Abstract
Radio-resistance becomes a large obstacle for effective cancer treatment. MicroRNAs (miRNAs) play important roles in response to radiation. However, the underlying mechanism of miR-630 on the radio-resistance of human glioma is less elucidated. In this study, we found that miR-630 was downregulated in glioma cell lines after radiation. MiR-630 inhibition enhanced the survival fraction, cell number in S stage and colony formation ability in glioma cells after radiation, while miR-630 overexpression resulted in inverse effects. By detecting the molecular mechanism of miR-630, we validated that CDC14A was a direct target of miR-630 and miR-630 suppressed CDC14A protein level. CDC14A overexpression can attenuate the inhibitory roles of miR-630 in survival fraction and cell proliferation. Finally, in vivo study demonstrated that miR-630 inhibition increased the volumes of xenografts bearing with glioma cells after radiation. In conclusion, our data indicate that anti-miR-630 enhances the radio-resistance of human glioma cells by targeting CDC14A, implying that miR-630 may act as a novel therapeutic target for enhancing the radiation efficiency on glioma patients.
Keywords: microRNA, miR-630, glioma, radio-resistance, CDC14A
Introduction
Glioma is the most common primary central nervous system tumor and accounts for the majority of malignant brain tumors in human [1,2]. The outcome of glioma is diverse due to the different histological types and grades. Radiotherapy is one of the most used methods for cancer treatment, including glioma [3,4]. However, glioma patients develop resistance to radiation, resulting in the low efficiency of radiotherapy. Therefore, understanding the underlying molecular mechanism of radio-resistance of glioma will be crucial for the development of novel target for glioma patients and the therapeutic strategies.
MicroRNAs (miRNAs) are a family of small, non-protein-coding RNA molecules that act as post-transcriptional regulators of gene expression through the direct binding to the 3’untranslated region (3’UTR) of target mRNAs, leading to the cleavage of target mRNAs or the repression of translation [5-8]. MiRNAs are involved in diverse biological processes, including cell proliferation and apoptosis [9]. Accumulating evidence indicates that miRNAs are deregulated in glioma and related to tumorigenesis [10-12]. Recent studies demonstrate that miRNAs are related to the radio-sensitivity of glioma cells. For instance, miR-181a increases the radio-sensitivity of glioma cells by downregulating Bcl-2 [13]. MiR-26a sensitizes glioblastoma cells to radiation by targeting of ataxia-telangiectasia mutated [14]. MiR-21 inhibition enhances the radio-sensitivity of glioma cells through the inhibition of PI3K/AKT pathway [15]. MiR-630 has been reported to be as a modulator of cisplatin-induced cell death [16,17]. It also serves as a prognostic biomarker of renal cell carcinoma [18] and gastric cancer [19]. However, the molecular mechanism as to how miR-630 modulates the radio-resistance of glioma cells is not fully understood.
Cell division cycle protein 14 A (CDC14A) gene is located in chromosome 1p21 and belongs to the dual specificity protein tyrosine phosphatase family. It is an important cell cycle regulatory phosphatase and affects the expression of cell-cycle related proteins including p53. CDC14A overexpression enhances the cell cycle progression and cell proliferation [20]. In addition, CDC14A plays an important role in regulating the cell cycle by downregulating CDC25 activity at the G2/M transition [21]. A previous study also demonstrates that CDC14A can interact and dephosphorate the ser315 of p53 in vivo and is involved in carcinogenesis [22].
In the current study, we found that miR-630 expression was reduced after radiation. MiR-630 inhibition increased the radio-resistance of glioma cells, G1/S transition and cell colony-forming ability, while miR-630 overexpression resulted in inverse effects. CDC14A was identified to be a direct target of miR-630 and CDC14A overexpression restored the effect of miR-630 on radio-sensitivity and proliferation. Finally, we found that miR-630 inhibition enhanced the radio-resistance of glioma cells in vivo. The results suggest that miR-630 may serve as a novel therapeutic target for modifying the efficiency of radiotherapy in glioma patients.
Materials and methods
Cell culture and transfection
Human glioma cell lines BT325, U373, U87 and U251 were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 1% mixture of penicillin (100 U/ml) and streptomycin (100 µg/ml), and maintained in a humidified atmosphere at 37°C with 5% CO2. The cell transfection was performed using LipofectamineTM 2000 (Invitrogen) according to the manufacturer’s instructions. MiR-630 mimics or miR-630 ASO (antisense oligonucleotides) were introduced into the cells at a final concentration of 50 nM and 100 nM, respectively.
