ABSTRACT
Despite the recent success of newly developed direct-acting antivirals against hepatitis C, the disease continues to be a global health threat due to the lack of diagnosis of most carriers and the high cost of treatment. The heterodimer formed by glycoproteins E1 and E2 within the hepatitis C virus (HCV) lipid envelope is a potential vaccine candidate and antiviral target. While the structure of E1/E2 has not yet been resolved, partial crystal structures of the E1 and E2 ectodomains have been determined. The unresolved parts of the structure are within the realm of what can be modeled with current computational modeling tools. Furthermore, a variety of additional experimental data is available to support computational predictions of E1/E2 structure, such as data from antibody binding studies, cryo-electron microscopy (cryo-EM), mutational analyses, peptide binding analysis, linker-scanning mutagenesis, and nuclear magnetic resonance (NMR) studies. In accordance with these rich experimental data, we have built an in silico model of the full-length E1/E2 heterodimer. Our model supports that E1/E2 assembles into a trimer, which was previously suggested from a study by Falson and coworkers (P. Falson, B. Bartosch, K. Alsaleh, B. A. Tews, A. Loquet, Y. Ciczora, L. Riva, C. Montigny, C. Montpellier, G. Duverlie, E. I. Pecheur, M. le Maire, F. L. Cosset, J. Dubuisson, and F. Penin, J. Virol. 89:10333–10346, 2015, https://doi.org/10.1128/JVI.00991-15). Size exclusion chromatography and Western blotting data obtained by using purified recombinant E1/E2 support our hypothesis. Our model suggests that during virus assembly, the trimer of E1/E2 may be further assembled into a pentamer, with 12 pentamers comprising a single HCV virion. We anticipate that this new model will provide a useful framework for HCV envelope structure and the development of antiviral strategies.
IMPORTANCE One hundred fifty million people have been estimated to be infected with hepatitis C virus, and many more are at risk for infection. A better understanding of the structure of the HCV envelope, which is responsible for attachment and fusion, could aid in the development of a vaccine and/or new treatments for this disease. We draw upon computational techniques to predict a full-length model of the E1/E2 heterodimer based on the partial crystal structures of the envelope glycoproteins E1 and E2. E1/E2 has been widely studied experimentally, and this provides valuable data, which has assisted us in our modeling. Our proposed structure is used to suggest the organization of the HCV envelope. We also present new experimental data from size exclusion chromatography that support our computational prediction of a trimeric oligomeric state of E1/E2.
KEYWORDS: hepatitis C virus, E1/E2 heterodimer, computational structural biology, Rosetta program, Martini force field
INTRODUCTION
Biomedical advances have rendered hepatitis C virus (HCV) infection highly treatable, and yet the disease remains a global health threat due to a lack of diagnosis of carriers and the high cost of treatment. One hundred fifty million people have been estimated to be infected with the virus, and many more are at risk for infection (http://www.who.int/mediacentre/factsheets/fs164/en/). A better understanding of the HCV envelope, which is responsible for hepatocyte attachment and fusion, could aid in the development of a vaccine and/or new treatments for this disease.
The HCV virion consists of a positive-sense single-stranded RNA molecule surrounded by a capsid composed of the core protein. This nucleocapsid structure is enveloped by two glycoproteins, E1 and E2, which are embedded in a host-derived lipid membrane (reviewed in reference 1). There is also an associated layer of serum apolipoproteins outside the HCV virion, which is believed to facilitate viral entry and secretion and which gives HCV a lower buoyant density than those of other viruses (2–4). The structural proteins that comprise the HCV virion are core, E1, and E2. Additional proteins encoded by the viral RNA genome include p7 (whose presence in the virion is disputed), six nonstructural proteins (5), and a possible F protein (encoded by an overlapping reading frame) (6). E1 and E2 comprise amino acid residues 192 to 383 and 384 to 746, respectively, of the genotype 1a H77 viral polyprotein (7).
In the viral life cycle, E1/E2 is known to have least two major roles. First, E1/E2 is responsible for viral entry via interactions with several cellular receptors, including CD81 (8). Second, E1/E2 facilitates fusion with the endosomal membrane once the virus has been internalized by clathrin-mediated endocytosis (8).
It has been demonstrated that a large component of the neutralizing antibody (Ab) response in HCV-infected patients is directed toward epitopes that interfere with the interaction of E1/E2 and CD81 (9, 10). A number of binding sites on E1/E2 for neutralizing monoclonal antibodies have been identified based on epitope mapping and structural studies. These sites have been independently classified as domains A to E (11–16), epitopes I and II (10), or antigenic regions 1 to 5 (17, 18), where some binding sites belonging to the three different sets are practically identical. An improved understanding of the structure of E1/E2 could be valuable in enabling the development of a vaccine against HCV. For example, the AR4A and AR5A monoclonal antibodies (although not prevalent in infected patients) are very broadly neutralizing and potent (18). Importantly, the AR4A and AR5A epitopes lie at the E1/E2 interface and require residues of both glycoproteins for their binding. Increased structural insight into E1/E2 contact sites may provide an opportunity for the generation of envelope proteins with improved immunogenicity. In addition, antiviral drugs could be designed to inhibit cell entry or fusion by targeting E1/E2.
Partial crystal structures of the E1 and E2 ectodomains have been determined (19, 20), although the proteins' transmembrane regions and multiple N-glycosylation sites (5 on E1 and 11 on E2) have impeded crystallization of the full-length proteins and of the heterodimer. These ectodomain crystal structures reveal the first 79 residues of the 192 residues belonging to E1 and the first 262 residues of the 363-residue E2 protein, with some flexible loops and N-terminal hypervariable domain 1 of E2 omitted. The two existing partial crystal structures of E2, reported under PDB accession numbers 4MWF (19) and 4WEB (21), determined for the two different viral genotypes H77 and J6, respectively, are quite similar to one another (root mean square deviation [RMSD] of 3.9 Å), with the exception of the disulfide-bonding network. The structures of the remaining parts of E1 and E2, the structure of the heterodimer, and the higher-order oligomeric interactions of E1 and E2 are unknown, although many experimental studies have revealed important pieces of information. Previously, homologies between HCV E1 and E2 and class II fusion proteins were used to create models of both the E2 structure (22, 23) and the E1/E2 heterodimeric structure (24). However, publication of the crystal structures revealed that these models were poor representations of the E2 and E1/E2 structures, since the crystal structures were too compact to make up class II fusion proteins. Using the partial crystal structures of E1 and E2 and other experimental data, we have applied computational methods to obtain a new model of the remaining parts of E1 and E2. Our model provides insights into how the glycoproteins may interact as a single heterodimer and how they may assemble into a higher-order oligomer.
Our computational approach makes use of the Rosetta computer program (25) for ab initio structure prediction and for docking. Rosetta employs a combination of knowledge-based and physics-based energy functions and an efficient Monte Carlo sampling protocol. Side-chain conformations are optimized by using the Dunbrack rotamer library (26). Rosetta can accurately predict structures of small, globular, soluble proteins or of small simple membrane proteins containing up to 100 residues (27). Moreover, during modeling in Rosetta, known portions of a structure can be held rigid while extensions are folded, a useful feature for problems such as the one that we address here, where a protein has been partially crystallized. The numbers of residues that are absent in the crystallized E1 construct (20) are 11 in a missing loop, 79 in the C-terminal part of the ectodomain (including stem residues), and 34 in the transmembrane helix. The residues missing from the E2 crystal structure reported under PDB accession number 4MWF (19) include 37 residues at the N terminus, a 39-residue loop and a few smaller loops, 74 C-terminal residues in the ectodomain and stem, and 27 residues in the transmembrane helix. Thus, the sizes of the missing regions of our target proteins are within the current limits of potential ab initio structure prediction in Rosetta.
When using Rosetta ab initio modeling, a model is selected based on the scoring of multiple decoys, and this selection process can be further assisted by available experimental structural data (27). Here, we make use of knowledge of the residues on both E1 and E2 needed for binding to the AR4A and AR5A antibodies (18); that is, structural models in which amino acids implicated in the epitopes for these antibodies were positioned very distant from each other were discounted. Although such residues identified via alanine scanning mutagenesis approaches do not always identify the correct binding residues (17, 19), we considered it likely that they would nonetheless be spatially close. We also use the fact that the transmembrane domains (TMDs) of both E1 and E2 are thought to consist of single-domain helices (28, 29), and we run Gromacs (30, 31) simulations to assist in assigning the relative orientations of these TMDs. Moreover, we make the assumption that residues C652 and C677 are disulfide bonded, as was previously hypothesized through experimental data (32).
We show that the shape of our predicted heterodimer is consistent with the formation of a trimeric arrangement, as was previously suggested by the finding from SDS-PAGE analysis that E1 forms trimers in the presence of E2 (33). We present experimental results demonstrating that this predicted trimeric state agrees with data obtained by size exclusion chromatography (SEC). We also present predictions based on in silico alanine scanning of residues involved in interactions between E1 and E2 that are important for heterodimerization.
RESULTS
Models of the N termini of E1 and E2.
We first established complete models of the ectodomains of the N terminus of E1 (nE1) and nE2 (residues 192 to 270 and 384 to 645, respectively), i.e., those parts for which partial crystal structures reported under PDB accession numbers 4UOI (20) and 4MWF (19), respectively, were obtained.
(i) Prediction of missing loops and the N terminus of the E2 crystal structure.
