt region (positions 2628 to 2743) immediately upstream of the CP ORF (construct 2) reduced CP levels by 73% (Fig. 1A and B). Deletion of nucleotides further upstream (Δ2581-2627) had no effect on CP expression (data not shown). To identify sequences/structures within the 116-nt region that were important for internal CP translation, additional deletions were generated. Constructs 1, 3, and 7 together deleted every portion of the 116-nt region (Fig. 1A). Constructs 1 (33 nt) and 7 (32 nt) maintained nearly WT levels of CP, while the largest deletion (construct 3; 52 nt) reduced expression by 11% (Fig. 1B and C). This result suggested either that no specific sequence within the 116-nt region was responsible for internal CP translation or that redundancies existed within the region that could compensate for specific deletions.
Additional deletions of different sizes were generated next to help to differentiate between these possibilities. Construct 6, which deleted 85 of the 116 nt, reduced CP expression by 69% (Fig. 1C). Constructs containing other large contiguous deletions (constructs 9 [106 nt] and 10 [84 nt]) produced similar reductions in CP levels (83% and 70%, respectively). Construct 12, which deleted 84 nt in two segments, had a similar negative effect on CP translation (74%). Deletion of the 5′ 64 nt within the 116-nt region (construct 8) reduced CP levels by a lesser amount (54%). Deletion of 43 nt in two segments (construct 13) reduced translation by only 18%, and deletions of <25 nt just upstream of the CP start codon (constructs 4 and 5) enhanced CP expression by ∼20%. These data suggested that primary RNA sequences (and their associated structures, if any) were not important for internal CP expression in WGE. Rather, there was an inverse correlation (R2 = 0.90) between the size of the deletion and CP expression (Fig. 1F). These results coincide with an earlier study of an unstructured dicistrovirus IRES that also found that larger deletions were detrimental for translation, whereas small deletions had only modest effects (30).
Four constructs were designed such that the p9 ORF, which partially overlaps and is out of frame with the CP ORF, was now in frame with the CP ORF. Surprisingly, strong expression of the p9-CP fusion protein was obtained from internal initiation at the p9 start codon in each of these constructs (constructs 4, 6, 9, and 11) (Fig. 1B and C, asterisks). The p9-CP fusion proteins migrated at their expected sizes and were expressed at levels similar to those of CP. Inspection of the sequence immediately upstream of the CP and p9 start codons revealed that the CP 116-nt region (CP-116) and a 113-nt region upstream of p9 (p9-113) are A-rich (33 and 40%, respectively) (Fig. 1G). Since construct 7 lacked the first 32 residues of CP-116 and maintained WT levels of translation, the percentage of adenylates increased to 41% within this 84-nt segment, which is similar to the percentage for p9-113.
CP-116 is mostly unstructured in TCV.
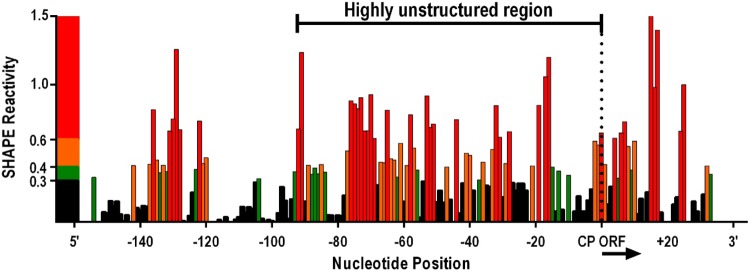
The RNA secondary structure folding algorithm tool mFold (31) predicted that the thermodynamic stability of CP-116 (ΔG = −0.81 kcal/mol) was substantially less than the stability of the upstream 116 nt (ΔG = −37.7 kcal/mol) and the downstream 116 nt (ΔG = −23.1 kcal/mol). When only the 84-nt region upstream of the CP ORF was subjected to mFold, the ΔG value decreased to −0.06 kcal/mol. To determine whether the folding prediction was correct for this region, RNA surrounding the CP translation start site was subjected to selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) RNA structure probing using full-length TCV gRNA. SHAPE uses chemical modification to determine the flexibility of individual nucleotides, as highly reactive residues are unstructured and their modification impedes primer extension by reverse transcriptase. SHAPE results indicated that over 50% of the residues in the 94-nt region immediately upstream of the CP start codon were moderately to highly flexible, suggesting that this region has few, if any, structured elements (Fig. 2). This highly unstructured region also extended 20 nt into the CP coding sequence.
FIG 2.
Structure of CP-116. SHAPE RNA structure probing was used to determine the flexibility of bases in and near CP-116 in full-length TCV gRNA. Normalized SHAPE reactivities are presented, with red, orange/green, and black representing high reactivity, moderate reactivity, and no reactivity, respectively. Band intensities from 8% denaturing polyacrylamide gels were determined using SAFA software (61). The highly unstructured region upstream of the CP ORF is indicated.