RNA extraction and quantitative real time PCR (qPCR)
Total RNAs (inclusive of miRNAs) were extracted from cells or xenografts tissues using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was transcribed from 500 ng of RNA using M-MLV reverse transcriptase and specific miR-630 primer. qPCR was performed using specific miR-630 primer and 2 × SYBR master mix in the ABI Step One Real-Time PCR system. Small nuclear RNA U6 served as an internal control to normalize miR-410 expression. The primers for reverse transcription and PCR were listed below: miR-630 RT primer: 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACACCTTC-3’. U6 RT primer: 5’-GTCGTATCCAGTGCAGG GTCCGAGGTATTCGCACTGGATACGACAAAAATAT-3’. miR-630 Forward: 5’-GTCAGCGCAGTATTCTGTAC-3’. U6 Forward: 5’-GTCAGCGCGTGCTCGCTTCG-3’. Common reverse primer: 5’-GTGCAGGGTCCGAGGT-3’.
Western blotting assay
Western blotting analysis was performed in lysates from transfected cells or xenografts tissues to assess CDC14A protein levels. Briefly, 30 µg of proteins were separated on a 10% SDS-PAGE and subsequently transferred to a PVDF membrane. After incubation in 5% milk for 1 h, the blots were incubated in the primary antibodies overnight at 4°C. Mouse monoclonal to CDC14A and rabbit polyclonal to GAPDH were used as primary antibodies. Finally, the blots were incubated in HRP-conjugated goat anti-mouse or anti-rabbit antibody for 1.5 hrs at room temperature. The blots were observed using the ECL Western blotting kit according to the manufacturer’s protocols. GAPDH served as a loading control to normalize CDC14A expression.
Radiation treatment and radio-sensitivity assay
Radiation was performed at room temperature using an irradiator at various doses (0, 4, 8 and 12 Gy). For detection of the radio-sensitivity, the transfected cells were seeded at a density of 2000 cells per well in 96-well plates followed by radiation on day 2. After exposure to radiation for 3 days, the radio-sensitivity was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. The absorbance on 570 nm was measured using a spectrophotometer. The survival fraction = (the viability of the cells exposed to indicated dose of radiation)/(the viability of cells without radiation).
Colony formation assay
The transfected cells were seeded in 12-well plate (200 cells/well) and exposed to 8 Gy of radiation on day 2 for colony formation assay. During the experiment, the medium was refreshed every 3 days. After incubation for approximately 14 days, the colonies were stained with 1% crystal violet and a minimum of 50 cells were counted.
Cell cycle analysis
The cell cycle was determined using flow cytometry. Briefly, the transfected cells were harvested 12 hrs post-radiation with a dose of 8 Gy for the detection of cells in G1, S and G2 stages using the cell cycle analysis kit (Genscript) according to the manufacturer’s instructions. Finally, the number of cells in G1, S and G2 stages was analyzed using FACS Calibur (BD Biosciences).
Luciferase reporter assay
CDC14A3’UTR containing putative miR-630 binding sequence was amplified and inserted downstream of a luciferase reporter gene, named wild type pmirGLO/CDC14A 3’UTR. In addition, the mutated CDC14A 3’UTR was generated using the QuikChange Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. For the luciferase assay, the glioma cells were seeded in 24-well plates and then co-transfected with miR-630 mimics and wild type or mutant CDC14A 3’UTR reporter. The luciferase intensity was measured 48 hrs post-transfection using a Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. The expression of Renilla luciferase was used as an endogenous control.
In vivo animal study
In this study, the NOD/SCID mice were 6~8 week-old. All procedures during animal experiments were approved and performed by the Tianjin Medical University’s Institutional Animal care and the Use Committee. The subcutaneous tumor growth assay was performed under specific pathogen-free conditions. Briefly, 1×106 glioma cells transfected with miR-630 antagomir (Ribobio, 50 nM) or control were suspended in 100 μl of serum-free medium and subcutaneously injected into the back of nude mice. The tumor volumes were measured by a slide caliper every 5 days and calculated using the formula (Length×Width 2)/2. To detect the tumor radio-sensitivity in vivo, the tumors were exposed to radiation with a dose of 8 Gy 10 days after injection. Mice were killed 30 days after injection. The xenografts were imaged and then harvested for the detection of miR-630 and CDC14A expression.