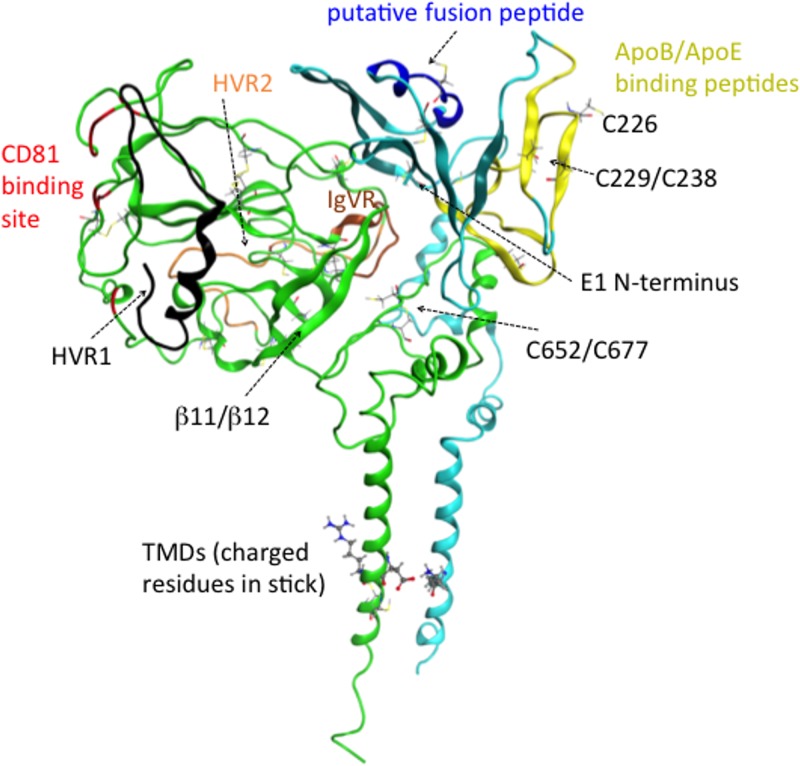
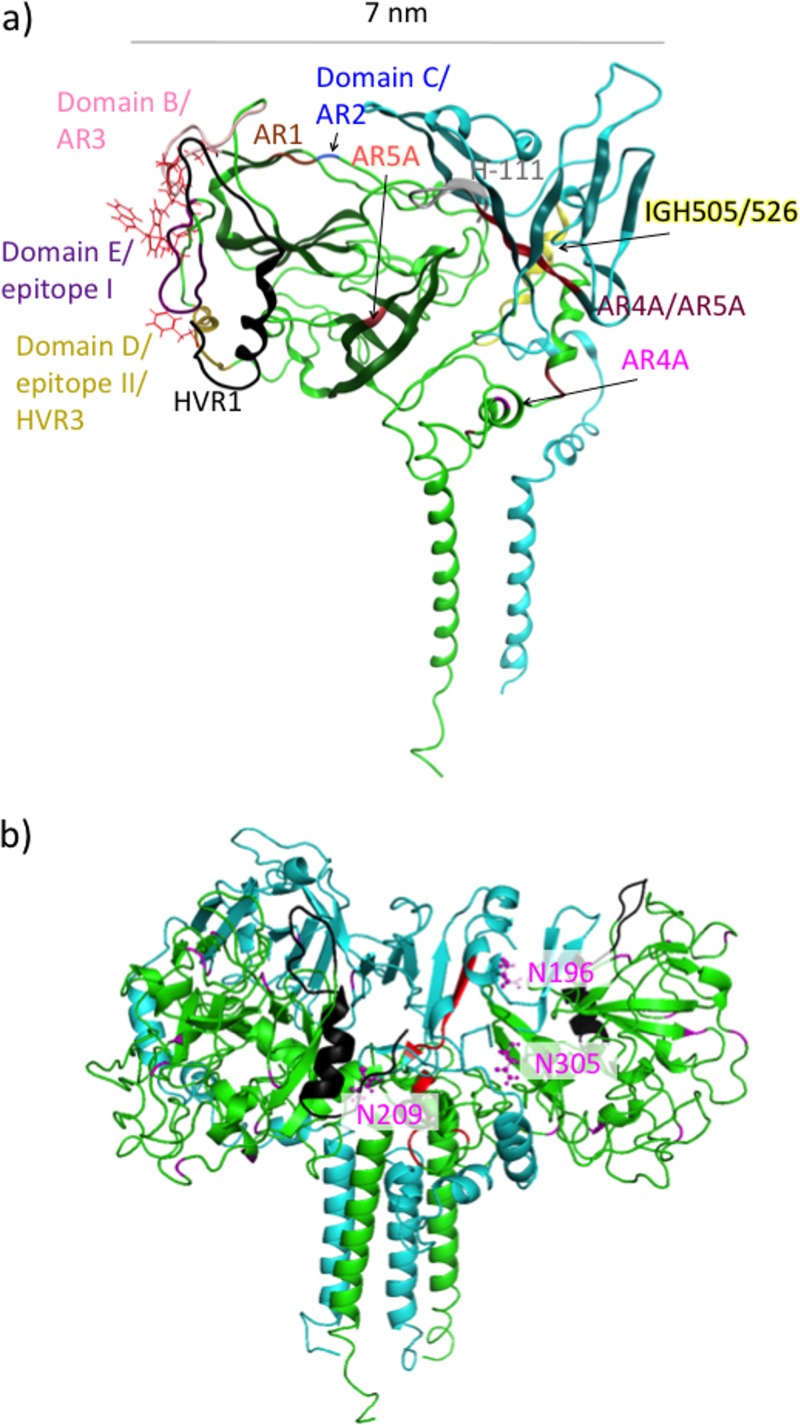
We based our structure of nE2 on the crystal structure reported under PDB accession number 4MWF (19). This crystal structure has two surfaces, one of which is more basic and contains a central β-sandwich forming an Ig-fold motif and the other of which is more hydrophobic and contains a second β-sheet roughly parallel to the first one that incorporates the lengthy β11/β12 strands (19, 21). We computationally predicted hypervariable region 1 (HVR1) (residues 384 to 411), HVR2 (residues 453 to 491), and part of the intergenomic variable region (IgVR) (residues 574 to 577), which are missing from the crystal structure; in addition, residues 412 to 420 and 586 to 596, which are missing, were also added. In our model, HVR1 and HVR2 are located on opposite sides of the CD81 binding site, and the loop corresponding to the part of IgVR that is missing from the crystal structure is positioned close to HVR2 (Fig. 1). Despite being absent in the crystal structures, the positions of these hypervariable regions were observed in negative-stain cryo-electron microscopy (cryo-EM) images of the E2 ectodomain bound to Fab AR3C (19), and these positions appear similar to those based on the predictions from our model.
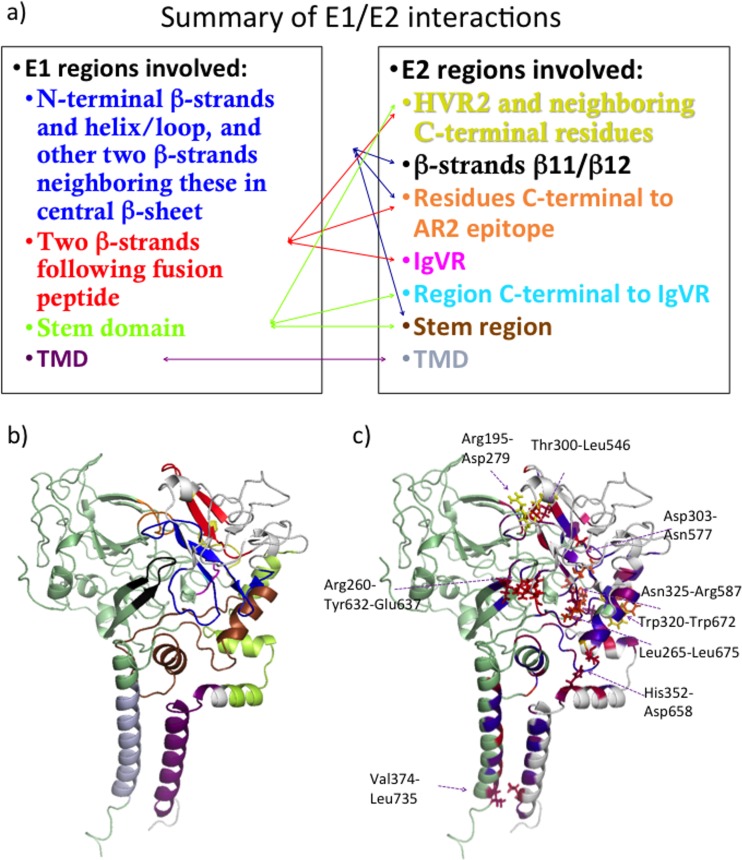
FIG 1.
Ribbon representation of our computational model of the E1/E2 heterodimer, with E1 in teal and E2 in green and with some of the regions discussed in the text highlighted by using color and labeling. Residues important for CD81 binding are shown in red, HVR1 is in black, HVR2 is in orange, IgVR is in brown, the ApoB/ApoE binding peptides are in yellow, and the putative fusion peptide is in dark blue. All cysteine residues are shown in a line representation.
(ii) Homology modeling of E1.
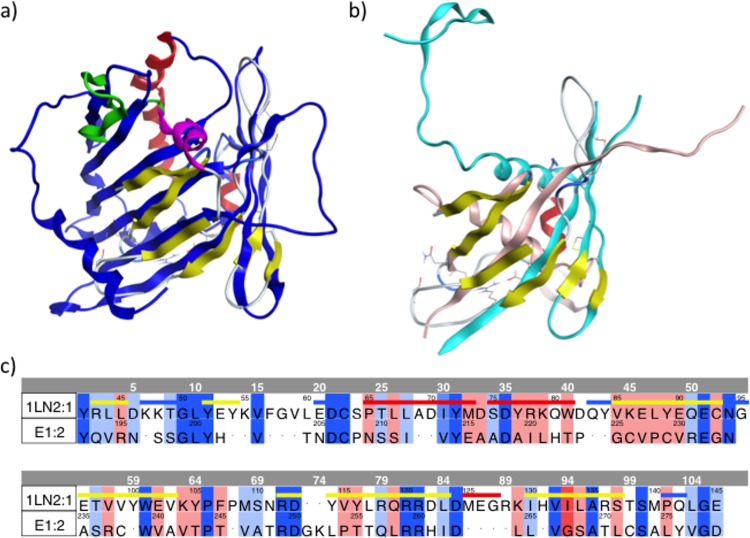
The prediction of the structure of the E1 ectodomain is complicated by the likelihood of domain swapping involving the N terminus of the crystal structure of nE1 reported under PDB accession number 4UOI (20). In the crystal structure, a central α-helix is flanked by a connected three-stranded β-sheet; the flexible region that could not be assigned is situated between the last two strands, β4 and β5, of this β-sheet. At the N-terminal end of this α-helix is the protein's N terminus, comprising a β-hairpin, which extends at a right angle from the central α-helix in a position that seems unlikely to be native. It was therefore suggested that the native E1 monomer may adopt a structure similar to that of a structural homologue that was identified for the crystallized E1 dimer, namely, phosphatidylcholine transfer protein (PDB accession number 1LN2 [34]) (20). In this proposed structure, the N terminus is suggested to fold back, forming a 5-stranded β-helix together with the three other β-strands (20). We therefore examined the sequence homology between E1 and this structural homologue and found that it extends approximately as far as the sequence of the crystallized construct. Based on our alignment between nE1 and the sequence reported under PDB accession number 1LN2, which is shown color-coded by similarity in Fig. 2c, we created a homology model for nE1 structured as a six-stranded β-sheet (Fig. 2a and b). The overall values for similarity and identity are, respectively, 45% and 26%. We note that a portion of the sequence of E1, namely, residues 219 to 230, completely lacks homology to the sequence alignment reported under PDB accession number 1LN2. However, high confidence may be placed in the β-strand structure that we have assigned to residues 227 to 230, since this segment contains a pair of cysteine residues connecting a disulfide-linked β-strand in the nE1 crystal structure (20).
FIG 2.
(a and b) Structural alignment of our nE1 homology model (yellow) with structures reported under PDB accession numbers 1LN2 (34) (dark blue) (a) and 4UOI (20) (monomers in teal and pink, respectively) (b). The α-helix and two more β-strands that were added to the C-terminal end of nE1 by Rosetta are shown in magenta and green, respectively, and a similarly positioned α-helix in the structure reported under PDB accession number 1LN2 is shown in red. (c) Sequence alignment of the nE1 sequence with the sequence corresponding to the structure reported under PDB accession number 1LN2, color-coded by similarity, with dark blue representing identity and red representing clashes. The thick colored line above the sequence reported under PDB accession number 1LN2 is color-coded by secondary structure, with blue representing turns, white representing loops, red representing helical regions, and yellow representing β-strands.
(iii) Association of nE1 and nE2.
In Fig. 3, the part of our model corresponding to nE2 that was taken from the crystal structure reported under PDB accession number 4MWF with missing loops modeled as described above and the part corresponding to nE1 based on homology to the sequence reported under PDB accession number 1LN2 are shown in the context of our full-length model. The relative orientations of nE1 and nE2 were determined from the Rosetta docking procedure, which locates the N-terminal β-strands of E1 and the β11/β12 strands of E2 at the E1/E2 interface. The prediction that the N-terminal part of E1 participates in this interaction is consistent with experimental data demonstrating that a recombinant protein corresponding to the first 47 residues of E1 binds to E2 (35).
FIG 3.
