Abstract
MicroRNAs (miRNAs) are small single-stranded RNAs that bind to the 3’UTR of the mRNAs of target genes. They can target multiple genes and regulate translation or degradation of the mRNA. miRNAs target genes in a tissue-specific manner, and the role of a particular miRNA varies according to tumor origin or even subtype within the same cancer. This study evaluated the effect of miR-21 expression in triple-negative breast cancer (TNBC) tissues and MDA-MB-468, a cell line derived from TNBC tissues. miR-21 was consistently upregulated in TNBC and MDA-MB-468 cells compared to normal tissues. Inhibition of miR-21 by miR-21 antisense oligonucleotides decreased the proliferation, viability, and invasiveness of MDA-MB-468 cells and enhanced apoptosis. Furthermore, we confirmed that PTEN was downregulated by miR-21 in MDA-MB-468 cells. The results indicated that PTEN may mediate the oncogenic properties of miR-21 in TNBC. In summary, miR-21 was upregulated in TNBC tissues and cells, and promoted the proliferation and invasion of MDA-MB-468 cells, but negatively regulated the expression of PTEN protein. Inhibition of miR-21 or overexpression of PTEN protein could be promising strategies for the treatment of patients with TNBC.
Keywords: Triple-negative breast cancer, microRNA-21, PTEN, proliferation, migration
Introduction
Triple-negative breast cancer (TNBC) is defined clinically as a heterogeneous subtype with the absence of three most common types of receptors known to promote breast cancer growth - the estrogen receptor, the progesterone receptor (PR), and epidermal growth factor receptor 2 (HER2) [1,2]. The TNBC subtype represents approximately 10~15% of all breast cancers, with a 5-year survival rate of less than 30% [3,4]. This type of breast cancer is clinically aggressive with a high rate of distant recurrence, poor differentiation, and poor prognosis [5]. Based on a large population study, women under 50 years of age had a higher prevalence of TNBC [6]. Many systematic approaches, such as radiation, chemotherapy, and surgery, have been combined into regimens for treating patients with TNBC. However, eradication of the cancer is still difficult because of the absence of targets for anti-HER2 and endocrine therapies. Tremendous effort is being dedicated to elucidate and identify new actionable therapeutic targets for better treatment efficacy.
miRNAs are small non-coding RNAs typically containing 17-26 nucleotides, which play important roles in various biological processes via sequence-specific regulation of gene expression at the post-transcriptional level [7]. Briefly, miRNAs regulate biological functions by binding to the 3’-untranslated region (3’-UTR) of their target mRNAs, subsequently leading to mRNA degradation when the miRNA is completely complementary to the target regions, or translational inhibition when partially complementary [8]. Because miRNAs are short and the targeting of 3’-UTR regions is not strictly complementary, a single miRNA may have hundreds or thousands of target genes, some of which may be causative factors in tumorigenesis [9]. Previous studies showed that microRNA-21 (miR-21) is upregulated in a variety of solid tumors, such as lung cancer, prostate cancer, pancreatic endocrine tumors, hepatocellular carcinoma, glioblastoma, breast cancer, and stomach carcinomas [10]. Growing evidence supports an oncogenic role of miR-21 in inhibiting tumor cell apoptosis and contributing to cancer growth [11]. Although miR-21 has been proposed as a potential novel diagnostic biomarker for breast cancer, the results of related studies remain inconsistent. For example, a Chinese population-based study conducted by Wang et al. suggested circulating miR-21 as a potential biomarker because it had high sensitivity (up to 80.0%) and high specificity (87.7%) for breast cancer detection [12]. Whereas Gao et al. showed that the diagnostic accuracy of single miR-21 was much lower with only 25.8% sensitivity [13]. Nevertheless, the regulatory effect of miR-21 remains poorly understood and requires further study. Therefore, we conducted a systematic analysis to evaluate the diagnostic value of miR-21 in TNBC.
miR-21 was higher in TNBC and MDA-MB-468 cells than in normal tissues. Inhibition of miR-21 decreased the proliferation, cell viability and invasion capability of MDA-MB-468 cells and enhanced the apoptosis. Furthermore, we confirmed that PTEN can be targeted by miR-21 in MDA-MB-468, which leads to the downregulation of PTEN, indicating PTEN may serve as mediator of the oncogenic property of miR-21 in TNBC.