Statistical analysis
All data were tested in triplicates and expressed as mean ± SD of values from at least three independent experiments. The statistical analysis was performed using the GraphPad Prism5 software and differences between two groups were tested using two-tailed Student’s t-test. The value of p<0.05 was considered to be statistically significant.
Results
MiR-630 is downregulated after radiation
To investigate the potential roles of miR-630 in glioma radio-sensitivity, we assessed miR-630 expression in a panel of glioma cell lines BT325, U87, U373 and U251 [23], some of which were reported to have radio-sensitivity. As shown in Figure 1A, we found that U87 and U373 had a lower level of miR-630 expression. In addition, we treated the above cells with 8 Gy of radiation and analyzed miR-630 expression. We found that miR-630 was decreased after radiation treatment compared to control (Figure 1B), suggesting that miR-630 may be involved in the cell response to radiation. We then detected the radio-sensitivity of BT325, U87, U373 and U251 cells without transfection of miR-630 using a MTT assay. As shown in Figure 1C, U87 and U373 cells had a higher survival fraction than U215 and BT325 cells. Moreover, we treated U87 and U373 cells with increasing doses of radiation and discovered that miR-630 was lower (Figure 1D). These results indicate that miR-630 expression is downregulated after radiation and its level in those cell lines may be positively correlated with their radio-sensitivity.
Figure 1.
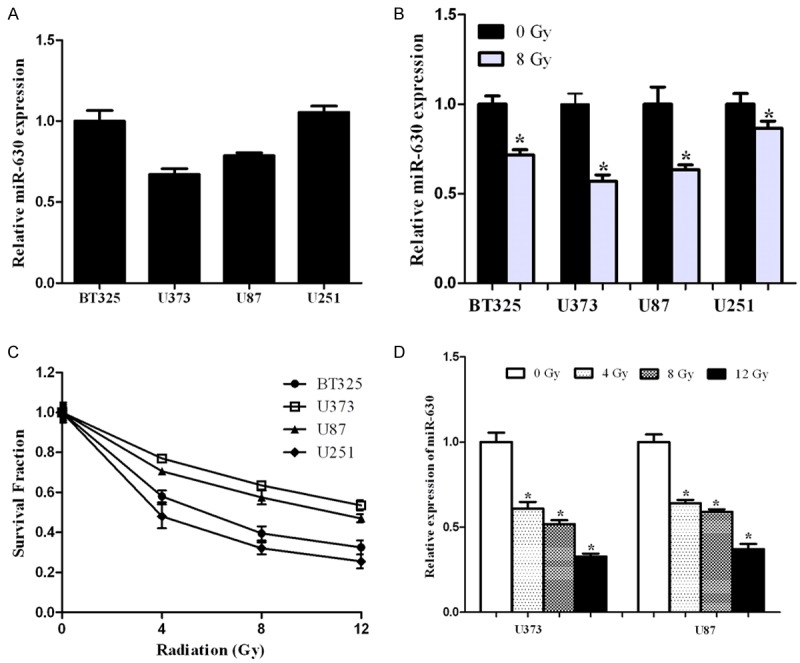
miR-630 was downregulated after radiation. A. miR-630 expression in BT325, U373, U87 and U251 cells was determined by qPCR. B. BT325, U373, U87 and U251 cells were exposed to 8 Gy of radiation and miR-630 expression was then determined by qPCR. C. BT325, U373, U87 and U251 cells were exposed to various doses of radiation (0, 4, 8 and 12 Gy) after transfection with miR-630 or control. The radio-sensitivity was then analyzed using MTT assay. D. BT325, U373, U87 and U251 cells were exposed to various doses of radiation (0, 4, 8 and 12 Gy) after transfection, and miR-630 expression was then determined by qPCR. U6 was used as an internal control. *P<0.05.
MiR-630 modulates the glioma cell radio-sensitivity and cell proliferation
We inhibited or overexpressed miR-630 by transient transfection with miR-630 ASO or miR-630 mimics, respectively. The cells were exposed to various doses of radiation and survival fraction was determined. As shown in Figure 2A, we found that the inhibition of miR-630 increased the survival fraction of U373 cells compared to control group, while miR-630 overexpression decreased the survival fraction. Similar results were observed in U87 cells (Figure 2B). Considering that cell cycle stage exerts important roles in the cell response to radio-sensitivity [24], we tested the effect of miR-630 on cell cycle after radiation using flow cytometry. As shown in Figure 2C and 2D, miR-630 inhibition increased the number of cells in S stage compared to control group.