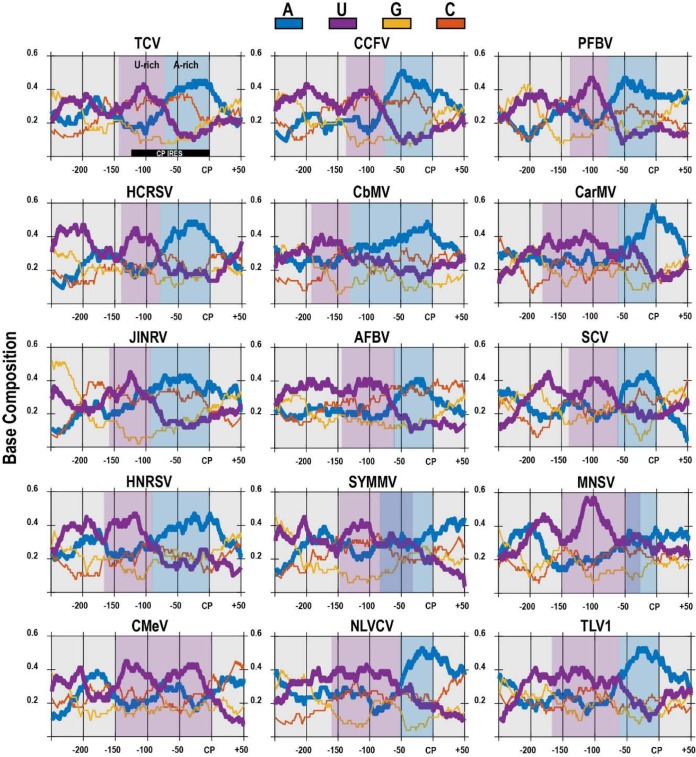
The sequence upstream of the CP ORF is poorly conserved among carmoviruses (data not shown). To determine if the base composition in the region is conserved, ntdensity sliding window analysis (50-nt window size) in the MATLAB Bioinformatics Toolbox (32) was used to analyze the base composition for 250 nt upstream and 50 nt downstream of the CP start codon (Fig. 3). For 14 of the 15 carmoviruses, a U-rich region (shaded purple) was followed by an A-rich region (shaded blue), with the A-rich region located within the ∼84-nt sequence suggested to be important for internal CP synthesis in TCV (construct 7) (Fig. 1). In addition, within these U- and A-rich regions, the level of guanylates was substantially reduced compared to that of cytidylates. Together, these results suggest that an unstructured, A-rich, guanylate-poor region may be responsible for internal initiation of CP synthesis from the gRNA.
FIG 3.
U- and A-rich regions upstream of carmovirus CP ORFs. The nucleotide density of the region 250 nt upstream to 50 nt downstream of the CP initiation site for 15 carmoviruses was determined using a 50-nt sliding window analysis. “CP” denotes the CP start codon. An A-rich region (shaded blue) was observed immediately upstream of the CP start site, whereas a U-rich region (shaded purple) was observed further upstream. An A/U-rich region is marked by overlapping blue and purple shading for SYMMV and MNSV. Viruses (accession numbers) used for this analysis were as follows: TCV (NC_003821), Cardamine chlorotic fleck virus (CCFV) (NC_001600), PFBV (NC_005286), HCRSV (NC_003608), Calibrachoa mottle virus (CbMV) (NC_021926), Carnation mottle virus (CarMV) (NC_001265), Japanese iris necrotic ring virus (JINRV) (NC_002187), Angelonia flower break virus (AFBV) (NC_007733), Saguaro cactus virus (SCV) (NC_001780), Honeysuckle ringspot virus (HNRSV) (NC_014967), Soybean yellow mottle mosaic virus (SYMMV) (NC_011643), Melon necrotic spot virus (MNSV) (NC_001504), Cowpea mottle virus (CMeV) (NC_003535), Nootka lupine vein clearing virus (NLVCV) (NC_009017), and Trailing lespedeza virus 1 (TLV1) (NC_015227).
CP-116 and p9-113 have IRES activity in heterologous reporter constructs.
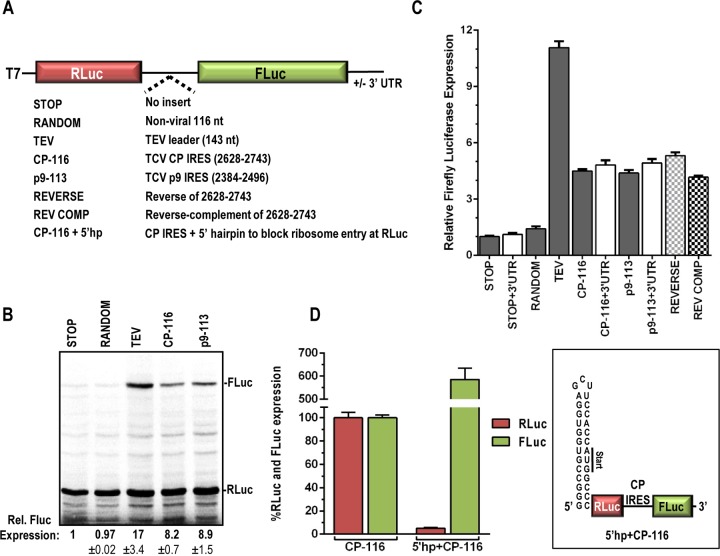
To determine if CP-116 and p9-113 are able to independently promote internal translation from the gRNA, CP-116 and p9-113 were inserted between Renilla (RLuc) and firefly (FLuc) luciferase ORFs in the presence and absence of the TCV 3′ UTR to measure IRES activity denoted by translation of FLuc (Fig. 4A). The TCV 3′ UTR was previously found to enhance translation from the 5′ end in reporter constructs in vivo, but it had no significant stimulatory effect in WGE (28). In contrast, the 3′ UTR of HCRSV enhanced the efficiency of the CP IRES in vitro (20). Additional constructs included those with (i) no insert (STOP), (ii) insertion of a 116-nt random sequence (23% A, 24% G, 26% U, and 27% C) (RANDOM), (iii) insertion of the reverse of the CP-116 sequence (REVERSE), and (iv) insertion of the reverse complement of the CP-116 sequence (REV COMP). In addition, the 143-nt leader sequence from Tobacco etch virus (TEV) was used as a positive control because it possesses strong translational enhancement and IRES activity (33, 34).
FIG 4.
Regions upstream of the CP and p9 ORFs have IRES activity in heterologous reporter constructs. (A) The TCV CP and p9 IRES were inserted between the Renilla (RLuc) and firefly (FLuc) luciferase ORFs to generate CP-116 and p9-113, respectively. Negative controls included no insert (STOP), insertion of a random 116-nt sequence (∼25% A/U/G/C; RANDOM), and insertion of the reverse of CP-116 (REVERSE) or the reverse complement (REV COMP). The TEV leader sequence was used as a positive control (TEV). (B) In vitro translation of selected reporter constructs. Data are means and standard deviations and are from two independent experiments. (C) Dual-luciferase assays were performed for at least 3 independent experiments performed in triplicate. Relative FLuc expression levels are presented as means with standard errors. White bars indicate constructs that contained the 3′ UTR of TCV. (D) The 29-base stable hairpin shown (right), incorporating an in-frame AUG, was introduced immediately upstream of RLuc in the CP-116 construct (5′hp+CP-116). RLuc and FLuc values were set at 100% for the CP-116 construct. Data are from two independent experiments with 8 replicates.