Materials and methods
Study population
The study design was approved by the Institutional Review Board of The Affiliated Zhongshan Hospital of Dalian University and written informed consent was obtained from all participants. TNBC tissues and adjacent normal tissues were collected from TNBC patients who were undergoing surgical resection at The Affiliated Zhongshan Hospital of Dalian University. Patients who met the following two criteria were included: 1) having had a history of chemotherapy, radiotherapy, or other treatments before the surgery; or 2) having other inflammatory diseases. From January 2011 to December 2012, 42 patients (22 males and 20 females) ranging from 25 to 67 years old were recruited. TNBC tissues and adjacent normal breast tissues (≥5 cm away from the tumor sites) were obtained after surgical resection and subsequently verified by two experienced pathologists in our hospital.
Cell lines and major reagents
The TNBC cell line MDA-MB-468 was purchased from the Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences. The following culture medium was used: fetal bovine serum (FBS), Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium (DMEM/F12). Lipofectamine™ 2000 (Gibco, Grand Island, NY, USA) was used for transfections. miRNA isolation and quantitative reverse transcription polymerase chain reaction (qRT-PCR) were performed with the TaqMan miRNA Isolation Kit, TaqMan microRNA Assay Kit, and TaqMan Universal PCR Master Mix (Applied Biosystems, Waltham, MA, USA). The miR-21 inhibitor and non-targeting sequences (negative control) were designed and synthesized by Santa Cruz Biotechnologies (Dallas, TX, USA). Primary antibodies for Western blot included rabbit anti-human PTEN polyclonal antibody and mouse anti-human β-actin monoclonal antibody (Invitrogen, Carlsbad, CA, USA). The secondary antibodies were purchased from Dakocytomation (Carpinteria, CA, USA) and protein quantification was performed with protein extraction and quantification kits (Bio-Rad, Hercules, CA, USA).
Cell culture
MDA-MB-468 cells were cultured in DMEM/F12 medium with 10% FBS and incubated in a humidified chamber at 37°C and 5% CO2. Cell growth curves were calculated by daily counts of cells under an inverted microscope. Cells were trypsinized at 70-80% confluence every 4 days and passaged in fresh medium.
qRT-PCR analysis for detection of miR-21 expression
MDA-MB-468 cells were seeded into 6-well plates at a density of 3×105 cells/ml in each well. The miR-21 inhibitor or negative control were transfected into cells using Lipofectamine™ 2000 (Invitrogen) following the manufacturer’s instructions. DMEM/F12 medium with 10% FBS then was added to the transfected cells and the untransfected control cells, and the cultures were incubated for 48 h. RNA was extracted from each sample using the TaqMan microRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR assays were performed using the Bio-Rad CFX-96 system and SYBR Premix ExTaq kit (Takara, Dalian, China). Endogenous U6 snRNA was used as an internal reference to allow comparison of miR-21 expression between samples.
MTT assay for cell viability detection
MDA-MB-468 cells were seeded into 96-well culture plates at a concentration of 3×105 cells/ml per well. MTT solution (0.5 mg/ml) was added into each well 48 hrs after seeding, followed by a 4-h incubation at 37°C and 5% CO2. Then, 100 μl sodium dodecyl sulfate (SDS) solution (20% concentration, 50% dimethyl formamide as the co-solvent) was added to each well and the plates were incubated for 24 h. Finally, OD570 values for each well were measured using a microplate reader (Bio-Tek, Winooski, VT, USA). In this study, each sample had six replicates, and every experiment was performed in triplicate.
Flow cytometry for measurement of cell proliferation
MDA-MB-468 cells were seeded into 6-well plates at a concentration of 3×105 cells/ml per well, washed twice with PBS 48 hrs after transfection, and then dissociated with trypsin. The dissociated cells were stained in the dark for 25 min at 4°C, then filtered to remove clumps. All of the treated cells were analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) and counted using CellQuest software (BD Biosciences). The cell proliferation capability was reflected by the proliferation index (PI), which was calculated by (S+G2M)/(G0G1+S+G2M) [5].