Figure 2.
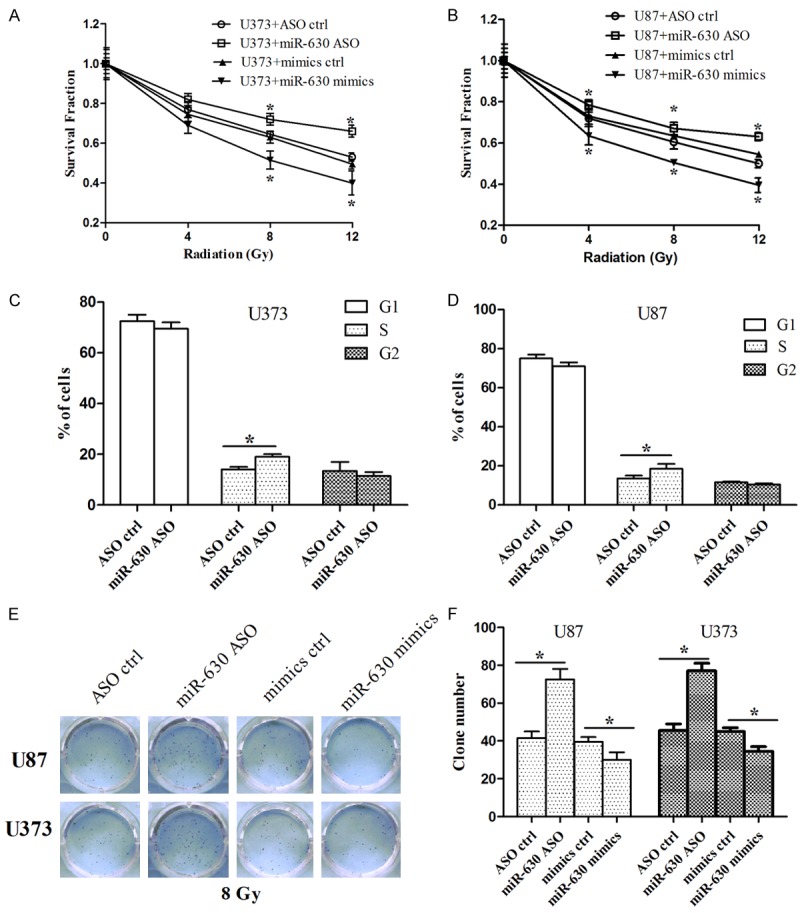
miR-630 modulated the radio-sensitivity, G1/S progression and colony formation in glioma cells after radiation. A, B. U373 and U87 cells were exposed to 8 Gy of radiation after transfection with miR-630 mimics or miR-630 ASO, and radio-sensitivity was then analyzed using MTT assay. C, D. U373 and U87 cells were exposed to 8 Gy of radiation after transfection with miR-630 ASO, and number of cells in G1, S and G2 stage was measured using flow cytometry. D, E. U373 and U87 cells were exposed to 8 Gy of radiation after transfection with miR-630 mimics or miR-630 ASO, and cell proliferation was then analyzed using colony formation assay. *P<0.05.
In addition, we performed colony formation assay to detect the effect of miR-630 on cell proliferation after radiation, which is considered as a canonical method to determine radio-sensitivity [25]. As shown in Figure 2E and 2F, we found that miR-630 inhibition increased the colony number of U87 and U373 cells, while miR-630 overexpression resulted in opposite effects. Taken together, these results indicate that inhibition of miR-630 enhanced the radio-resistance of glioma cells by promoting G1/S transition and colony-forming ability.
CDC14A is a direct target of miR-630
By using the miRanda and TargetScan algorithms, we chose CDC14A as a putative target of miR-630 (Figure 3A). We found that CDC14A protein level was reduced after the cells were transfected with miR-630 mimics compared to control group (Figure 3B). To validate the direct interaction between miR-630 and CDC14A, a luciferase reporter construct was created. The CDC14A 3’UTR containing miR-630 binding site was subcloned downstream of a luciferase reporter gene in parallel with a mutated CDC14A 3’UTR shown in Figure 3A. The cells were co-transfected with miR-630 and either wild type or mutated CDC14A 3’UTR reporter, and subjected to luciferase assay. As shown in Figure 3C, we found that miR-630 reduced the luciferase intensity of CDC14A 3’UTR, while a mutation within the binding site abolished the inhibitory role of miR-630 in CDC14A 3’UTR. Taken together, these results suggest that miR-630 downregulates CDC14A expression through the direct binding to its 3’UTR.