Dual-luciferase RNA transcripts were subjected to translation in WGE, and FLuc levels were determined by both gel electrophoresis and luciferase assays. TEV generated a 17-fold increase in FLuc levels compared to those with the STOP negative control in the gel assay, with an 11-fold increase in FLuc activity. In contrast, the 116-nt random sequence (RANDOM) generated background levels of FLuc (Fig. 4B and C). CP-116 and p9-113 enhanced FLuc activity 8.2- and 8.9-fold, respectively, by the gel assay and 4.5-fold by the luciferase assay (Fig. 4B and C). The presence of the TCV 3′ UTR had no significant effect on FLuc activity in vitro, which is similar to what was previously found when it was added to reporter constructs containing the TCV 5′ UTR and assayed in WGE (28). The REVERSE construct, which maintained a base composition identical to that of CP-116 but differed in its primary sequence, also generated a similar increase in FLuc activity (Fig. 4C). Interestingly, the reverse-complement sequence of CP-116 (REV COMP) had activity equivalent to that of the CP IRES as well. These results concur with earlier findings with TuMV, where the IRES reverse-complement sequence maintained activity in reporter assays (13). This suggests that an A/U-rich base composition is favorable for internal initiation, possibly due to the inherent lack of structure associated with A/U-rich sequences.
To determine if an accessible 5′ end is required for internal initiation, a stable hairpin was introduced immediately upstream of the RLuc ORF in the CP-116 construct. The hairpin was designed to include an in-frame AUG. Addition of the stable hairpin reduced RLuc expression over 20-fold while enhancing FLuc expression over 6-fold (Fig. 4D). This suggests that the CP IRES is in competition with the 5′ end for the translation machinery and does not require a free 5′ end for IRES activity. Together, these results suggest that regions immediately upstream of the CP and p9 ORFs possess IRES activity and that, at least for CP-116, the activity is independent of primary sequence and likely dependent on a lack of structure and/or base composition.
Ribosome scanning in the CP IRES.
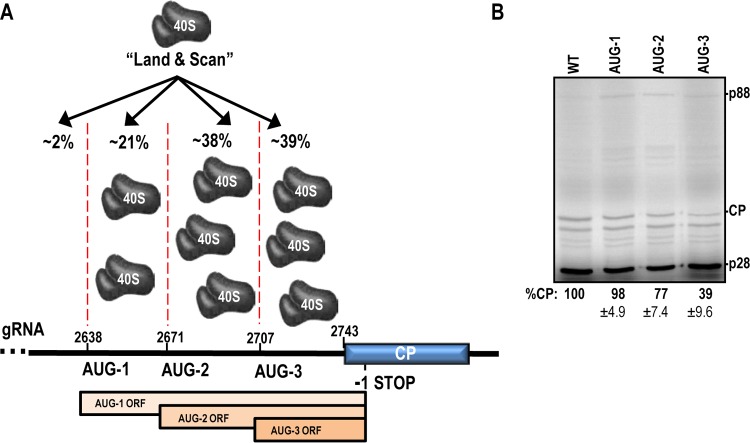
To determine if the unstructured CP IRES is used as a “landing pad” for ribosomes to enter and then scan to the CP initiation codon, −1 frame initiation codons were generated in the CP IRES region within full-length gRNA. The context of the −1 frame start codons (GAAAUGG) was designed to be identical to the strong context of the CP start codon to reduce the possibility of leaky scanning (3). Figure 5 shows that AUG-1, located 106 nt upstream of the CP start codon, had little or no effect on CP translation, suggesting that ∼98% of ribosomes enter downstream, within the more unstructured sequence. Addition of AUG-2 73 nt upstream of the CP start site decreased CP translation by 21%, suggesting that approximately 21% of ribosomes enter between 73 and 100 nt upstream of the CP ORF and then scan to the initiation codon. AUG-3, located 37 nt upstream of the initiation codon, reduced CP translation by 61%. This suggests that around 38% of ribosomes enter between 37 and 73 bases upstream of CP and that a similar number are recruited within 37 nt of the start codon. It is worth noting that translation of any of the upstream ORFs could block translation initiation within the CP IRES, making it difficult to obtain a precise value for the amount of ribosomes recruited to a particular region.
FIG 5.
Ribosome recruitment to the CP IRES. (A) The “land and scan” model of initiation suggests that 40S ribosomal subunits land upstream of the start codon before scanning in the 5′-to-3′ direction. Three AUG codons were individually introduced into the −1 frame upstream of the CP in full-length TCV genomic RNA. Translation initiating at a −1 frame AUG terminates within the CP coding region (labeled “−1 STOP”). The percentage of ribosome entry within each region is from the data in panel B. The ORFs for expression from AUG-1 to -3 are shown. (B) In vitro translation of AUG-1 to -3 mutants. CP expression data are shown as mean percentages of the WT level and standard deviations. Data from three independent experiments were normalized to p28 levels.
CP IRES activity is enhanced by addition of eIF4G.