Flow cytometry for detection of cell apoptosis
Forty-eight hours after transfection with the miR-21 inhibitor or negative control, MDA-MB-468 cells were washed once or twice with phosphate-buffered saline (PBS) and incubated with Annexin V-FITC and propidium iodide staining solutions in the dark for 15 min at room temperature. Cell clumps were removed using mesh filters, and samples were analyzed by flow cytometry (BD Biosciences, USA). Cell counts were performed with CellQuest software (BD Biosciences), and data were analyzed using Macquit software.
Transwell assay to evaluate cell invasion capability
Twenty-four hours after transfection with the miR-21 inhibitor or negative control, MDA-MB-468 cells were seeded onto a Matrigel-coated membrane matrix (BD Biosciences) for another 24 h. After washing twice with PBS, the cells were stained with 0.1% crystal violet. Stained cells that invaded through the matrix and penetrated the polycarbonate membrane were counted in eight randomly selected fields under a microscope.
Verification of PTEN as a miR-21 target gene by luciferase assay
The luciferase reporter plasmid was constructed as follows: the pGL3-Luciferase vector (Shanghai GenePharma Co., Ltd., China) digested with Xba I and a chemically synthesized 3’ UTR sequence of the target gene PTEN was inserted that was complementary to the predicted miR-21 binding site. After uniformly seeding MDA-MB-468 cells into 6-well culture plates at a density of 3×105 cells per well, the cells were cotransfected with the luciferase reporter plasmid along with the miR-21 inhibitor or negative control. Cells from the cotransfected group and the untransfected control group were collected 48 h later and processed using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Reporter gene activity was measured using a microplate reader (E2920, Promega, USA).
Western blot for detection of PTEN expression
Total proteins extracted from each group of cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad) that had been pre-blocked in blocking solution for 1 h at room temperature. The blots were incubated with rabbit anti-human PTEN polyclonal antibody (1:500 dilution; Invitrogen) and mouse anti-human β-actin monoclonal antibody (1:1,000 dilution; Abcam, Cambridge, UK) overnight at 4°C. The blots then were incubated with their corresponding IRDye 800-labeled secondary antibodies (1:2000; LI-COR Biosciences, Lincoln, NE, USA) overnight at 4°C. PTEN/β-actin grayscale ratios were used to evaluate relative levels of PTEN, and the Quantity One software (Bio-Rad) was used for protein band analysis.
Statistical analysis
All statistical analysis was performed using SPSS 17.0 (SPSS, Chicago, Illinois, USA). Data were reported as means ± SD. Student’s t-test was used to compare the differences between two groups (two-tailed), and Analysis of Variance (ANOVA) was used to evaluate the differences among three or more groups. P-values less than 0.05 indicated statistically significant differences.
Results
miR-21 expression was upregulated in TNBC tissues and MDA-MB-468 cells
The relative expression of miR-21 in TNBC and MDA-MB-468 cells was measured by qRT-PCR. Results showed that the expression of miR-21 was higher in MDA-MB-468 cells and TNBC tissues than in the adjacent normal breast tissues (P<0.01; Figure 1). The consistently higher expression of miR-21 in TNBC and MDA-MB-468 cells suggested that miR-21 may play a role in oncogenesis.
Figure 1.
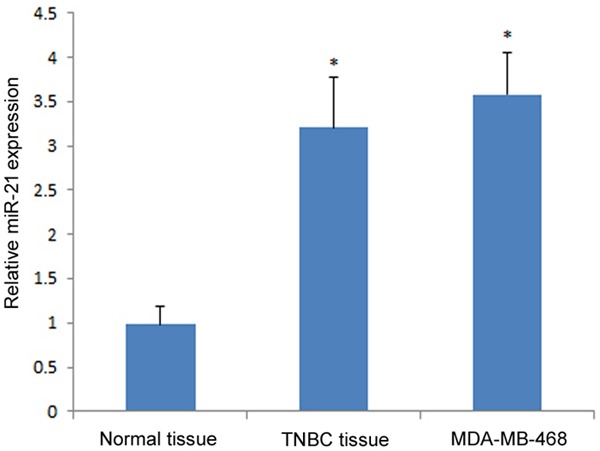
miR-21 expression in MDA-MB-468 cells and TNBC tissues. The relative miR-21 expression in both groups were compared with normal breast tissues. The asterisk *represents a significant difference between tumor tissue (or cell line) and normal tissue (P<0.05), and the error bar indicates the standard deviation.