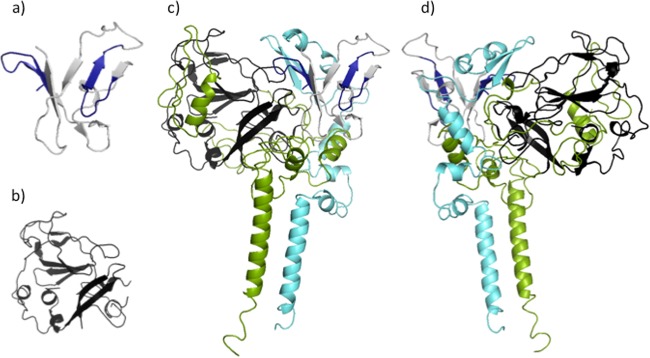
(a) nE1 model, with residues modeled after the structure reported under PDB accession number 4UOI (20) shown in dark blue and those based on homology to the structure reported under PDB accession number 1LN2 (34) in gray. (b) The part of our nE2 model taken from the crystal structure reported under PDB accession number 4MWF (19). (c and d) The same regions colored as described above for panels a and b are seen in the context of our full-length model of the E1/E2 heterodimer in a back view (c) and a front view (d).
We performed multiple coarse-grained simulations in the membrane environment using the Martini force field and selected two models in which the two proteins associate in a manner consistent with the experimental data for the antibody binding sites. We compare the orientations of these two models with the one from our docking in Fig. 4 and observe that the interface occupies similar locations in all three structures, although the relative orientations differ substantially.
FIG 4.
Comparison of E1/E2 interfaces and relative orientations of E1 obtained by protein-protein docking in Rosetta (orange ribbon) or by coarse-grained Gromacs simulations of the two proteins converging to form a heterodimer within a membrane environment (brown and purple ribbons). The position of E2 is the same for all models, and it is shown in yellow and in a surface representation. The viewpoint shown in panel b is rotated approximately 90° from that shown in panel a.
Full-length computed structural models of E1 and E2.
After the docking of nE1 and nE2, the structures of the remainders of the ectodomains, the stem regions, and TMDs were computed first for E2 and then for E1 in the presence of E2 (since the correct folding of E1 is believed to require the presence of E2 [36, 37]).
(i) E2.
In our computed structure of E2, the CD81 binding site is oriented in a lateral position in relation to the horizontal membrane (Fig. 1). The C-terminal region (residues 646 to 717) of the E2 ectodomain contains three α-helices, formed from residues 659 to 667, 693 to 701, and 713 to 717 and connected by unstructured loop regions. The latter two of these helices are consistent with the structure determined by nuclear magnetic resonance (NMR) for a peptide comprised of residues 684 to 719 that shows a helical conformation for residues 687 to 703 and a fraying helix for residues 706 to 714 (38). In our model, the helix comprising residues 713 to 717 forms an N-terminal extension of the helix making up E2's TMD. The disulfide bond between residues C652 and C677, which was enforced during our modeling, is shown in Fig. 1.
(ii) E1.
Based on the outcome of our Rosetta ab initio modeling, an α-helix (residue numbers 275 to 286) adjoins the E1 core at its C terminus. This α-helix is followed by two more β-strands (residue numbers 290 to 303), extending the β-sheet to a total of 8 strands. In light of this, it is interesting that the structural homologue reported under PDB accession number 1LN2 (34) has three β-strands in addition to those belonging to the region of homology to nE1 and forms a 9-strand beta barrel. However, in the case of the structure reported under PDB accession number 1LN2, these additional strands are positioned between the N-terminal strands and the remaining β-strands within the region of homology, unlike in our E1 model. Additionally, at the C terminus of the structure of the phosphatidylcholine transfer protein reported under PDB accession number 1LN2, there is an α-helix (34) positioned similarly to the α-helix belonging to E1 mentioned above (Fig. 2). It has been suggested that in the phosphatidylcholine transfer protein, this α-helix binds to the membrane and is involved in the conformational change of the protein allowing the binding or release of phosphatidylcholine (34). In our model, the corresponding α-helix stretches across the top of E1 on the surface of the protein rather than being close to the viral membrane. This suggests that this helix may be covered by associated lipoproteins. Cryo-EM images of lipoviral particles (LVPs) stained for apolipoproteins reveal that these are accessible outside the virion (39) and probably tend to mask E1/E2 from the host immune system (40–42).
The E1 α-helical region discussed above contains the putative virion fusion peptide at residues 276 to 286 (43–45). Previously, two sequences in E2 that exhibited homology to fusion peptides were identified (23, 46). However, a role in fusion was not supported for these E2 regions since they were located within E2's central β-sheet and CD81 binding domain, respectively (19, 21, 47). It is notable that the putative fusion peptide in E1 is exposed in our model, yet no neutralizing antibodies are known to target this peptide (10). This may be another indication that this peptide is concealed by apolipoproteins. In our model, the E1 binding peptides for the interaction with the apolipoproteins ApoB and ApoE (48) are located on the top of the structure (a plausible position for such binding), with the position of the putative fusion peptide being adjacent to these peptides (Fig. 1). Note that it has also been suggested that E2 plays a major role in the interaction between E1/E2 and ApoE. Coprecipitation analysis of lysates of cells transfected with HCV-derived constructs expressing E1/E2 shows that E2 binds ApoE, and this interaction depends strongly on the presence of the E2 TMD but more weakly on the presence of E1 (49). However, whether this direct E2/ApoE interaction is biologically relevant is disputable.
The stem region of our E1 model contains three α-helices, as is similarly the case for E2. The stem E1 α-helices occur at residues 315 to 324, 333 to 338, and 348 to 352. NMR was used previously to solve the structure of an E1 peptide spanning residues 314 to 342, which supported that the E1 stem region contained two α-helices at residues 319 to 323 and 329 to 338 (50).
Heterodimeric interface.
The parts of E1 and E2 predicted to be involved in heterodimerization and their network of interactions are specified in Fig. 5a and are illustrated in Fig. 5b. We performed in silico alanine scanning to estimate which residues in E1/E2 contribute most to heterodimerization. Positions of residues determined in this manner were visually inspected in our structural model, and some of them could be assigned to pairs of interacting residues. These pairs are given in Table 1, and their positions are illustrated in Fig. 5c. Note that our analysis selected only one interaction between the TMDs of E1 and E2 (Val 374/Leu 735). It is very likely that interactions between TMD residues are underestimated here since the potential term used to obtain these estimates was designed for interactions in aqueous solution (51, 52).
FIG 5.
(a and b) Interacting regions of E1 and E2 observed in our model of the heterodimer are shown schematically (a) and illustrated on our model colored as shown in panel a (b). (c) Pairs of residues identified by interface alanine scanning as making significant energetic contributions to the E1/E2 binding free energy are illustrated on our model. E1 is shown in light gray, and E2 is in pale green. Predicted E1/E2 interface residues for which in silico mutation to Ala significantly affected E1/E2 binding free energy are colored on a scale according to increased prediction score from blue to red to yellow (highest binding free energy difference).
TABLE 1.
Pairs of residues identified by interface alanine scanning as making significant energetic contributions to E1/E2 binding free energya
| Interaction | E1 residue(s) | ΔΔG for E1 (complex) | E2 residue(s) | ΔΔG for E2 (complex) |
|---|---|---|---|---|
| Hydrophobic | Leu 265 | 2.48 | Leu 675 | 1.12 |
| Trp 320 | 2.64 | Trp 672 | 3.82 | |
| Val 374 | 1.35 | Leu 735 | 1.08 | |
| Charged or polar | Arg 195, Asp 279 | 4.20, 4.04 | ||
| Arg 260 | 1.82 | Tyr 632, Glu 637 | 1.75, 1.40 | |
| Thr 300 | 1.43 | Leu 546 | 2.05 | |
| Asp 303 | 1.65 | Asn 577 | 2.50 | |
| Asn 325 | 0.80 | Arg 587 | 2.06 | |
| His 352 | 1.26 | Asp 658 | 1.41 |
The third and fifth columns give the changes in Rosetta scores for the E1/E2 interaction resulting from mutation of E1 and E2 residues, respectively, to Ala, compared to the E1/E2 interaction energy for the wild-type proteins. A positive value indicates that the mutation destabilizes the interaction between E1 and E2.
The E1 regions involved in heterodimerization in our model include the N-terminal β-strands and helix/loop (residues 192 to 215) and the two other β-strands neighboring them in the central β-sheet (residues 258 to 268), the two β-strands following the putative fusion peptide (residues 290 to 306), the stem domain (specifically residues 312 to 348), and the TMD (residues 352 to 378). It is known that the interactions between the TMDs of E1 and E2 are very significant for heterodimerization since E1 and E2 lacking these regions fail to heterodimerize (53). Each of the TMDs are believed to consist of a single α-helix (28, 29), and charged residues at their centers, viz., Lys 370 on E1 and Asp 728/Arg 730 on E2, are thought to play a role in their interactions (Fig. 1) (29, 54–56). There is also experimental evidence that residues of E1 that lie outside the TMD interact with E2. For example, N-terminal residues 201 to 206 on E1 have been found to be essential for the structure of the AR4A and AR5A antibody epitopes (18). Moreover, the N-terminal part of E1 (specifically the peptide comprised of residues 192 to 238) binds to E2 (35). A peptide corresponding to the remainder of the E1 ectodomain (residues 239 to 308) and most of the stem domain (residues 309 to 340) was also demonstrated to bind, albeit more weakly, to E2 (35). The stem regions of both the E1 and E2 proteins contain heptad repeat regions, which are known to have roles in cell entry based on the effects of mutations in these regions (45, 57). Previously, E1 heptad residues (A330, V333, L337, I340, I344, and M347) were mutated to either the helix-stabilizing or helix-breaking residue Ala or Pro, respectively. These mutations were shown to have little effect on heterodimerization (45). Nevertheless, some of the E1 heptad residues, specifically V333 and L337, contact E2 in our model, although they are predicted to form weak interactions. Another E1 residue, His 352, which lies in close proximity to the membrane, contacts E2 in our model and was previously shown to be important for E1/E2 heterodimerization (58).
In our model, E2 heterodimerization residues include those in HVR2 and the neighboring C-terminal region (residues 480 to 487), the region C terminal to the AR2 epitope (residues 541 to 548), IgVR (residues 574 to 579), the region immediately C terminal to IgVR (residue 587), β-strands β11/β12 (residues 630 to 638), the stem region (residues 651 to 703), and the TMD (residues 716 to 738). There are experimental data supporting the involvement of each of these regions in heterodimerization. Several residues were shown previously to be essential for heterodimerization, such as HVR2 (residues 461 to 481) (59), highly conserved residues 484 to 491 containing the “W489HY” sequence and flanking HVR2 (35), IgVR (residues 570 to 580) (59), and the TMD residues (29, 53–56).
The residues C terminal to the AR2 epitope belong to a set of E2 residues (residues 483 to 491 and 523 to 545) that, when mutated, were shown previously to substantially decrease the binding of the E1-specific antibody A7 (18), suggesting that these residues may be located at the E1/E2 interface. β11/β12 and the stem region are demonstrated to be close to the heterodimeric interface since mutation of residue 639 on the β12 strand or any of the stem residues 657 to 659 or 692 resulted in a significant reduction in the binding of the AR5A Ab. Furthermore, mutation of any of the same stem residues or D698 had a large negative impact on the binding of the AR4A Ab (18). Additional experimental data also support that the stem region of E2 forms important interactions with E1. First, a 5-residue insertion at residue 692, located at the pretransmembrane region of E2, did not alter E1 or E2 stability but had a specific impact on heterodimerization (as also did an insert at residue 587, C terminal to IgVR) (60). Second, by mutating residues in the heptad repeat regions of E2 to Ala or Pro (Leu 675, Ser 678, Leu 689, and Leu 692), these residues were identified as key determinants of heterodimerization. Ala substitutions were reported to be less severe than Pro substitutions, emphasizing the importance of the helical structure in this region (57). Another residue in this region, His 691, is also essential for heterodimerization (58). Data from our computational analysis concur that Leu 675, Ser 678, and His 691 within E2 all form interactions with E1 important for heterodimerization. The corresponding E1 residues forming these interactions are Leu 265 for Leu 675 (Table 1), Trp 353 for His 691, and Ile 262 for Ser 678.