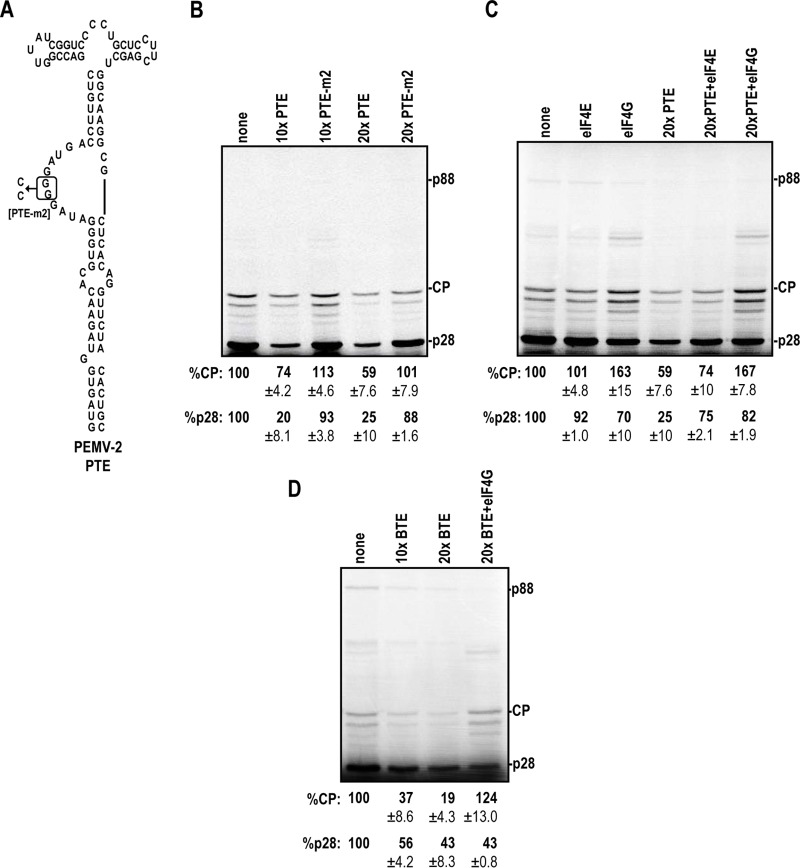
One of the 3′ CITEs of the umbravirus Pea enation mosaic virus RNA 2 (PEMV-2) is the Panicum mosaic virus-like translational enhancer (PTE), which is known to bind to eIF4E with high affinity (35) (Fig. 6A). Addition of the PTE in trans at 10× and 20× molar ratios compared to the level of TCV gRNA reduced CP expression by 26% and 41%, respectively (Fig. 6B). Alterations in the PTE guanylate-rich asymmetric loop that reduced eIF4E binding by >90% (PTE-m2) (35) eliminated the negative effect of the PTE on CP IRES activity (Fig. 6B). Addition of the PTE also reduced expression of the 5′-terminal ORF product p28.
FIG 6.
The CP IRES is stimulated by eIF4G. (A) The PEMV-2 PTE binds to eIF4E, which interacts with eIF4G when added in trans to WGE. PTE-m2 contains two point mutations that reduce eIF4E binding (35). (B) In vitro translation reactions in the presence of the PTE. The PTE was added at a 10× or 20× molar ratio to standard WGE reaction mixtures. (C) Recombinant eIF4E and eIF4G (200 pM) were added in trans in the presence or absence of 20× PTE. (D) The TNV-D BTE was added at 10× and 20× molar ratios in the absence and presence of 200 pM recombinant eIF4G. All data are from at least three independent experiments and are means and standard deviations.
eIF4E binds to the N terminus of eIF4G and is present in large excess compared to eIF4G in WGE, as assayed by Western blotting (Karen Browning, personal communication). Thus, the addition of the PTE should effectively sequester both eIF4E and eIF4G, since all eIF4G is assumed to be bound by eIF4E (36). To determine if the reduction in CP expression was due to PTE sequestration of eIF4E and/or eIF4G, recombinant Arabidopsis thaliana eIF4E and eIF4G were added to TCV gRNA in WGE either alone or in the presence of the PTE to determine the impact on translation. Addition of eIF4E to gRNA had no effect on translation of CP, whereas adding eIF4G enhanced expression of CP over WT levels by 63% (Fig. 6C). Addition of eIF4G also reduced p28 synthesis by 30%, which may reflect its heterologous origin and suboptimal interactions with other initiation factors needed for translation at the 5′ end. Addition of recombinant eIF4E in the presence of the PTE had a slight positive effect on CP expression, whereas addition of eIF4G enhanced CP levels to 167% of the WT level, which is similar to the observation for addition of eIF4G in the absence of the PTE (Fig. 6C). To directly test whether eIF4G was required for efficient internal initiation, the Tobacco necrosis virus D (TNV-D) translational enhancer (BTE), which specifically binds to eIF4G (37, 38), was added to WGE along with TCV gRNA. BTE at 10× and 20× reduced CP expression by 63% and 81%, respectively (Fig. 6D). The observed inhibition was rescued by adding recombinant eIF4G in trans, restoring CP expression to higher-than-WT levels. These results strongly suggest that eIF4G, not eIF4E, is important for CP IRES function.
Expression of TCV CP from gRNA in vivo.