Inhibition of miR-21 decreased cell viability and proliferation of MDA-MB-468
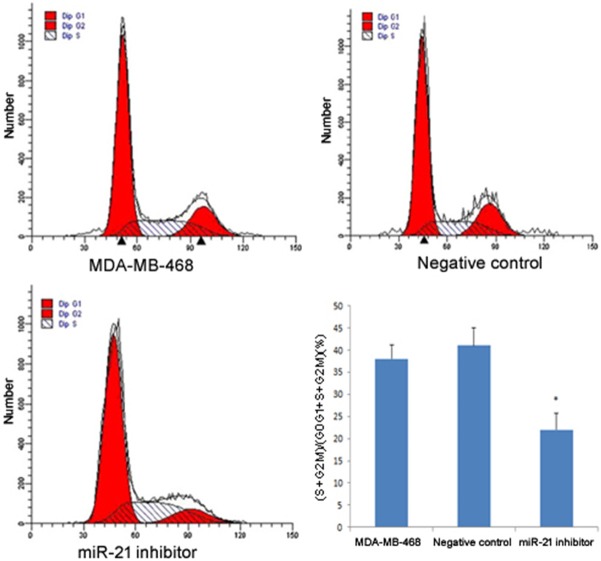
The effect of miR-21 on cell viability and proliferation was investigated by transfection of the miR-21 inhibitor into MDA-MB-468 cells. MTT assay results showed that the miR-21 inhibitor transfection group had reduced cell viability compared to the untransfected control group and the negative control group (P<0.05), which indicated that miR-21 promoted cell viability of MDA-MB-468 (Figure 3). Flow cytometry analysis showed similar results where the miR-21 inhibitor transfected group had a lower PI than the untransfected control group and the negative control group (P<0.05; Figure 4). There was no significant difference in PI between the untransfected control group and the negative control group.
Figure 3.
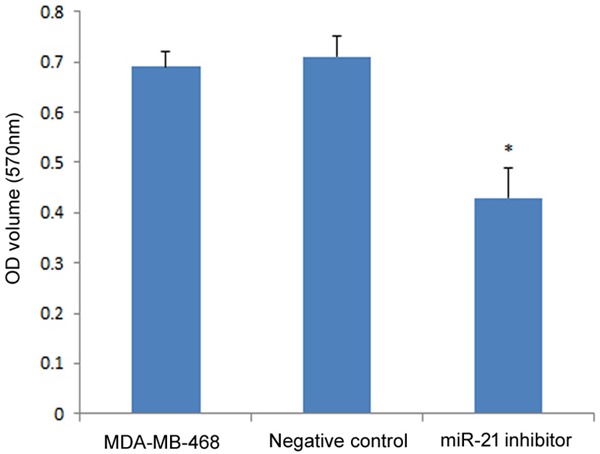
Effect of miR-21 on MDA-MB-468 cell viability measured by MTT assay. The OD values of both groups were compared to the negative control. The asterisk *represents a significant difference between the miR-21 inhibitor transfected group and normal tissue (P<0.05), and the error bars indicate standard deviation.
Figure 4.
Effect of miR-21 on cell proliferation by flow cytometry analysis. The histograms show the results of flow cytometry analysis; red represents the number of cells at the G0-G1 and G2-M phases, and the shadow represents the number of cells in S phase. The bar plot shows the proliferation index. Student’s t-test was used to compare the difference between the miR-21 inhibitor transfected group and negative control. The asterisk* indicates a significant difference (P<0.05), and the error bars indicate standard deviation.
Relative miR-21 expression in each group was measured by qRT-PCR. Compared to the untransfected control group (MDA-MB-468) and negative control group, miR-21 expression was significantly lower in the miR-21 inhibitor transfection group (P<0.01; Figure 2), which confirmed that the miR-21 inhibitor significantly suppressed miR-21 expression in MDA-MB-468 cells.
Figure 2.