Higher-order oligomerization. (i) Molecular mechanics.
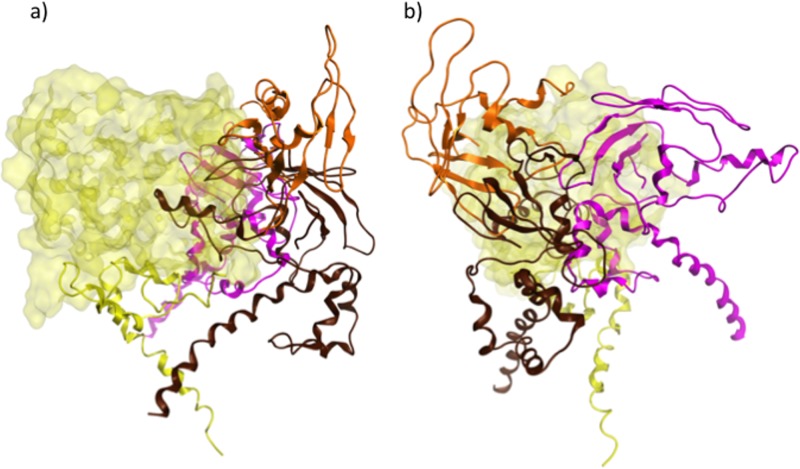
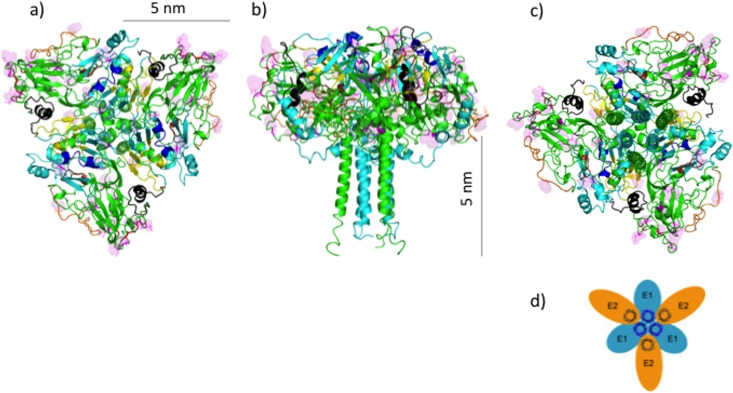
In a previous study, the oligomeric structure of E1/E2 was investigated by SDS-PAGE of purified HCV virions under reducing conditions at 37°C. These experiments showed that E1 forms non-disulfide-linked trimers when coexpressed with E2 and that the TMDs of E1 form interactions essential for this trimerization (33). This suggested to those authors that E1/E2 might form a trimer with the TMDs of E1 at the center (Fig. 6d). We attempted to fit together replicas of our predicted heterodimer in compliance with this model using molecular mechanics. In Fig. 6, our model of E1/E2 is shown to fit into a trimeric arrangement consistent with this prediction. It is likely that the trimeric organization with all TMDs at the center could contribute to viral fusion mediated by the heterodimers by exerting force on the viral membrane.
FIG 6.
(a to c) Ribbon representations of the predicted trimer of heterodimers formed by E1/E2, viewed from the top (a), side (b), and bottom (c) in relation to the viral membrane being positioned below the ectodomains. The putative fusion peptides and apolipoprotein binding peptides are shown in dark blue and yellow, respectively, on E1, which is mainly in teal. E2 is shown mainly in green, with CD81 binding residues in red, HVR1 in black, HVR2 in orange, and IgVR in brown. Sites of N-glycosylation are shown in line mode and in a surface representation and are in magenta. (d) Model proposed by Falson et al. in which E1/E2 is arranged as a trimer of heterodimers, with E1 components interacting with one another at the center of the trimer through their TMDs (33).
In addition to forming trimers, there is evidence that the E1/E2 heterodimers can be arranged into higher-order oligomeric structures. Specifically, cryo-EM was used to visualize HCV-like particles formed in a baculovirus system, and a 30-Å-resolution reconstruction was created. Based on the obtained images of these particles, E1/E2 has an arrangement with 2-, 3-, and 5-fold symmetry axes (61) in the baculovirus system. Accordingly, it is encouraging that our modeled E1/E2 heterodimer can be arranged into a pentamer of trimers, possibly corresponding to the observed 5-fold symmetry axis (Fig. 7). The 2-fold axis seen in the cryo-EM reconstruction (61) would, in this case, correspond to the junction of two E1/E2 trimers into a hexamer. The 3-fold axis indicates an arrangement of the pentamers of trimers into a higher-order trimer and suggests that the HCV-like particle visualized by cryo-EM, as well as possibly the HCV virion, may take the form of a pentactic dodecamer, i.e., a composition of 12 replicas of our pentamer. However, it is also arguable that the HCV virion does not have a symmetrical glycoprotein envelope since data from previous studies indicate that there may be a low abundance of envelope glycoproteins in the HCV virion, based on immunostaining analyses of virions of HCV (39) (similar to data for the pestivirus bovine viral diarrhea virus [BVDV] [62]). Moreover, images from Piver et al. (63) show that the HCV virion is an LVP of variable size with an irregularly shaped nucleocapsid surrounded by an external crescent containing the apolipoproteins. It may that the HCV glycoprotein envelope is simply deformable, leading to the irregular shapes, and the size diversity might be due to differing amounts of lipids and apolipoproteins coating the glycoprotein envelope. On the other hand, it is also possible that the nucleocapsid of HCV is coated with a mixture of lipids, apolipoproteins, and only a small number of envelope glycoproteins (E1/E2).
FIG 7.
Model of the pentamer formed by the trimer of E1/E2 heterodimers, viewed from the top in relation to the viral membrane being positioned below the ectodomains. A hexamer forming part of this pentamer is shown here in a ribbon representation, with E1 shown in teal and E2 shown mainly in green, except that CD81 binding residues are in red (shown as balls), HVR1 is in black, HVR2 is in orange, and IgVR is in brown. Cysteine residues are shown in line mode, and sulfur atoms belonging to C304 and C486 are depicted as yellow and orange balls, respectively.
According to the predicted quaternary structure in Fig. 6 and 7, the relative positions of various regions of E1 and E2 can be identified. The CD81 binding site is located close to the corners of the trimer of heterodimers, and these sites occupy positions near the center of our predicted pentamer of trimers. In our hexamer (i.e., the dimer of trimers in Fig. 7), HVR2 from one E2 monomer is positioned close to HVR1 from another monomer and is flanked on another side by IgVR. This observation is supported by experimental data suggesting that HVR1, HVR2, and IgVR are structurally linked in the virion (64). For example, antibodies that bind epitope I in E2 have been shown to exhibit improved binding to E2 and an improved ability to inhibit E2-CD81 interactions when any one of these hypervariable regions was deleted. Furthermore, inhibition of the E2-CD81 interaction was additive if two or more of the hypervariable regions were deleted (64). In addition, antibodies that recognize an epitope that included HVR1 residues were shown to have a reduced affinity for E1/E2 when IgVR was deleted but not when IgVR and HVR2 were deleted simultaneously (64). A possible explanation for this observation is that IgVR may be important for constraining the position of HVR2 in wild-type E2. Therefore, in its absence, HVR2 may reposition, which may have an impact on the accessibility of HVR1.
We examined potential disulfide linkages in our predicted quaternary structure. In our model, 2 of 8 conserved cysteine residues in E1 and 16 of 18 conserved cysteine residues in E2 are linked by intramolecular disulfide bridges. In E2, these disulfide bridges include those that were observed in the crystal structure reported under PDB accession number 4MWF (19) and that were retained during our modeling. We note that data from other experiments reported previously (21, 32) suggested alternative linkages, as well as the C652-C677 linkage (32). Our model shows only one disulfide bond in E1 between C229 and C238, which was identified in the partial E1 crystal structure. The model also shows that C272 from nE1 is very close to C281 in the putative fusion peptide of E1, suggesting a potential for interactions. However, Li et al. (65) noted that it is unlikely that these two residues are disulfide linked, since E1/E2 from HCV pseudoparticles (HCVpp) with the mutations C272A and C281A did not migrate differently on reducing or nonreducing SDS-PAGE gels compared to wild-type E1/E2. It is unclear based on experimental data in the literature which, if any, disulfide linkages are formed between E1 and E2 within the heterodimer and in the higher-order structure. Most intracellular E1/E2 heterodimers are noncovalently linked (66), and there are not known to be any disulfide bridges between E1 and E2 within the heterodimer on the surface of the virus. However, experimental data indicated that the majority of E1/E2 found in the virion exists as disulfide-linked complexes with molecular masses of at least 440 kDa (cf. roughly 30/70 kDa for the glycosylated E1/E2 heterodimer) (66). In our model, C304 in E1 and C486 in E2 appear to be close enough to form a potential disulfide bond between two different heterodimers (Fig. 7).
(ii) Size exclusion chromatography and Western blotting.
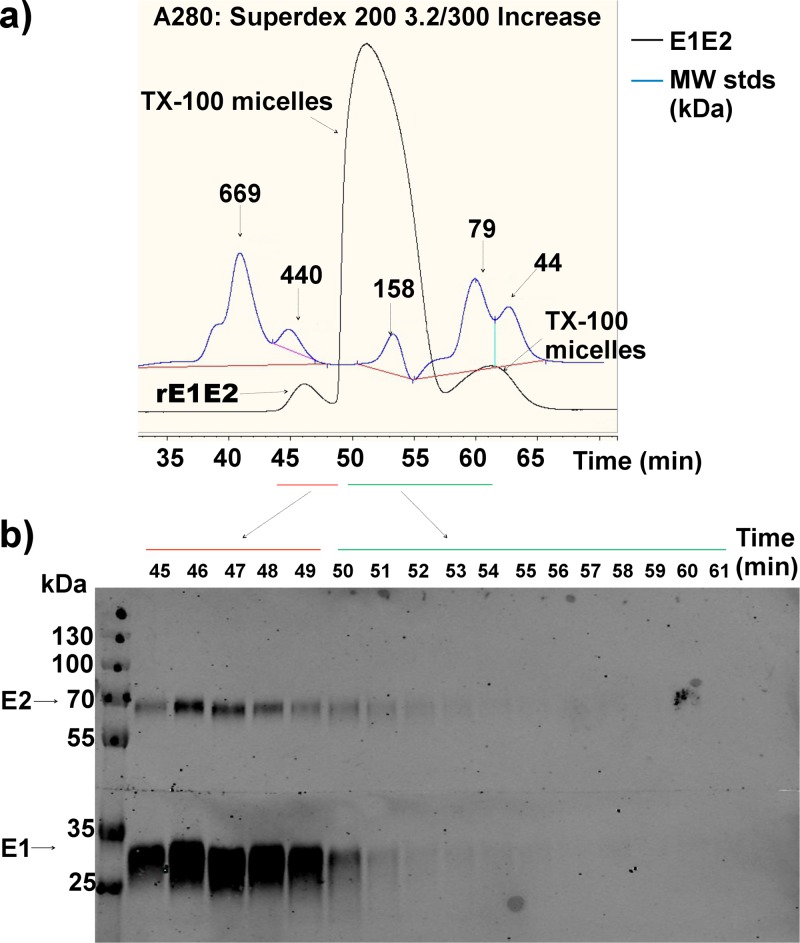
To further investigate the oligomeric arrangement of E1/E2, we performed SEC on the intracellular E1/E2 (H77c) heterodimer purified from the lumen of the endoplasmic reticulum following the transduction of a lentiviral vector into a CHO cell clone. The UV absorption spectrum at 280 nm displays three peaks (Fig. 8a). The earliest of these peaks corresponds to an analyte with an average molecular mass of 367 kDa, and E1/E2 was detected in this fraction by Western blotting (Fig. 8b). Next, there is a large peak at a molecular mass of 100 to 200 kDa and a later peak at a lower molecular mass. These peaks were found to contain little or no E1/E2 by Western blotting. A separate run of the E1/E2 purification buffer plus 10% Triton X-100 alone showed a profile matching the second and third peaks (not shown), suggesting that these fractions are Triton X-100 micelles. Since glycosylated E1/E2 has a molecular mass of ∼100 kDa, the analyte molecular mass of 367 kDa is consistent with a trimer of E1/E2, as was suggested to form by our modeling.