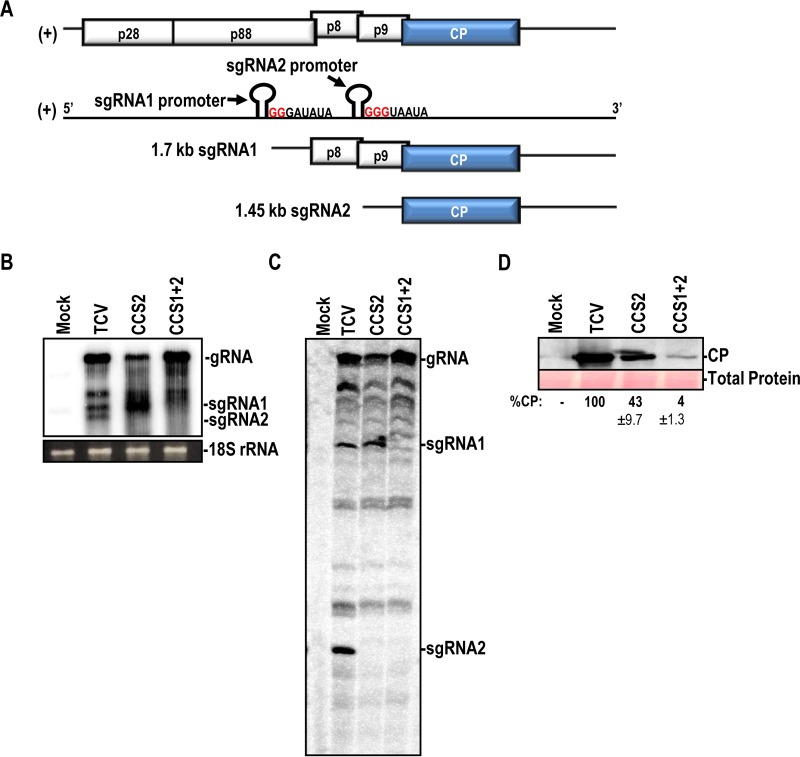
Although CPs of two carmoviruses were previously shown to be expressed from the gRNA in vitro (20, 21), no in vivo evidence of CP expression from infectious gRNA was presented. To determine if the TCV CP is expressed from gRNA in vivo, constructs were generated to prevent synthesis of sgRNA2 (CCS2) or sgRNA1 and sgRNA2 (CCS1+2) (Fig. 7A). CCS1+2 gRNA contained GG2331CC and GGG2606UUC mutations in the sgRNA1 and sgRNA2 carmovirus consensus sequence [G2–3(U/A)3–7], which is a motif present at the 5′ termini of carmovirus gRNAs and sgRNAs (Fig. 7A) (39). Twenty-four hours after inoculation of Arabidopsis protoplasts, WT and CCS1+2 gRNAs accumulated to high levels, with CCS2 gRNA accumulating to a lower level (Fig. 7B). CCS2 eliminated detection of sgRNA2, and sgRNA1 and sgRNA2 were not detected for CCS1+2, as assayed by Northern blotting (Fig. 7B). To ensure that trace amounts of sgRNA were not present, primer extension reactions were also performed using total RNA from infected cells. As shown in Fig. 7C, sgRNA primer extension products for CCS1 and CCS2 were not detected (Fig. 7C). CCS2 gRNA expressed 43% of the WT level of CP, and CCS1+2 expressed 4% of the WT CP level (Fig. 7D), suggesting that CP was being synthesized in vivo from both sgRNA1 (if present) and the gRNA. CP expression from leaky scanning of sgRNA1 has previously been observed for both PFBV and TNV-D (21, 40). Interestingly, gRNA accumulation was not affected by the CCS1+2 mutations (Fig. 7B). However, when a termination codon was introduced into the CP ORF just downstream of the initiation codon or when a mutation was generated in the CP ORF that prevented synthesis of a functional CP, gRNA accumulation decreased by about 60% in protoplasts (41, 42). We speculate that the 4% CP level observed in the absence of the sgRNAs was sufficient to allow for WT-level RNA accumulation in protoplasts.
FIG 7.
TCV CP is expressed from gRNA in vivo. (A) Genomic organization of TCV showing the locations of mutations in the carmovirus consensus sequence. CCS1 contains a GG2331CC mutation, and CCS2 contains a GGG2606UUC mutation. (B) Northern blot for detection of TCV gRNA and sgRNAs at 24 hpi. (C) Primer extension reaction of total RNA from TCV-infected protoplasts. Exposing the screen for three times longer also did not reveal any products corresponding to the sgRNAs. (D) Western blot of CP in TCV-infected protoplasts for WT and CCS mutant constructs. All images are representative of experiments performed in duplicate.
DISCUSSION
Internal initiation of translation allows for cap-independent translation of 5′-proximal ORFs and for translation of internal ORFs. For carmoviruses, translation of CP from the gRNA would allow for early synthesis of the CP silencing suppressor before its sgRNA2 template is available. We have found that a 116-nt region immediately upstream of the CP ORF has a low level of IRES activity that is sufficient for accumulation of normal levels of TCV gRNA in protoplasts. Within this 116-nt region, an unstructured sequence of about 84 nt was important for optimal IRES activity in WGE, using full-length gRNA as the template (Fig. 1). The CP IRES was unstructured as determined by SHAPE structure probing (Fig. 2), and it did not contain specific sequences or elements responsible for IRES activity. The lack of a requirement for specific sequences/structures is based on the orientation independence of CP-116 in promoting translation in bicistronic assays (both REVERSE and REV COMP) (Fig. 4) and on the absence of a major effect on CP synthesis when templates contained small deletions throughout the region. Similarly, the IRES region of TuMV was functional when the IRES was replaced by the reverse complement sequence (13). The high degree of conservation of A/U-rich and G-poor sequences upstream of the CP ORF in carmoviruses (Fig. 3) suggests that nucleotide composition may also be a factor in TCV IRES activity. However, since this sequence is also a portion of the p9 coding region, it is currently unknown whether this specific sequence composition reflects a codon requirement.
Earlier work using the carmovirus HCRSV showed that the 5′ and 3′ halves of the CP IRES maintained nearly WT levels of activity (20). These results can now be explained by our finding that unstructured regions of suitable length can function as IRES. In HCRSV, the 3′ UTR enhanced CP IRES activity 4-fold in bicistronic assays, and this was dependent on a 6-nt sequence within the 3′ UTR (20). HCRSV has a PTE-type 3′ CITE (43) that has been proposed to engage in a long-distance interaction with the 5′-end sequence (15), and the 6-nt sequence is in the required internal asymmetric loop that binds to eIF4E. The carmovirus PFBV also has a PTE-type 3′ CITE (43); however, there was no effect of the PFBV 3′ UTR on CP IRES activity in vitro (21). Since 3′ CITEs may not function out of context in reporter assays in vitro, despite having demonstrated importance in vivo (44), lack of involvement of the 3′ UTR in vitro does not necessarily reflect a lack of importance in vivo. In the present study, the TCV 3′ UTR also had no synergistic effect on CP IRES activity in reporter assays. Interestingly, the same CP IRES sequence within an sgNRA2 template had synergistic interactions with the 3′ UTR (45). However, we observed a lack of interplay between the TCV gRNA 5′ UTR and 3′ UTR when we used luciferase reporter constructs in WGE (28). Thus, it remains open whether the TCV 3′ UTR has a beneficial effect on the CP IRES in vivo.