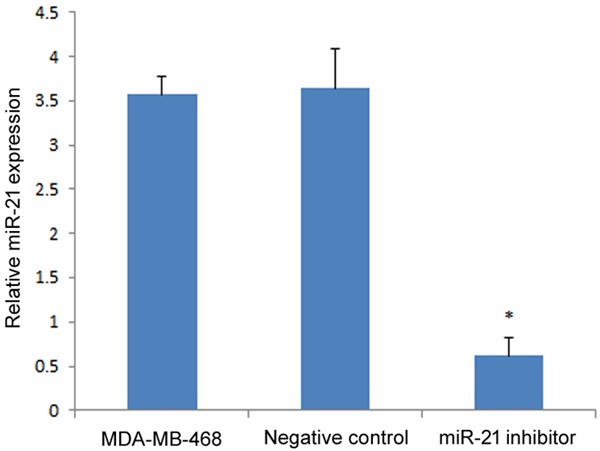
miR-21 expression in MDA-MB-468 cells after transfection with the miR-21 inhibitor. The relative expression of both groups were compared to the negative control group. The asterisk* represents a significant difference between the miR-21 inhibitor transfected group and negative control (P<0.05), and the error bars indicate the standard deviation.
Inhibition of miR-21 promoted cell apoptosis
Apoptosis is an important mechanism through which the number of cells are maintained in a stable status. However, apoptosis of cancer cells frequently is suppressed, which leads to their uncontrolled growth. To investigate whether miR-21 could suppress apoptosis of TNBC, the miR-21 inhibitor was transfected into MDA-MB-468 cells. Flow cytometry analysis showed that the miR-21 inhibitor transfection group had a significantly higher percentage of apoptotic cells compared to the untransfected control group and the negative control group (P<0.05; Figure 5), which suggested that inhibition of miR-21 expression could promote cell apoptosis.
Figure 5.
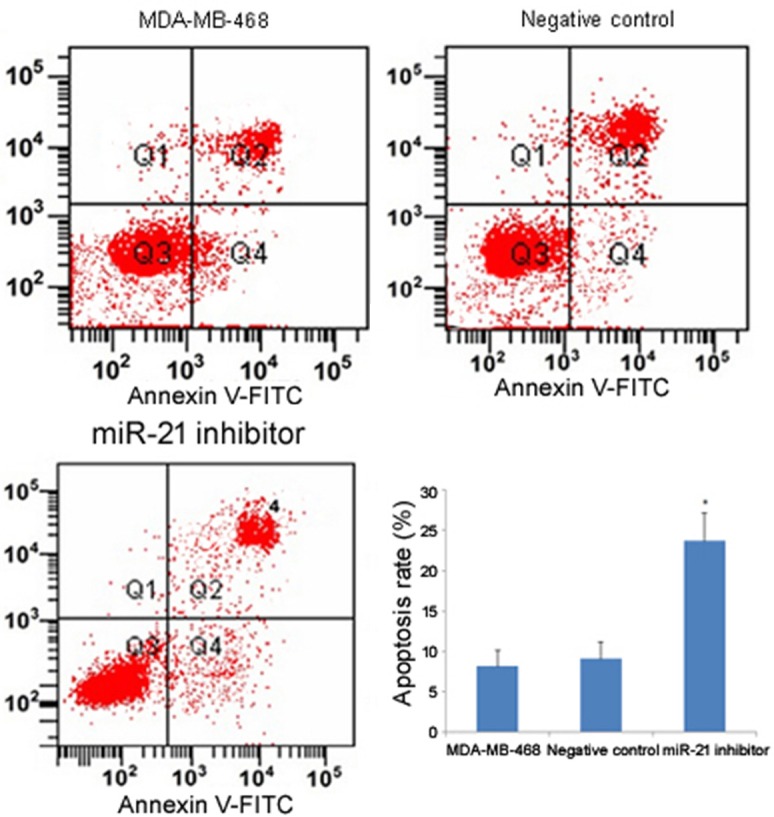
Effect of miR-21 on cell apoptosis by Annexin V staining and flow cytometry analysis. For the 2-D histograms, the upper left quadrant (Q1) represents necrosis, upper right (Q2) represents late apoptosis, and lower right (Q4) represents early apoptosis. The bar plot shows the apoptosis rate of each group. The asterisk *indicates a significant difference (P<0.05), and the error bars indicate standard deviation.