FIG 8.
SEC of purified recombinant E1E2 (H77) (a) and Western blot analysis of the fractions of the collected eluate (b). TX100, Triton X-100; MW, molecular weight (in thousands).
Functional aspects of E1/E2 structure. (i) Antibodies.
Since E1/E2 is exposed on the surface of the HCV virion, it is the major target of the host humoral immune response, and both E1/E2 and E2 are being investigated as vaccine candidates (67). In Fig. 9a, the binding sites for the major classes of neutralizing antibodies are shown with respect to our model. It is encouraging that all of these sites appear to be accessible for antibody binding in our model. Many of these antibodies neutralize the binding of HCV to the host receptor CD81, which is required for cell entry (9, 10). Residues belonging to the CD81 binding site (19, 68), viz., Y527, W529, G530, and D535 from the CD81 binding loop of E2 that connects the inner and outer sheets of the β-sandwich; W420, L427, and N428 belonging to the N terminus of the structure reported under PDB accession number 4MWF; and F442, are also shown in Fig. 9a. AR3, epitope I, and epitope II all overlap the CD81 binding site. The AR2 and AR1 antibodies do not directly interact with the CD81 binding site, but it is possible that their binding may result in a conformational change in E2 that affects its interaction with CD81. Currently, the precise mechanism for neutralization mediated by these antibodies is not known. AR4A and AR5A are both potent, broadly neutralizing antibodies that target the heterodimeric interface of E1/E2, which includes conserved residues implicated in viral fusion with the endosomal membrane.
FIG 9.
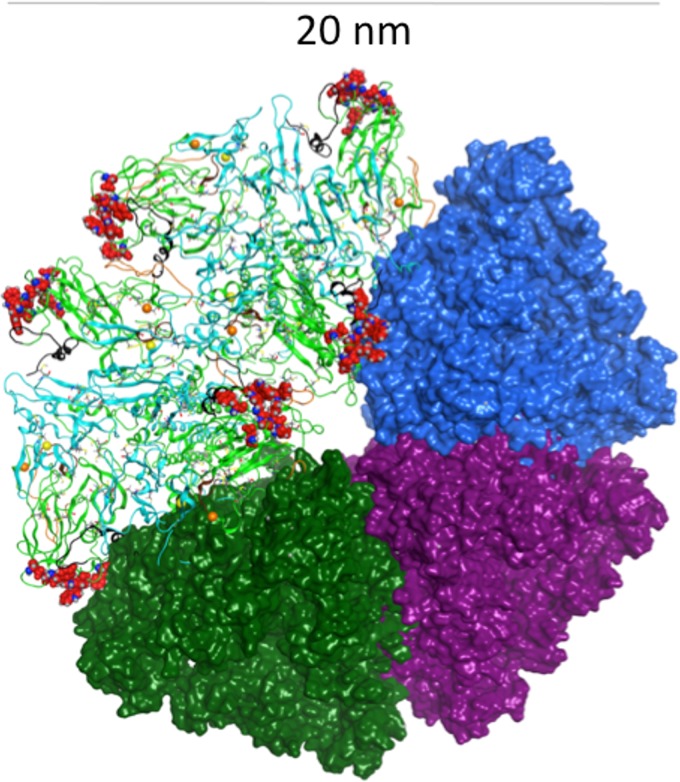
Ribbon representations of our computational model of the E1/E2 heterodimer, with E1 shown in teal and E2 shown in green. (a) The binding epitopes of classes of neutralizing antibodies are highlighted by using color and labeling. Residues important for CD81 binding are shown in red and in a line representation. Arrows representing suggested angles of approach are shown for the AR4A and AR5A antibodies. (b) Residues important for binding by the AR4A antibody are shown highlighted in red on one (right-hand side) of two adjacent E1/E2 heterodimers structured as part of a trimer of heterodimers. Three sites of N-linked glycosylation belonging to E1, which are close to the antibody binding site, are shown in magenta and in a ball-and-stick representation.
The binding site shown in Fig. 9a and b for the AR4A antibody is less accessible than those for the other antibodies, which correlates with the previously reported observation that E1/E2 elicits less AR4A-type antibodies than other antibodies (69). Although the AR4A and AR5A antibodies share many residues important for their binding, they do not compete with each other for binding to E1/E2. AR5A has been shown to compete with CBH-7, one of the antibodies targeting AR2 at residues 541 to 549 on E2 (18). Thus, it is likely that some of the residues important for AR4A and AR5A binding are not contact residues but rather conformational determinants of their respective epitopes (18).
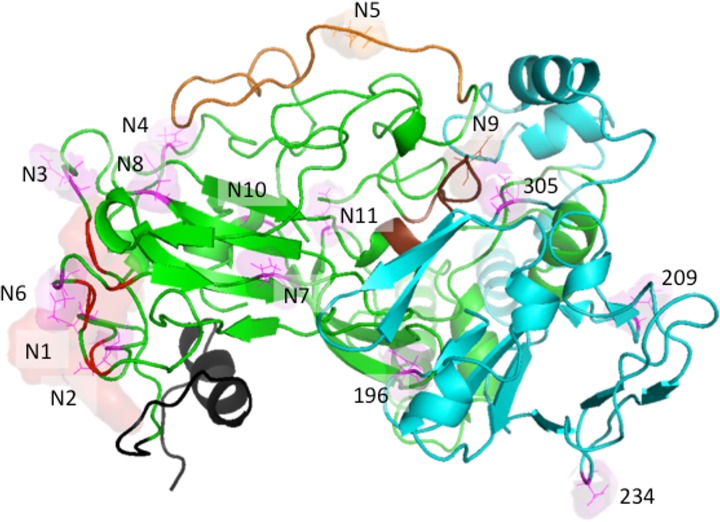
The N-linked glycosylation sites of E1/E2 are positioned in our model in locations that are not buried in any of the predicted oligomeric interfaces between the proteins or in the interface between E1/E2 and the membrane (Fig. 7). It may be seen in Fig. 9b that the N-linked glycosylation site at N209 is predicted to occupy a position that would potentially obscure the AR4A binding site. N196 and N305 are also depicted in Fig. 9b. It has been reported that simultaneous mutations of both of these residues (but not single mutations) to Ala substantially disrupt the binding of AR4A and AR5A to E1/E2 (18). The N209A single mutation has little effect on the binding of either Ab (18). However, it is possible that the N209 glycan may have a kinetic effect on neutralization by the antibodies. In Fig. 10, the N-linked glycosylation sites N1 and N2, both located in epitope I, and N6, located in AR3, are observed to shield the CD81 binding site in our model. Mutation of each of these three glycans has been shown experimentally to increase the sensitivity of the virus to antibody-mediated neutralization (70). Glycans at N4 and N11 have similar effects on antibody binding (70). In the context of our pentameric model, N4 appears to obscure the CD81 binding site. Since N11 is distal from the CD81 binding site, glycosylation at N11 likely impacts E2 folding.
FIG 10.
Locations of N-linked glycosylation sites on E1/E2, shown in line and surface representations, viewed from the top in relation to the viral membrane being positioned below the ectodomains. E1 is shown in teal, and E2 is shown mainly in green, with CD81 binding residues in red (a surface representation is also shown), HVR1 in black, HVR2 in orange, and IgVR in brown. N-glycosylation sites belonging to E2 are labeled N1 to N11, while those belonging to E1 are labeled by residue numbers.
The N terminus of HVR1 is exposed on the surface of our E1/E2 model, which supports a role in the antigenicity and/or accessibility of neutralizing antibodies. Indeed, previous studies have shown that HVR1 can interfere with the binding of antibodies to E2 (64, 71), and HCV pseudoparticles lacking HVR1 have been reported to exhibit increased sensitivity to a range of neutralizing monoclonal antibodies that are directed to diverse E2 epitopes (72). This observation is surprising since these antibody binding sites are not all clustered into one region. One possible explanation is that the surface of the wild-type HCV virion is rather rigid, with the monoclonal Ab binding sites being too constricted to interact easily with Abs; the removal of HVR1 may leave more room within the lattice of interacting proteins forming the surface of the virion for Ab binding domains to undergo minor conformational changes required for binding.
(ii) Fusion.
Based on homology to other flaviviruses, HCV was originally thought to have a class II fusion mechanism (22, 23). However, the crystal structure of nE2 indicated a compact globular structure that was quite dissimilar from class II fusion proteins (19, 21). Class II fusion proteins are composed of domains I, II, and III with mainly β-sheet structures. Domains II and III fold over the central N-terminal domain (domain I) to form a hairpin that is essential for merging the host and viral membranes. Domain II has an elongated structure and contains the fusion loop (73, 74). In our model, the E1/E2 heterodimer has a compact form, indicating, in agreement with data from previous reports (20, 47, 75), that neither E1 nor E2 is a class II fusion protein. It has been suggested that E1 and E2 together may comprise a structure that resembles a class II fusion protein (47). However, since there is no region with structural similarity to domain II from class II fusion proteins, it is more likely that E1/E2 employs a novel fusion mechanism.
It is possible that the lipoproteins associated with the HCV virion may play a role in the fusion mechanism, since hydrophobic interactions of the endosomal membrane with virus-associated apolipoproteins and/or lipids may help to explain how E1/E2 may initiate viral fusion when it does not have an elongated domain containing the fusion peptide to extend toward the endosomal membrane as in class I and class II fusion proteins (73, 74). For example, in class I fusion proteins, during the fusion process, the fusion peptide is projected in some cases up to 100 Å toward the host membrane (74), and in class II fusion proteins, domains I and II go from lying parallel to the viral surface to extending in a perpendicular manner toward the endosomal membrane (74). In the case of HCV, if the fusion peptide is covered with apolipoproteins, or lipids only as in systems such as HCVpp, where apolipoproteins are absent, they could potentially participate in fusion by bringing the fusion peptide into contact with the endosomal membrane since they would be expected to form favorable interactions with the lipids belonging to the host membrane. In this case, no outward extension of the fusion loop would be required. However, this speculative hypothesis requires experimental support. It is also possible that E1, which resembles a phosphatidylcholine transfer protein, might play a role in lipid transfer that could serve to merge the two membranes.