For HCRSV and PFBV, small segments of RNA primary sequence were important for CP IRES activity. In HCRSV, an 18-nt sequence in a predicted stem-loop within the 5′ half of the CP IRES was required for maximum activity, as substitutions (including compensatory mutations) resulted in a 5-fold decrease in CP expression (20). Base substitutions within a small hairpin in the computer-predicted structure of the PFBV CP IRES reduced CP levels by 35% and also were not compensatory, leading the authors to suggest that the primary sequence was important for activity (21). In contrast, every residue of the CP-116 sequence could be deleted, with small deletions (<52 nt) causing a <11% drop in CP expression. The CP-116 sequence could also be reversed or reverse complemented and retain WT levels of activity, confirming that the primary sequence is not important. Since no specific sequence is required for CP IRES activity, there is no pairing with 18S rRNA through a Shine-Dalgarno-like sequence as has been suggested for some picornaviral IRES (46). Our results strongly suggest that the CP IRES functions by maintaining unstructured regions of RNA that can initiate low levels of translation by acting as a landing pad for ribosomes followed by scanning to the initiation codon (Fig. 5).
When present in modified gRNA dual-reporter constructs, HCRSV and PFBV CP IRES were active in mammalian cells and when agroinfiltrated into plants, respectively, as deletions within the IRES led to reduced expression of the 3′-proximal reporter gene. Conservative substitutions within the CP IRES in PFBV also led to a 20-fold reduction in lesions on inoculated leaves, suggesting that the IRES is required for optimal infectivity in vivo and highlighting the biological significance of low-level IRES activity (21). In the current study, the carmovirus consensus sequence for both sgRNAs was mutated such that no detectable sgRNA1 or sgRNA2 was observed during infection. Thus, the small amount of CP present (4% of the WT level) was likely expressed from the gRNA (Fig. 7). Interestingly, despite only 4% expression of CP, TCV gRNA accumulated to WT levels, suggesting that WT-level gRNA accumulation is possible when a low level of CP is expressed from the gRNA. TCV gRNA normally accumulates to rRNA levels after 24 h postinoculation (hpi). Thus, 4% expression of CP from the extraordinary amount of gRNA results in high levels of CP even in the absence of sgRNAs. Altering only sgRNA2 (CCS2) led to increased synthesis of sgRNA1, as found previously (47). Whether this increase was responsible for the decrease in TCV accumulation that was observed in protoplasts is not known.
The short, unstructured CP IRES of TCV shares characteristics with some potyviral IRES located at the 5′ end of the gRNA. For example, the TEV leader is 143 nt long and A-rich, and it lacks substantial amounts of guanylates (43% A and 8% G). The TCV CP IRES shares similar attributes, with a length of ∼84 nt (from construct 7) (Fig. 1) and a base composition of 41% A and 10% G. The TEV leader has a weak predicted secondary structure (ΔG = −5.69 kcal/mol), as does the 84-nt TCV region (−0.06 kcal/mol). It is interesting that the TCV CP IRES serves a triple function that also includes being a portion of the leader sequence for sgRNA2 and the coding sequence for p9.
Internal initiation of translation was also observed for the p9 movement protein in vitro, and the 113-nt region upstream of p9 possessed IRES activity in bicistronic reporter assays. The TCV p9 IRES lies within the coding region for the upstream p8 movement protein, and p9 is thought to be expressed only by leaky scanning of sgRNA1 when the start codon of p8 is bypassed (48). It is possible that internal initiation of p9 may complement leaky scanning in the context of either gRNA or sgRNA1.
The roles of any initiation factors in CP IRES activity had not previously been defined. In this study, the roles of eIF4E and eIF4G were examined. eIF4G was necessary for high-level expression, since CP levels were reduced by over 80% in the presence of the eIF4G-sequestering BTE from TNV-D (Fig. 6). These results support earlier findings showing that TCV replication was inefficient in Arabidopsis protoplasts lacking eIF4G, potentially due to the observed decrease in CP production (49). eIF4E was dispensable, as the PEMV-2 PTE added in trans was deleterious for translation only in the absence of added eIF4G (Fig. 6). Similarly, eIF4G binds to the TEV leader and is the only eIF required for translation of the TEV genome (50, 51). There are two isoforms of eIF4G in plants, and while the second isoform, eIFiso4G, was not tested, previous studies have shown that eIFiso4G preferentially enhances translation of capped mRNAs and is not involved in noncanonical translation initiation (50, 52). Further studies using depleted extracts will be required to determine the role of additional initiation factors in CP IRES activity.
The short, unstructured IRES characterized in this study may represent a cross-kingdom mechanism for low-level translation initiation. IRES-directed translation is an alternative strategy used by some cellular mRNAs during stress or cell cycle progression, albeit at <2% activity compared to that of cap-dependent translation (53, 54). As with the CP IRES, cellular mRNA IRES are generally short and unstructured (55) and serve as the leader sequence of the mRNA, much like the CP IRES is the leader of sgRNA2. In support of this, the crTMV CP IRES displayed cross-kingdom activity in plant, yeast, and human cell-based assays (16).
In conclusion, we have characterized the CP IRES of TCV and found a remarkable lack of sequence and structural requirements for activity. Interestingly, a recent analysis of >100 positive-sense single-stranded RNA virus gRNAs found short sequences (174 nt) with IRES activity throughout the coding regions of uncapped viruses (56). Thus, simple, unstructured IRES have the potential to add an additional level of translational control for viral genomes that are hampered by limited coding potential.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
All TCV constructs used in this study were derived from pTCV66, which contains a cDNA copy of the TCVm isolate downstream of a T7 promoter (57). Site-directed mutagenesis was performed using Q5 DNA polymerase (New England BioLabs) and partially overlapping primers as previously described (58). All constructs were propagated in Escherichia coli DH5α, and all mutations were confirmed by sequencing.
In vitro translation using WGE.