Inhibition of miR-21 suppressed cell invasion
Cancer cell invasion is an event that precedes metastasis, leading to more advanced stages of cancer that are refractory to many therapies. To investigate the role of miR-21 in cancer invasion, a transwell migration assay was performed. The transfection of the miR-21 inhibitor into MDA-MB-468 cells prevented the cells from penetrating the transwell chambers (P<0.05; Figure 6). This result indicated that cell invasiveness could be suppressed by inhibition of miR-21 expression.
Figure 6.
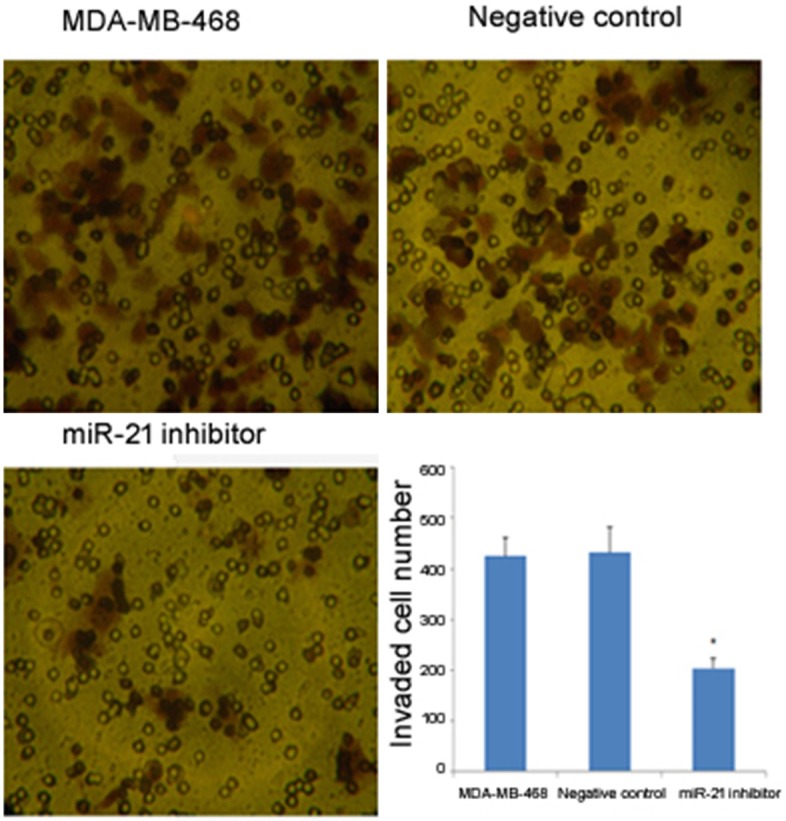
The effect of miR-21 on cell invasion detected by transwell migration assay. The photomicrographs illustrate the number of cells that penetrated through the membrane, which correlates with invasion capability. The bar plot shows the numbers of cells that penetrated through the membrane in the transwell migration assay. The asterisk* indicates a significant difference (P<0.05), and the error bars indicate standard deviation.
miR-21 functions through targeting PTEN
Given that miRNAs affect the expression of mRNA via binding to its 3’-UTR, the role of miR-21 is critically related to the function of target genes. However, because one miRNA may target multiple genes, it has been difficult to identify the effector. Because PTEN has frequently been reported as a target of miR-21, the role of miR-21 in PTEN expression in TNBC needs further study. Whether miR-21 could inhibit PTEN expression was examined by luciferase assay. The results showed that luciferase activity was significantly decreased in the miR-21 inhibitor transfected group compared to the untransfected control group and the negative control group (P<0.05; Figure 7). Moreover, PTEN protein expression was significantly upregulated in the miR-21 inhibitor transfected group compared to the two control groups (Figure 8), which suggested that the expression of PTEN could be upregulated by the inhibition of miR-21.
Figure 7.