Conformational changes in E1/E2 prior to fusion in the endosome are believed to take place on a slow time scale of one or more hours (76). Isomerizations of the disulfide linkages in E1/E2 are thought to be required for cell attachment and entry, since entry is eliminated by a sulfhydryl-alkylating agent that covalently modifies free Cys residues that are, presumably, involved in catalyzing disulfide isomerization (77). In support of such a role of free Cys residues, it has been demonstrated that other viral fusion proteins undergo a reduction of a labile disulfide bond between another Cys residue and a CXXC motif, which activates fusion (78, 79). A functionally similar mechanism may occur for HCV fusion. For example, E1 contains a highly conserved C226(V/L)PC protein disulfide isomerase (PDI) motif (77). C226 in E1 may be one of the free cysteines required for cell entry that is observed in our model. In this scenario, C226 would most likely form a new disulfide bridge with C229 while simultaneously catalyzing the dissociation of the disulfide linkage between C229 and C238 (Fig. 1). This may result in an important conformational change in the central β-sheet of E1. In our model, the α-helix in E1 containing the putative fusion peptide is contacted on one side by two β-strands at its C terminus, which itself is stabilized by contacts with E2. On another side, this α-helix interacts with two strands from the central β-sheet of E1, namely, β4 and one of the two β-strands replacing β5 of the crystal structure reported under PDB accession number 4UOI in our model. Isomerization or pH-dependent structural changes in E1 and/or E2 in the endosome could contribute to the restructuring of E1/E2 required for bridging the length between the viral and host membranes. For example, it is likely that the protonation of H445 in E2 by exposure to the acidic environment of the endosome helps to promote structural changes important for fusion, since the H445R/K mutations have been found to enhance fusion (58). In our model, the stem domains of E1 and E2 are positioned beneath their ectodomains. When the fusion peptide in E1 is embedded into the host membrane, the stem regions of E1 and E2 are likely to participate by docking onto the ectodomains and helping to pull the TMDs and the viral membrane closer to the host membrane, as is the case for class II fusion proteins (57).
DISCUSSION
We have drawn upon computational methods to predict a full-length model of the E1/E2 heterodimer based on the known partial crystal structures of the envelope glycoproteins E1 and E2. E1/E2 has been widely studied experimentally, and previous studies have given us valuable data to assist in our modeling. Negative-stain cryo-EM imaging has revealed a rough outline of the positions of HVR1 and HVR2 and of the C-terminal region of the E2 ectodomain (19). HVR2 and the E2 stem region presumably make significant interactions with E1, but experimental methods have not been successful in providing structural information on the orientation of E1 and E2 within the heterodimer or in relation to the viral membrane. Experimental tests of our computational prediction should therefore be useful for determining the positions of the partial crystal structures within the quaternary structure as well as providing valuable insights into the reliability of the computational techniques that we have employed.
In our modeling, we based the structure of nE1 on homology to the phosphatidylcholine transfer protein (PDB accession number 1LN2), according to suggestions made previously by El Omari et al. (20). When Rosetta ab initio modeling was used to extend this structure in the presence of E2, the structural similarity between the two proteins was found to extend even further than the previously observed structural homology between the crystal structures reported under PDB accession numbers 1LN2 and 4UOI (20) and also further than the sequence homology between the two proteins. A helical region containing the putative fusion loop belonging to E1 extends across the top of our model, bearing similarity to a long α-helix existing in the structure reported under PDB accession number 1LN2, which has been proposed to be membrane proximal (34). Since the putative fusion peptide is on the outer surface of E1/E2 in our model, this may indicate that it is buried by contacts with virion-associated apolipoproteins.
Our proposed heterodimeric structure is plausible in consideration of regions of E1 and E2 that are known experimentally to be important for their interaction (Table 2). The docking of nE1 and nE2 was performed without bias in the sense that the lowest-scoring model was selected in a blind docking procedure. When further modeling of the E2 stem domain was performed, key interacting residues for the AR4A and AR5A antibodies could be assigned coordinates that were in relatively close proximity to each other. Moreover, our model correctly predicts that the stem regions each contain two α-helices, consistent with data from previous NMR studies (38, 50). Based on our model of the E1/E2 heterodimer, molecular mechanics was applied to determine the oligomeric organization of the HCV virion. The predicted trimeric arrangement of the E1/E2 heterodimer is in agreement with data from a previous study (33) that suggested a trimeric arrangement. Our preliminary SEC data are also consistent with E1E2 existing as a trimer, but it will be necessary in future work to confirm its molecular weight using multiangle laser light scattering in combination with SEC (SEC-MALLS).
TABLE 2.
Corroboration between experimental data and our computational model of E1/E2
| Criterion and residue(s) | Protein | Correspondence with expt | Experimental method | Reference(s) |
|---|---|---|---|---|
| Criteria used as constraints during modeling | ||||
| 192–270 | E1 | Homology model based on structures reported under PDB accession no. 4UOI and 1LN2 | Crystallography | 20, 34 |
| 421–645 | E2 | Same as structure reported under PDB accession no. 4MWF | Crystallography | 19 |
| C652, C677 | E2 | Disulfide bonded | Ab binding patterns upon mutation | 32 |
| 352–378, 716–738 | E1/E2 | Single-domain helices | NMR; fluorescent imaging of epitope-tagged C termini | 28, 29 |
| K370, D728 | E1/E2 | Salt bridge between TMDs of E1 and E2 | Mutational analysis | 29, 54–56 |
| Criteria used to guide de novo model selection | ||||
| 201–206, 639, 657–659, 692, 698 | E1/E2 | Residues are proximal to each other | Required for binding to E1/E2 by AR4A and/or AR5A Abs | 18 |
| 315–324, 333–338, 693–701, 713–717 | E1/E2 | Two α-helices exist in each of E1 and E2 stem domains | NMR studies of peptides | 38, 50 |
| Agreement with expt observed after modeling | ||||
| 192–238 | E1 | Near E1/E2 interface | Peptide binds to E2 | 35 |
| 271–304 | E1 | Extension of structural homology of E1 to structure reported under PDB accession no. 1LN2 | Crystallography | 34 |
| H352 | E1 | Important for heterodimerization | Mutational analysis | 58 |
| 480–487, 574–579 | E2 | HVR2 (residues 461–481), residues 484–491, and IgVR (residues 570–580) are essential for heterodimerization | Studies of deletion constructs | 35, 59 |
| 541–548 | E2 | Near E1/E2 interface | Mutations of residues 523–545 affect binding to E1/E2 by E1-specific Ab A7 | 18 |
| 587, 651–703 | E2 | 5-residue insertions at residue 587 or 692 affect heterodimerization | Linker-scanning mutagenesis | 60 |
| L675, S678, H691 | E2 | L675, S678, and H691 are all determinants of heterodimerization | Mutational analysis | 57 |
MATERIALS AND METHODS
Computational methods.
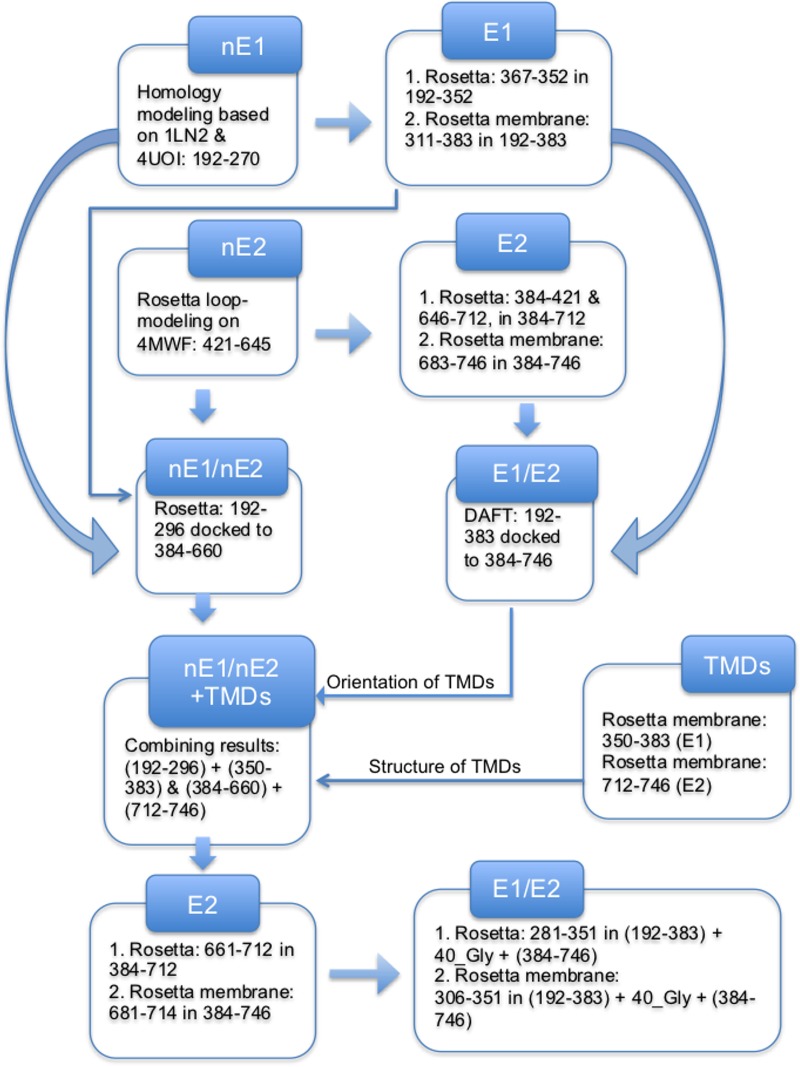
Our computational procedure used to model E1/E2 is summarized in Fig. 11 and described below.
FIG 11.
Flowchart illustrating the computational procedure used to obtain the final model of E1/E2. For Rosetta ab initio modeling or Rosetta membrane ab initio modeling, we indicate the residues being modeled and the context in which these residues were modeled with all other residues being fixed.
Loop modeling of the nE2 crystal structure and homology modeling of nE1.
We first determined full-length models of the N-terminal regions of E1 and E2 (H77 strain), nE1 and nE2, corresponding to the sequences of the crystallized constructs. In the case of nE2, we used loop modeling to complete the structure reported under PDB accession number 4MWF, and in the case of nE1, we created a model based partly on the crystal structure reported under PDB accession number 4UOI and partly on a previously identified structural homolog reported under PDB accession number 1LN2, as we describe in detail below.
The loops missing from the nE2 crystal structure reported under PDB accession number 4MWF (19) were predicted by using the Robetta Protein Structure Prediction Server (http://robetta.bakerlab.org/), which uses loop modeling to rebuild missing parts and then runs full-atom refinement. Note that the structure of the crystallized portion of nE2 may be expected to differ somewhat within the context of E1/E2 compared to that in the crystallized ectodomain. However, the computational methods for de novo structure prediction in Rosetta are not expected to be sufficiently powerful to improve upon the structure of E2 within E1/E2 over what is known from the crystal structure, and so the crystallized portion of nE2 was held fixed during our modeling.
nE1 presents a greater challenge since it appears to be domain swapped (80) in the crystal structure reported under PDB accession number 4UOI (20). We performed sequence alignment between nE1 and the homologous crystal structure reported under PDB accession number 1LN2 (34) in the Molecular Operating Environment (MOE) program (81) and used it to thread homologous regions of nE1 onto four β-strands from the structure reported under PDB accession number 1LN2 (Fig. 2); two of these β-strands match the positions of β3 and β4 in the structure reported under PDB accession number 4UOI, while β5 of the structure reported under PDB accession number 4UOI was divided into two β-strands in our model. The N-terminal β-hairpin β1/β2 was then threaded in MOE onto a position matching the position of strands β3/β4 in the homodimeric E1 partner protein in the structure reported under PDB accession number 4UOI, extending our model to a six-stranded β-sheet (the alignment for threading was chosen based on auto-structural homology).
Preliminary models of C-terminal ectodomains, stem regions, and TMDs of E1 and E2.