RNA was synthesized from SmaI-linearized TCV plasmids by use of bacteriophage T7 RNA polymerase. Five microliters of WGE (Promega) was programmed with 1 μg of in vitro-synthesized RNA in a 10-μl reaction volume. After 90 min, translation reaction mixtures were resolved by 10% SDS-PAGE. Gels were dried and exposed to a phosphor screen, and [35S]Met-labeled proteins were visualized by autoradiography. CP band intensities were quantified using Multi Gauge software (Fujifilm) and normalized to the levels of p28, unless noted otherwise.
Dual-luciferase assays.
The construct p2Luc, which contains a multiple-cloning site between Renilla and firefly luciferase genes, has been described previously (59). For all constructs, the corresponding IRES region was amplified by PCR and cloned into SalI and BamHI restriction sites located between the two luciferase reporter ORFs. The TCV 3′ UTR was amplified by PCR and inserted immediately downstream of the firefly luciferase gene by use of PmlI and EagI restriction sites. The REVERSE and REV COMP sequences of CP-116 (engineered to lack start codons) were synthesized commercially (Integrated DNA Technologies) and inserted into the SalI and BamHI restriction sites of p2Luc. Plasmids lacking the TCV 3′ UTR were linearized with PmlI, and plasmids containing the 3′ UTR were linearized with EagI. All constructs were confirmed by sequencing of the region of interest. For each assay, in vitro translation reactions were performed as described above, and the reaction mixtures were diluted 1:20 in 1× phosphate-buffered saline (PBS) after 90 min. Twenty microliters of each reaction mixture was analyzed by using a dual-luciferase assay system (Promega) following the manufacturer's protocol and a Modulus microplate multimode reader (Turner Biosystems).
SHAPE RNA structure probing.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) was performed using previously described methods (60). Briefly, full-length TCV RNA transcripts were modified with either dimethyl sulfoxide (DMSO) (negative control) or N-methylisatoic anhydride (NMIA) for 40 min and then subjected to reverse transcription using Superscript III (Invitrogen) and 32P-labeled oligonucleotides complementary to TCV positions 2819 to 2842 and 2753 to 2775. Primer extension products were resolved by 8% denaturing PAGE and visualized using a Typhoon phosphorimager (GE Healthcare). Band intensities were determined using semiautomated footprinting analysis (SAFA) software (61). For each position, the signal of the DMSO reaction was subtracted from that of the NMIA reaction, and net reactivities were ranked. The top 1% were discarded as outliers, and the net reactivity was normalized to the top 10% of highest-intensity values. Bases covered by both oligonucleotides differed by <0.2 SHAPE reactivity units.
Transfection of Arabidopsis protoplasts and blotting.
Protoplasts derived from Arabidopsis thaliana (Col-0 strain) calluses were used for transfections as previously described (62). Briefly, 15 μg of TCV gRNA was transfected into 7 × 106 cells in a 60-mm petri dish by use of polyethylene glycol. Samples were placed in the dark at 22°C for 24 h and then divided for Northern and Western blotting. Total RNA was isolated using RNA extraction buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 100 mM NaCl, 1% SDS), followed by phenol-chloroform extraction and ethanol precipitation. For Northern blotting, 1 μg of total RNA was resolved in a 1.2% nondenaturing agarose gel and transferred to a nylon membrane by capillary transfer. Three different [α-32P]dCTP-labeled DNA probes targeting the subgenomic portion of the TCV genome were used for hybridization at 50°C. Primer extension reactions were performed using SuperScript III (Invitrogen) and 1 μg of total RNA with a reverse primer binding at position 2711 of the TCV genome. Blots were imaged using a FLA-5100 fluorescence image analyzer (Fujifilm). For Western blotting, total protein lysates were isolated by resuspending 3.5 × 106 cells in protein analysis buffer (30 mM Tris-HCl [pH 6.8], 5% glycerol, 2.5% β-mercaptoethanol, 1.5% SDS). Lysates were resolved by 10% SDS-PAGE. A semidry transfer method was used to transfer proteins to a 0.45-μm nitrocellulose membrane by use of a Trans-Blot SD apparatus (Bio-Rad Laboratories). Total protein was stained using 0.2% (wt/vol) Ponceau S and imaged for loading controls. After blocking, a primary polyclonal antibody (rabbit anti-CP) was used at a 1:2,000 dilution. A horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody was used at a 1:10,000 dilution. Enhanced chemiluminescence was detected using the SuperSignal West Pico substrate (Thermo Scientific).
ACKNOWLEDGMENTS
We thank all members of the Simon lab for helpful discussions and Karen Browning for eIF4E and eIF4G.
This work was supported by an award from the National Institutes of Health to J.M. (grant F32GM119235) and by awards from the National Institutes of Health (grant R21AI117882) and the National Science Foundation (grant MCB-1411836) to A.E.S.