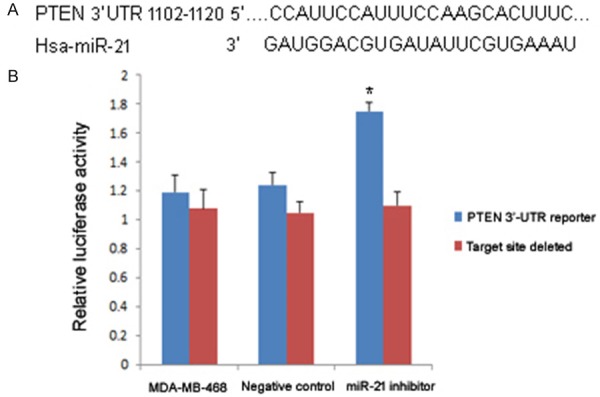
Relative luciferase activity after miR-21 inhibitor transfection. A: Schematic representation of the PTEN 3’-UTR showing the putative miR-21 target site; B: Relative luciferase activity of each group. The asterisk* indicates a significant difference (P<0.05), and the error bars indicate standard deviation.
Figure 8.
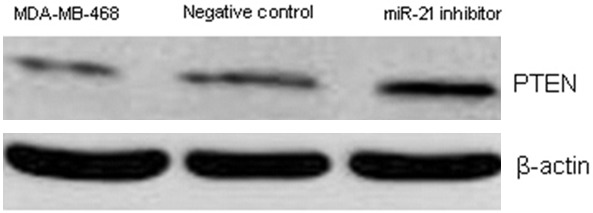
Comparison of PTEN protein expression after miR-21 inhibitor transfection by Western blot.
Discussion
Multiple lines of evidence have demonstrated that miR-21 is an important regulator of development and progression in a variety of tumors [14]. Because miRNAs are more resistant to RNAse, miR-21 was proposed as a potential biomarker for clinical diagnosis of TNBC [15]. Multiple reports indicated that anti-mRNA antisense oligonucleotides might be useful for biological therapy of clinical diseases. Song et al. reported that ASO against miR-21 modulated the radiosensitivity of cervical cancer cells [16]. Shang et al. showed that miR-21 ASO inhibited the proliferation of glioblastoma by downregulating miR-21 expression [17]. Consistently, miR-21 ASO used in this study downregulated the expression of miR-21 in TNBC cells. Furthermore, the viability and proliferation of TNBC cells were also significantly reduced. These results provided support a previous report from Dong et al., which demonstrated that downregulation of the miR-21 expression reduced the growth of TNBC cells [18]. These findings suggested that miR-21 could be a valuable therapeutic target in TNBC.
PTEN, regarded as a tumor suppressor gene, is involved in the regulation of many basic cellular functions, such as cell cycle progression, cell migration, cell invasion, and cell proliferation [19]. Downregulation of PTEN exerted a neuroprotective effect through enhancement of Akt activation or inhibition of NR2B-containing NMDA receptors [20]. Wang et al. reported that activation of the PTEN/PI3K/pAKT pathway was a potential predictor for positive prognosis in TNBC [21]. In this study, we found that miR-21 ASO inhibited the expression of PTEN and reduced the metastatic capability of TNBC cells. Furthermore, the transduction of AKT and ERK-signaling pathway were elevated, which was consistent with the results of Setia et al. [22]. Khotskaya et al. further demonstrated that inhibition of the Akt/mTOR/S6K1 axis inhibited the migration of TNBC cells [23]. In addition, there is evidence that PTEN is also involved in the regulation of VEGF expression, which is a well-known protein essential in carcinogenesis [24]. Based on these findings, we postulate that miR-21 ASO could reverse the expression of PTEN, which would directly influence the transduction of the AKT and ERK pathways.
Conclusions
In summary, the present study showed that miR-21 ASO effectively reduced the expression of miR-21, promoted the proliferation and invasion of MDA-MB-468 cells, and negatively regulated PTEN protein expression and AKT and ERK pathway transduction. These results suggest that miR-21 might be a potential therapeutic target that could be useful for predicting the clinical outcome of TNBC.
Disclosure of conflict of interest
None.