The remaining C-terminal regions of E1 and E2 and the N-terminal region of E2 missing from the crystal structure, including HVR1, were predicted in the context of the crystal structure-based models described above by using ab initio modeling with the Rosetta program (25, 82). A topology broker (83) was used to designate the previously determined parts to be kept rigid while terminal regions of the proteins were predicted. Three- and nine-residue fragment libraries corresponding to the primary sequences of E1 and E2 were obtained from the Robetta fragment server. Robetta's fragments are created in accordance with secondary-structure predictions using four different methods: PsiPred (84), SAM-T99 (85, 86), and Jufo/Jufo3D (87). Fragments were then assembled by a Monte Carlo fragment insertion protocol, which favors insertions leading to geometries with good scores. Low-resolution models were first built by using a centroid energy function, which were later refined by using an all-atom energy function (27). A total of 100,000 models were generated, and the lowest-scoring models were subsequently clustered. Best-scoring representatives were found to belong to highly populated clusters, adding to their reliability, since in general, large clusters have a higher probability of containing native-like models. Thus, these representatives were selected to represent E1 and E2 ectodomains in the next steps.
Stem regions and TMDs were then predicted in the context of previously elucidated N-terminal parts of E1 and of E2, which were held rigid, by using the Rosetta membrane ab initio program (88–90). For these simulations, a span file, defining membrane residues, was generated by using the Octopus prediction server (91) and then manually edited to reflect the single-domain topologies of both the E1 and E2 TMDs and used as the input in predictions of lipophilicity information by run_lips in the Rosetta membrane program (88–90). A total of 10,000 models were built for each of the two proteins, the top-scoring structures were clustered, and the best models were chosen based on score, cluster size, and manual inspection.
Association of E1 and E2.
Global docking in Rosetta (92) was applied to determine the heterodimeric association of the E1 and E2 ectodomains (residues 192 to 296 and 384 to 660, respectively). We ran 100,000 decoys and then selected the one with the lowest score. We note that out of the top 10 best-scoring models, this model alone satisfied our pass/fail criteria of the proximity of AR4A/AR5A binding residues (18). The stability of this low-scoring model was confirmed by performing local docking of the E1 and E2 ectodomains around this docking pose using small perturbations of up to 8° and 20 Å and verifying that the resulting 10 lowest-scoring poses were almost identical to the original one.
Modeling and orientation of E1 and E2 TMDs.
We ran coarse-grained molecular dynamics (MD) simulations to study the association of E1 and E2 in the membrane environment and to find the positions of the E1 and E2 ectodomains relative to the TMDs. DAFT (93) is a set of scripts designed to automate MD simulations of protein-protein associations within the membrane in the Gromacs program with the coarse-grained Martini force field (94–96). After assignment of secondary structure using the dssp program (97), 200 systems of randomly oriented E1/E2 heterodimers were set up with DAFT by using an initial separation of 2.5 nm and a periodic box with a dimension perpendicular to the membrane of 5 nm. We did not apply an elastic network (98), as is often used to constrain secondary structure during Martini simulations, since much of the secondary structure of E1/E2 is unknown. The environment, consisting of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer and Martini water, was created by using insane (99). Simulations of 1,000 ns were performed on each of these systems by using a 20-fs time step in Gromacs 5.0 (30, 31). The resulting models were visually examined for the juxtaposition of E1 and E2 TMDs and for the proximity of AR4A/AR5A binding residues (residues 201 to 206, 639, 657 to 659, 692, and 698), which led to the selection of the four best models; these models could be grouped into two clusters by visual inspection.
Since the two interacting TMDs are relatively short, we assumed that these TMDs should be positioned roughly perpendicular to the membrane. We note that although this perpendicular orientation was not observed in the Gromacs simulations, we believe that this was an artifact related to the inherent imprecision of our coarse-grained simulations. Moreover, the simulations allowed the E1 and E2 molecules to relax into energetically favorable orientations in the membrane. We back-mapped the best E1/E2 models from the Gromacs simulations, i.e., converted the coarse-grained systems to all-atom representations (100), and then structurally aligned these all-atom models to the original all-atom E1/E2 models to facilitate visual inspection, since the coarse-grained simulations had disordered parts of the known secondary structure. Thus, by examining the positions of the ectodomains from the Gromacs simulations relative to the lipid membrane, by making the assumption of perpendicular orientations of the TMDs in the membrane, and by making the additional assumption that the TMDs are oriented relative to one another in such a way that residues K370 and D728 interact, we arrived at a proposed orientation of the TMDs relative to the ectodomains. With this prediction, after rebuilding the structures of the TMDs of E1 and E2 as separate domains by running the Rosetta ab initio program and selecting the best of 10,000 models, we fixed the positions of the TMDs of E1 and E2 relative to the ectodomains.
Final full-length models of E1 and E2.
After having decided upon fixed positions of the TMDs relative to the E1 and E2 ectodomains, and having docked the ectodomains to each other, we rebuilt full-length models of E2 and then of E1 in the presence of E2. A total of 100,000 models of E2 were again built by ab initio modeling in Rosetta (25, 82), clustered, and filtered based on the experimental criteria related to antibody binding sites. During these simulations, a topology broker was again used to hold previously determined regions rigid (83); to enforce the proximity of residues C652 and C677, which have been predicted to be disulfide linked (32); as well as to define constraints on internal coordinates of N- and C-terminal residues of the TMD in order to fix its orientation relative to the ectodomain. Finally, 10,000 models of the interfacial/stem region were run by using Rosetta membrane ab initio modeling (88–90), and the lowest-scoring structure was selected. Because this model contained an unclosed loop, loop modeling by Cyclic Coordinate Descent (CCD) (101) followed by a relaxation step was performed to repair the chain break.
For E1 to be modeled in the presence of E2, a linker of 40 Gly residues was then added, connecting the C terminus of E1 to the N terminus of E2 in the MOE program (81). A total of 100,000 models were built by ab initio modeling in Rosetta (25, 82), while E2 and the previously determined N-terminal part of E1 were held rigid. The lowest-scoring model was selected, and the interfacial/stem region of E1 was then rebuilt by using Rosetta membrane ab initio modeling (88–90) by selecting from 10,000 models. Finally, in order to estimate the contributions of each interface residue to the binding free energy of E1 and E2, we relaxed our final model in Rosetta and then submitted it to the Robetta Computational Interface Alanine Scan Server (51, 52).
Higher-order oligomeric structure of E1/E2.
The MOE program was used to position replicas of our predicted E1/E2 heterodimer side by side in such a way that they fit into a trimer and also to fit replicas of the trimer into a pentamer.
Experimental methods.
Full-length recombinant E1/E2 (H77c) (amino acids 192 to 746) was purified from a transduced CHO cell clone as described previously (102). Purified E1/E2 (5 μg) was loaded onto a Superdex 200 Increase 3.2/300 column (GE Healthcare) in GWB (Galanthus nivalis agarose [GNA] wash buffer; 10 mM sodium phosphate, 80 mM NaCl, 0.1% Triton X-100 [pH 6.8]), and the UV absorbance was measured at 280 nm. The molecular weight was determined according to standards using a Gel Filtration Calibration HMW (high molecular weight) kit (GE Healthcare) by averaging over a sample size of 3. For Western blotting, the E1 and E2 proteins were detected with the A4 and H52 antibodies, respectively.
ACKNOWLEDGMENTS
H.F. thanks Rocco Moretti for assistance with Rosetta and Philip Winter for technical computer help. This research was enabled by the use of computing resources provided by WestGrid and Compute Canada.
We are grateful for funding support from a Canadian Excellence Research Chair (CERC) grant held by M.H.
REFERENCES
- 1.Freedman H, Logan MR, Law JL, Houghton M. 2016. Structure and function of the hepatitis C virus envelope glycoproteins E1 and E2: antiviral and vaccine targets. ACS Infect Dis 2:749–762. doi: 10.1021/acsinfecdis.6b00110. [DOI] [PubMed] [Google Scholar]
- 2.Thomssen R, Bonk S, Propfe C, Heermann KH, Kochel HG, Uy A. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol 181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 3.Thomssen R, Bonk S, Thiele A. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med Microbiol Immunol 182:329–334. [DOI] [PubMed] [Google Scholar]
- 4.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voisset C, Dubuisson J. 2004. Functional hepatitis C virus envelope glycoproteins. Biol Cell 96:413–420. doi: 10.1016/j.biolcel.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J 20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. 1991. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A 88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball JK, Tarr AW, McKeating JA. 2014. The past, present and future of neutralizing antibodies for hepatitis C virus. Antiviral Res 105:100–111. doi: 10.1016/j.antiviral.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sautto G, Tarr AW, Mancini N, Clementi M. 2013. Structural and antigenic definition of hepatitis C virus E2 glycoprotein epitopes targeted by monoclonal antibodies. Clin Dev Immunol 2013:450963. doi: 10.1155/2013/450963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keck ZY, Op De Beeck A, Hadlock KG, Xia J, Li TK, Dubuisson J, Foung SK. 2004. Hepatitis C virus E2 has three immunogenic domains containing conformational epitopes with distinct properties and biological functions. J Virol 78:9224–9232. doi: 10.1128/JVI.78.17.9224-9232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keck ZY, Li TK, Xia J, Bartosch B, Cosset FL, Dubuisson J, Foung SK. 2005. Analysis of a highly flexible conformational immunogenic domain A in hepatitis C virus E2. J Virol 79:13199–13208. doi: 10.1128/JVI.79.21.13199-13208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SK. 2012. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog 8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keck ZY, Li TK, Xia J, Gal-Tanamy M, Olson O, Li SH, Patel AH, Ball JK, Lemon SM, Foung SK. 2008. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol 82:6061–6066. doi: 10.1128/JVI.02475-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, Xia J, Patel AH, Bukh J, Foung SK. 2013. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol 87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck ZY, Angus AG, Wang W, Lau P, Wang Y, Gatherer D, Patel AH, Foung SK. 2014. Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412-423. PLoS Pathog 10:e1004297. doi: 10.1371/journal.ppat.1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 18.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Omari K, Iourin O, Kadlec J, Sutton G, Harlos K, Grimes JM, Stuart DI. 2014. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat Commun 5:4874. doi: 10.1038/ncomms5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yagnik AT, Lahm A, Meola A, Roccasecca RM, Ercole BB, Nicosia A, Tramontano A. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355–366. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayak A, Pattabiraman N, Fadra N, Goldman R, Kosakovsky Pond SL, Mazumder R. 2015. Structure-function analysis of hepatitis C virus envelope glycoproteins E1 and E2. J Biomol Struct Dyn 33:1682–1694. doi: 10.1080/07391102.2014.967300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simons KT, Kooperberg C, Huang E, Baker D. 1997. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 26.Shapovalov MV, Dunbrack RL Jr. 2011. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender BJ, Cisneros A III, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, DiMaio F, Meiler J, Moretti R. 2016. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry 55:4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocquerel L, Op de Beeck A, Lambot M, Roussel J, Delgrange D, Pillez A, Wychowski C, Penin F, Dubuisson J. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J 21:2893–2902. doi: 10.1093/emboj/cdf295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Op De Beeck A, Montserret R, Duvet S, Cocquerel L, Cacan R, Barberot B, Le Maire M, Penin F, Dubuisson J. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J Biol Chem 275:31428–31437. doi: 10.1074/jbc.M003003200. [DOI] [PubMed] [Google Scholar]
- 30.Hess B, Kutzner C, van der Spoel D, Lindahl E. 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 31.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25. [Google Scholar]