Figure 3.
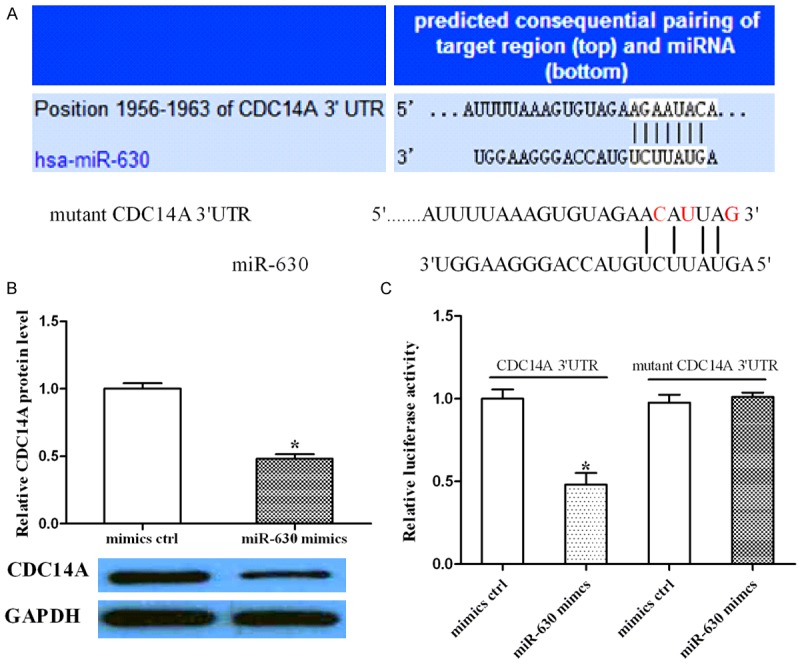
miR-630 downregulated CDC14A expression via direct binding to the 3’UTR of CDC14A. A. Sequence alignment of CDC14A 3’UTR and miR-630. A mutation was generated within the binding sites (red). B. The cells transfected with miR-630 mimics or control were subjected to Western blotting to analyze CDC14A expression. GAPDH was used as a loading control. C. The cells were co-transfected with miR-630 and wild type or mutated CDC14A 3’UTR, and luciferase assay was performed to assess the effect of miR-630 on CDC14A 3’UTR intensity. *P<0.05.
Restoration of CDC14A expression attenuates the effect of miR-630 on radio-sensitivity and cell proliferation
To determine whether the effect of miR-630 on radio-sensitivity and cell proliferation was mediated by the suppression of CDC14A, we co-transfected U373 or U87 cells with miR-630 and the CDC14A-overexpressed vector. CDC14A expression was confirmed by Western blotting assay (Figure 4A, 4B). MTT assay showed that overexpression of CDC14A increased the survival fraction that was inhibited by miR-630 (Figure 4C, 4D). Cell cycle analysis and colony formation assay suggested that CDC14A overexpression increased the number of cells in S stage and colony number that were suppressed by miR-630, respectively (Figure 4E, 4H). Similar results were shown in U373 and U87 cells. These data indicate that miR-630 modulates cell radio-sensitivity and proliferation by downregulating CDC14A.
Figure 4.