References
- 1.Sharma S, Barry M, Gallagher DJ, Kell M, Sacchini V. An overview of triple negative breast cancer for surgical oncologists. Surg Oncol. 2015;24:276–283. doi: 10.1016/j.suronc.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Fausto P, Barni S. Benefit of tamoxifen in estrogen receptor positive DCIS of the breast. Gland Surg. 2012;1:3–4. doi: 10.3978/j.issn.2227-684X.2012.04.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashid OM, Takabe K. The evolution of the role of surgery in the management of breast cancer lung metastasis. J Thorac Dis. 2012;4:420–424. doi: 10.3978/j.issn.2072-1439.2012.07.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Kahtani E, Xu Z, Al Rashaed S, Wu L, Mahale A, Tian J, Abboud EB, Ghazi NG, Kozak I, Gupta V, Arevalo JF, Duh EJ. Vitreous levels of placental growth factor correlate with activity of proliferative diabetic retinopathy and are not influenced by bevacizumab treatment. Eye (Lond) 2016 doi: 10.1038/eye.2016.246. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD, Pietenpol JA, Hortobagyi GN, Symmans WF, Ueno NT. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533–5540. doi: 10.1158/1078-0432.CCR-13-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B, Yu Q, Li H, Guo X, He X. Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int J Mol Med. 2014;33:43–50. doi: 10.3892/ijmm.2013.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia L, Zhang D, Qi X, Ma B, Xiang Z, He N. Identification of the conserved and novel miRNAs in Mulberry by high-throughput sequencing. PLoS One. 2014;9:e104409. doi: 10.1371/journal.pone.0104409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans. 2014;42:1135–1140. doi: 10.1042/BST20140142. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One. 2014;9:e100949. doi: 10.1371/journal.pone.0100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClure C, Brudecki L, Ferguson DA, Yao ZQ, Moorman JP, McCall CE, El Gazzar M. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect Immun. 2014;82:3816–3825. doi: 10.1128/IAI.01495-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138:1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Zhang Q, Xu J, Guo L, Li X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res. 2013;25:743–748. doi: 10.3978/j.issn.1000-9604.2013.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13:580–590. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millward-Sadler SJ, Costello PW, Freemont AJ, Hoyland JA. Regulation of catabolic gene expression in normal and degenerate human intervertebral disc cells: implications for the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2009;11:R65. doi: 10.1186/ar2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song L, Liu S, Zhang L, Yao H, Gao F, Xu D, Li Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016;37:12161–12168. doi: 10.1007/s13277-016-5073-3. [DOI] [PubMed] [Google Scholar]
- 17.Shang C, Guo Y, Hong Y, Liu YH, Xue YX. MiR-21 up-regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting FASLG. Mol Biol Rep. 2015;42:721–727. doi: 10.1007/s11033-014-3820-3. [DOI] [PubMed] [Google Scholar]
- 18.Dong G, Liang X, Wang D, Gao H, Wang L, Wang L, Liu J, Du Z. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31:57. doi: 10.1007/s12032-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Li Y, Wang P, Liang H, Cui M, Zhu M, Guo L, Su Q, Sun Y, McNutt MA, Yin Y. PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 2015;25:1189–1204. doi: 10.1038/cr.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teimourian S, Moghanloo E. Role of PTEN in neutrophil extracellular trap formation. Mol Immunol. 2015;66:319–324. doi: 10.1016/j.molimm.2015.03.251. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Zhang C, Chen K, Tang H, Tang J, Song C, Xie X. ERbeta1 inversely correlates with PTEN/PI3K/AKT pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res Treat. 2015;152:255–269. doi: 10.1007/s10549-015-3467-3. [DOI] [PubMed] [Google Scholar]
- 22.Yi YW, Hong W, Kang HJ, Kim HJ, Zhao W, Wang A, Seong YS, Bae I. Inhibition of the PI3K/AKT pathway potentiates cytotoxicity of EGFR kinase inhibitors in triple-negative breast cancer cells. J Cell Mol Med. 2013;17:648–656. doi: 10.1111/jcmm.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khotskaya YB, Goverdhan A, Shen J, Ponz-Sarvise M, Chang SS, Hsu MC, Wei Y, Xia W, Yu D, Hung MC. S6K1 promotes invasiveness of breast cancer cells in a model of metastasis of triple-negative breast cancer. Am J Transl Res. 2014;6:361–376. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H, Han C, Lu D, Wu T. miR-17-92 cluster promotes cholangiocarcinoma growth: evidence for PTEN as downstream target and IL-6/Stat3 as upstream activator. Am J Pathol. 2014;184:2828–2839. doi: 10.1016/j.ajpath.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]