- 32.Castelli M, Clementi N, Sautto GA, Pfaff J, Kahle KM, Barnes T, Doranz BJ, Dal Peraro M, Clementi M, Burioni R, Mancini N. 2014. HCV E2 core structures and mAbs: something is still missing. Drug Discov Today 19:1964–1970. doi: 10.1016/j.drudis.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falson P, Bartosch B, Alsaleh K, Tews BA, Loquet A, Ciczora Y, Riva L, Montigny C, Montpellier C, Duverlie G, Pecheur EI, le Maire M, Cosset FL, Dubuisson J, Penin F. 2015. Hepatitis C virus envelope glycoprotein E1 forms trimers at the surface of the virion. J Virol 89:10333–10346. doi: 10.1128/JVI.00991-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. 2002. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol 9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 35.Yi M, Nakamoto Y, Kaneko S, Yamashita T, Murakami S. 1997. Delineation of regions important for heteromeric association of hepatitis C virus E1 and E2. Virology 231:119–129. doi: 10.1006/viro.1997.8516. [DOI] [PubMed] [Google Scholar]
- 36.Michalak JP, Wychowski C, Choukhi A, Meunier JC, Ung S, Rice CM, Dubuisson J. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol 78(Part 9):2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 37.Patel J, Patel AH, McLauchlan J. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58–68. doi: 10.1006/viro.2000.0693. [DOI] [PubMed] [Google Scholar]
- 38.Albecka A, Montserret R, Krey T, Tarr AW, Diesis E, Ball JK, Descamps V, Duverlie G, Rey F, Penin F, Dubuisson J. 2011. Identification of new functional regions in hepatitis C virus envelope glycoprotein E2. J Virol 85:1777–1792. doi: 10.1128/JVI.02170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. 2013. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meunier JC, Engle RE, Faulk K, Zhao M, Bartosch B, Alter H, Emerson SU, Cosset FL, Purcell RH, Bukh J. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A 102:4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J, Bartenschlager R, Lavillette D, Cosset FL. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem 281:18285–18295. doi: 10.1074/jbc.M602706200. [DOI] [PubMed] [Google Scholar]
- 42.Voisset C, Op de Beeck A, Horellou P, Dreux M, Gustot T, Duverlie G, Cosset FL, Vu-Dac N, Dubuisson J. 2006. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J Gen Virol 87:2577–2581. doi: 10.1099/vir.0.81932-0. [DOI] [PubMed] [Google Scholar]
- 43.Flint M, Thomas JM, Maidens CM, Shotton C, Levy S, Barclay WS, McKeating JA. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol 73:6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Berna AJ, Moreno MR, Guillen J, Bernabeu A, Villalain J. 2006. The membrane-active regions of the hepatitis C virus E1 and E2 envelope glycoproteins. Biochemistry 45:3755–3768. doi: 10.1021/bi0523963. [DOI] [PubMed] [Google Scholar]
- 45.Drummer HE, Boo I, Poumbourios P. 2007. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol 88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 46.Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol 80:7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabahi A, Uprichard SL, Wimley WC, Dash S, Garry RF. 2014. Unexpected structural features of the hepatitis C virus envelope protein 2 ectodomain. J Virol 88:10280–10288. doi: 10.1128/JVI.00874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazumdar B, Banerjee A, Meyer K, Ray R. 2011. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology 54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JY, Acosta EG, Stoeck IK, Long G, Hiet MS, Mueller B, Fackler OT, Kallis S, Bartenschlager R. 2014. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol 88:12422–12437. doi: 10.1128/JVI.01660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spadaccini R, D'Errico G, D'Alessio V, Notomista E, Bianchi A, Merola M, Picone D. 2010. Structural characterization of the transmembrane proximal region of the hepatitis C virus E1 glycoprotein. Biochim Biophys Acta 1798:344–353. doi: 10.1016/j.bbamem.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Kortemme T, Baker D. 2002. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci U S A 99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kortemme T, Kim DE, Baker D. 2004. Computational alanine scanning of protein-protein interfaces. Sci STKE 2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 53.Vieyres G, Dubuisson J, Pietschmann T. 2014. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses 6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciczora Y, Callens N, Montpellier C, Bartosch B, Cosset FL, Op de Beeck A, Dubuisson J. 2005. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J Gen Virol 86:2793–2798. doi: 10.1099/vir.0.81140-0. [DOI] [PubMed] [Google Scholar]
- 55.Ciczora Y, Callens N, Penin F, Pecheur EI, Dubuisson J. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J Virol 81:2372–2381. doi: 10.1128/JVI.02198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummer HE, Poumbourios P. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J Biol Chem 279:30066–30072. doi: 10.1074/jbc.M405098200. [DOI] [PubMed] [Google Scholar]
- 58.Boo I, teWierik K, Douam F, Lavillette D, Poumbourios P, Drummer HE. 2012. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem J 443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- 59.McCaffrey K, Gouklani H, Boo I, Poumbourios P, Drummer HE. 2011. The variable regions of hepatitis C virus glycoprotein E2 have an essential structural role in glycoprotein assembly and virion infectivity. J Gen Virol 92:112–121. doi: 10.1099/vir.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 60.Rychlowska M, Owsianka AM, Foung SK, Dubuisson J, Bienkowska-Szewczyk K, Patel AH. 2011. Comprehensive linker-scanning mutagenesis of the hepatitis C virus E1 and E2 envelope glycoproteins reveals new structure-function relationships. J Gen Virol 92:2249–2261. doi: 10.1099/vir.0.034314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X, Qiao M, Atanasov I, Hu Z, Kato T, Liang TJ, Zhou ZH. 2007. Cryo-electron microscopy and three-dimensional reconstructions of hepatitis C virus particles. Virology 367:126–134. doi: 10.1016/j.virol.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 62.Callens N, Brugger B, Bonnafous P, Drobecq H, Gerl MJ, Krey T, Roman-Sosa G, Rumenapf T, Lambert O, Dubuisson J, Rouille Y. 2016. Morphology and molecular composition of purified bovine viral diarrhea virus envelope. PLoS Pathog 12:e1005476. doi: 10.1371/journal.ppat.1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piver E, Boyer A, Gaillard J, Bull A, Beaumont E, Roingeard P, Meunier JC. 11 October 2016. Ultrastructural organisation of HCV from the bloodstream of infected patients revealed by electron microscopy after specific immunocapture. Gut doi: 10.1136/gutjnl-2016-311726. [DOI] [PubMed] [Google Scholar]
- 64.Alhammad Y, Gu J, Boo I, Harrison D, McCaffrey K, Vietheer PT, Edwards S, Quinn C, Coulibaly F, Poumbourios P, Drummer HE. 2015. Monoclonal antibodies directed toward the hepatitis C virus glycoprotein E2 detect antigenic differences modulated by the N-terminal hypervariable region 1 (HVR1), HVR2, and intergenotypic variable region. J Virol 89:12245–12261. doi: 10.1128/JVI.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li HF, Huang CH, Ai LS, Chuang CK, Chen SS. 2009. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J Biomed Sci 16:89. doi: 10.1186/1423-0127-16-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol 84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houghton M. 2011. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev 239:99–108. doi: 10.1111/j.1600-065X.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 68.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong JA, Bhat R, Hockman D, Logan M, Chen C, Levin A, Frey SE, Belshe RB, Tyrrell DL, Law JL, Houghton M. 2014. Recombinant hepatitis C virus envelope glycoprotein vaccine elicits antibodies targeting multiple epitopes on the envelope glycoproteins associated with broad cross-neutralization. J Virol 88:14278–14288. doi: 10.1128/JVI.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Helle F, Vieyres G, Elkrief L, Popescu CI, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prentoe J, Verhoye L, Velazquez Moctezuma R, Buysschaert C, Farhoudi A, Wang R, Alter H, Meuleman P, Bukh J. 20 November 2015. HVR1-mediated antibody evasion of highly infectious in vivo adapted HCV in humanised mice. Gut doi: 10.1136/gutjnl-2015-310300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prentoe J, Velazquez-Moctezuma R, Foung SK, Law M, Bukh J. 9 August 2016. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology doi: 10.1002/hep.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kielian M. 2006. Class II virus membrane fusion proteins. Virology 344:38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 74.Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol 4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Modis Y. 2014. A novel membrane fusion protein family in Flaviviridae? Trends Microbiol 22:176–182. doi: 10.1016/j.tim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol 80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fraser J, Boo I, Poumbourios P, Drummer HE. 2011. Hepatitis C virus (HCV) envelope glycoproteins E1 and E2 contain reduced cysteine residues essential for virus entry. J Biol Chem 286:31984–31992. doi: 10.1074/jbc.M111.269605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallin M, Ekstrom M, Garoff H. 2006. Receptor-triggered but alkylation-arrested Env of murine leukemia virus reveals the transmembrane subunit in a prehairpin conformation. J Virol 80:9921–9925. doi: 10.1128/JVI.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li K, Zhang S, Kronqvist M, Wallin M, Ekstrom M, Derse D, Garoff H. 2008. Intersubunit disulfide isomerization controls membrane fusion of human T-cell leukemia virus Env. J Virol 82:7135–7143. doi: 10.1128/JVI.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan AG, Miller MT, Marcotrigiano J. 2015. HCV glycoprotein structures: what to expect from the unexpected. Curr Opin Virol 12:53–58. doi: 10.1016/j.coviro.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chemical Computing Group Inc. 2013. Molecular Operating Environment (MOE). Chemical Computing Group Inc., Montreal, QC, Canada. [Google Scholar]
- 82.Rohl CA, Strauss CE, Misura KM, Baker D. 2004. Protein structure prediction using Rosetta. Methods Enzymol 383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 83.Porter JR, Weitzner BD, Lange OF. 2015. A framework to simplify combined sampling strategies in Rosetta. PLoS One 10:e0138220. doi: 10.1371/journal.pone.0138220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 85.Karplus K, Barrett C, Hughey R. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 86.Karplus K, Karchin R, Barrett C, Tu S, Cline M, Diekhans M, Grate L, Casper J, Hughey R. 2001. What is the value added by human intervention in protein structure prediction? Proteins 45(Suppl 5):86–91. doi: 10.1002/prot.10021. [DOI] [PubMed] [Google Scholar]
- 87.Meiler J, Baker D. 2003. Coupled prediction of protein secondary and tertiary structure. Proc Natl Acad Sci U S A 100:12105–12110. doi: 10.1073/pnas.1831973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yarov-Yarovoy V, Schonbrun J, Baker D. 2006. Multipass membrane protein structure prediction using Rosetta. Proteins 62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barth P, Schonbrun J, Baker D. 2007. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci U S A 104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barth P, Wallner B, Baker D. 2009. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci U S A 106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- 92.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. 2003. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol 331:281–299. doi: 10.1016/S0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 93.Wassenaar TA, Pluhackova K, Moussatova A, Sengupta D, Marrink SJ, Tieleman DP, Bockmann RA. 2015. High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. J Chem Theory Comput 11:2278–2291. doi: 10.1021/ct5010092. [DOI] [PubMed] [Google Scholar]
- 94.Marrink SJ, de Vries AH, Mark AE. 2004. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B 108:750–760. doi: 10.1021/jp036508g. [DOI] [Google Scholar]
- 95.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. 2007. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 96.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. 2008. The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput 4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 97.Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 98.Periole X, Cavalli M, Marrink SJ, Ceruso MA. 2009. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J Chem Theory Comput 5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 99.Wassenaar TA, Ingolfsson HI, Bockmann RA, Tieleman DP, Marrink SJ. 2015. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 100.Wassenaar TA, Pluhackova K, Bockmann RA, Marrink SJ, Tieleman DP. 2014. Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theory Comput 10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 101.Wang C, Bradley P, Baker D. 2007. Protein-protein docking with backbone flexibility. J Mol Biol 373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 102.Logan M, Law J, Wong JA, Hockman D, Landi A, Chen C, Crawford K, Kundu J, Baldwin L, Johnson J, Dahiya A, LaChance G, Marcotrigiano J, Law M, Foung S, Tyrrell L, Houghton M. 19 October 2016. Native folding of a recombinant gpE1/gpE2 heterodimer vaccine antigen from a precursor protein fused with Fc IgG. J Virol doi: 10.1128/JVI.01552-16. [DOI] [PMC free article] [PubMed] [Google Scholar]