REFERENCES
- 1.Haghighat A, Sonenberg N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem 272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 4.Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson RJ. 2005. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 33:1231–1241. [DOI] [PubMed] [Google Scholar]
- 6.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 8.Jackson RJ, Kaminski A. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 9.Filbin ME, Kieft JS. 2009. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieft JS. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Roberts R, Rakotondrafara AM. 2015. The role of the 5′ untranslated regions of Potyviridae in translation. Virus Res 206:74–81. doi: 10.1016/j.virusres.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Zeenko V, Gallie DR. 2005. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem 280:26813–26824. doi: 10.1074/jbc.M503576200. [DOI] [PubMed] [Google Scholar]
- 13.Basso J, Dallaire P, Charest PJ, Devantier Y, Laliberte JF. 1994. Evidence for an internal ribosome entry site within the 5′ non-translated region of turnip mosaic potyvirus RNA. J Gen Virol 75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- 14.Gallie DR, Tanguay RL, Leathers V. 1995. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene 165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 15.Simon AE, Miller WA. 2013. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorokhov YL, Skulachev MV, Ivanov PA, Zvereva SD, Tjulkina LG, Merits A, Gleba YY, Hohn T, Atabekov JG. 2002. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci U S A 99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorokhov YL, Ivanov PA, Komarova TV, Skulachev MV, Atabekov JG. 2006. An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. J Gen Virol 87:2693–2697. doi: 10.1099/vir.0.82095-0. [DOI] [PubMed] [Google Scholar]
- 18.Qu F, Ren T, Morris TJ. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol 77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas CL, Leh V, Lederer C, Maule AJ. 2003. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33–41. doi: 10.1016/S0042-6822(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 20.Koh DC, Wong SM, Liu DX. 2003. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J Biol Chem 278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Miragall O, Hernandez C. 2011. An internal ribosome entry site directs translation of the 3′-gene from Pelargonium flower break virus genomic RNA: implications for infectivity. PLoS One 6:e22617. doi: 10.1371/journal.pone.0022617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merai Z, Kerenyi Z, Kertesz S, Magna M, Lakatos L, Silhavy D. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J Virol 80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, Voinnet O. 2010. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev 24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Zhang X, Singh J, Li D, Qu F. 2012. Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J Virol 86:6847–6854. doi: 10.1128/JVI.00497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 26.Baumberger N, Baulcombe DC. 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A 102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stupina VA, Yuan X, Meskauskas A, Dinman JD, Simon AE. 2011. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J Virol 85:4638–4653. doi: 10.1128/JVI.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhlmann MM, Chattopadhyay M, Stupina VA, Gao F, Simon AE. 2016. An RNA element that facilitates programmed ribosomal readthrough in Turnip crinkle virus adopts multiple conformations. J Virol 90:8575–8591. doi: 10.1128/JVI.01129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terenin IM, Dmitriev SE, Andreev DE, Royall E, Belsham GJ, Roberts LO, Shatsky IN. 2005. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol Cell Biol 25:7879–7888. doi: 10.1128/MCB.25.17.7879-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henson R, Cetto L. 2005. The MATLAB bioinformatics toolbox, p 4:4.8:105 In Jorde L, Little P, Dunn M, Subramaniam S (ed), Encyclopedia of genetics, genomics, proteomics and bioinformatics. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 33.Carrington JC, Freed DD. 1990. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol 64:1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niepel M, Gallie DR. 1999. Identification and characterization of the functional elements within the tobacco etch virus 5′ leader required for cap-independent translation. J Virol 73:9080–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Treder K, Miller WA. 2009. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem 284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarun SZ Jr, Sachs AB. 1997. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol 17:6876–6886. doi: 10.1128/MCB.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen R, Miller WA. 2004. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol 78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraft JJ, Treder K, Peterson MS, Miller WA. 2013. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res 41:3398–3413. doi: 10.1093/nar/gkt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan HC, Carpenter CD, Simon AE. 2000. Requirement of a 5′-proximal linear sequence on minus strands for plus-strand synthesis of a satellite RNA associated with turnip crinkle virus. Virology 268:355–363. doi: 10.1006/viro.1999.0154. [DOI] [PubMed] [Google Scholar]
- 40.Chkuaseli T, Newburn LR, Bakhshinyan D, White KA. 2015. Protein expression strategies in Tobacco necrosis virus-D. Virology 486:54–62. doi: 10.1016/j.virol.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 41.Manfre AJ, Simon AE. 2008. Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology 379:161–167. doi: 10.1016/j.virol.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Wu B, Oliveri S, Mandic J, White KA. 2010. Evidence for a premature termination mechanism of subgenomic mRNA transcription in a carmovirus. J Virol 84:7904–7907. doi: 10.1128/JVI.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chattopadhyay M, Shi K, Yuan X, Simon AE. 2011. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of Saguaro cactus virus. Virology 417:113–125. doi: 10.1016/j.virol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cimino PA, Nicholson BL, Wu B, Xu W, White KA. 2011. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog 7:e1002423. doi: 10.1371/journal.ppat.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu F, Morris TJ. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J Virol 74:1085–1093. doi: 10.1128/JVI.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheper GC, Voorma HO, Thomas AA. 1994. Basepairing with 18S ribosomal RNA in internal initiation of translation. FEBS Lett 352:271–275. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Carpenter CD, Simon AE. 1999. Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology 253:327–336. doi: 10.1006/viro.1998.9538. [DOI] [PubMed] [Google Scholar]
- 48.Li W-Z, Qu F, Morris TJ. 1998. Cell-to-cell movement of Turnip crinkle virus is controlled by two small open reading frames that function in trans. Virology 244:405–416. doi: 10.1006/viro.1998.9125. [DOI] [PubMed] [Google Scholar]
- 49.Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. 2004. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallie DR. 2001. Cap-independent translation conferred by the 5′ leader of Tobacco etch virus is eukaryotic initiation factor 4G dependent. J Virol 75:12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray S, Yumak H, Domashevskiy A, Khan MA, Gallie DR, Goss DJ. 2006. Tobacco etch virus mRNA preferentially binds wheat germ eukaryotic initiation factor (eIF) 4G rather than eIFiso4G. J Biol Chem 281:35826–35834. doi: 10.1074/jbc.M605762200. [DOI] [PubMed] [Google Scholar]
- 52.Gallie DR, Browning KS. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- 53.Qin X, Sarnow P. 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 54.Jackson RJ. 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 5:a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia X, Holcik M. 2009. Strong eukaryotic IRESs have weak secondary structure. PLoS One 4:e4136. doi: 10.1371/journal.pone.0004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 57.Oh JW, Kong Q, Song C, Carpenter CD, Simon AE. 1995. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol Plant Microbe Interact 8:979–987. doi: 10.1094/MPMI-8-0979. [DOI] [PubMed] [Google Scholar]
- 58.Liu H, Naismith JH. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 61.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCormack JC, Simon AE. 2006. Callus cultures of Arabidopsis. Curr Protoc Microbiol Chapter 16:Unit16D.11. doi: 10.1002/9780471729259.mc16d01s00. [DOI] [PubMed] [Google Scholar]