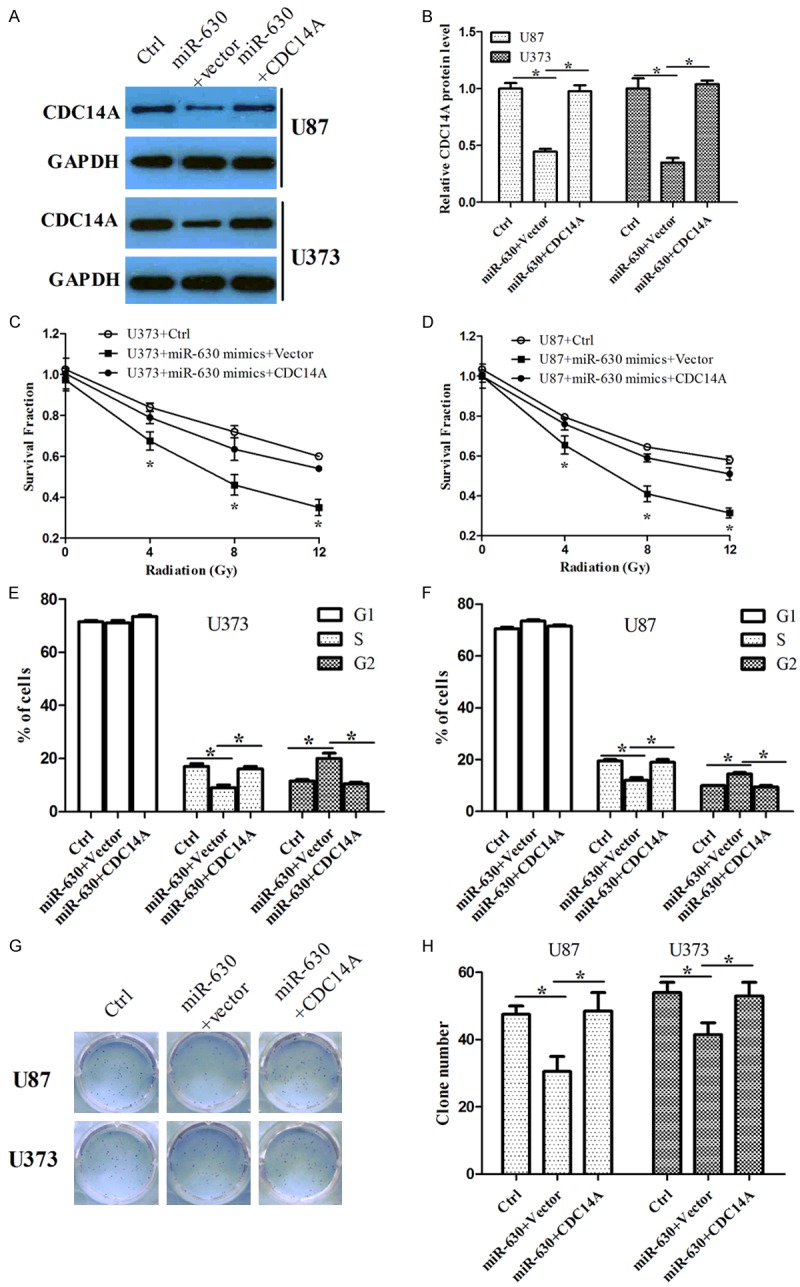
Overexpression of CDC14A attenuated the inhibitory roles of miR-630 in cell response to radiation, G1/S progression and colony formation in glioma cells after radiation. (A, B) U387 and U87 cells were transfected either with miR-630 alone or miR-630 and empty vector together or miR-630 and CDC14A-overexpressed vector, and CDC14A protein level was analyzed using Western blotting assay. GAPDH was used as a loading control. The graph (B) represented the relative CDC14A protein level. (C, D) U387 and U87 cells were exposed to 8 Gy of radiation after transfection either with miR-630 alone or miR-630 and empty vector or miR-630 and CDC14A-overexpressed vector, and radio-sensitivity was then analyzed using MTT assay. (E, F) Cell cycle analysis was determined in cells treated as above using flow cytometry. (G, H) Cell proliferation was analyzed in cells treated as above using colony formation assay. *P<0.05.
MiR-630 inhibition enhances the radio-resistance of glioma cells in vivo
Finally, we tried to determine whether miR-630 inhibition enhances the radio-resistance of glioma cells in vivo. The subcutaneous tumor was exposed to 8 Gy of radiation at 10 days after injection with miR-630 antagomir (an inhibitor of endogenous miR-630) or control. The volumes of xenografts were measured every 5 days until the mice were killed at day 30. As shown in Figure 5A and 5B, we found that the volumes of xenografts derived from cells with miR-630 antagomir were larger than that of control group. In addition, we quantified miR-630 expression in xenografts tissues and found that miR-630 expression was significantly reduced in xenografts from miR-630 antagomir-transfected cells (Figure 5C), indicating that the injection in the mice was convincible. CDC14A expression was also examined by Western blotting assay, suggesting that CDC14A protein level was higher in the xenografts tissues derived from the cells with miR-630 antagomir compared to that of control group (Figure 5D). Taken together, these data indicate that miR-630 inhibition enhances the radio-resistance of glioma cells in vivo by upregulating CDC14A.
Figure 5.
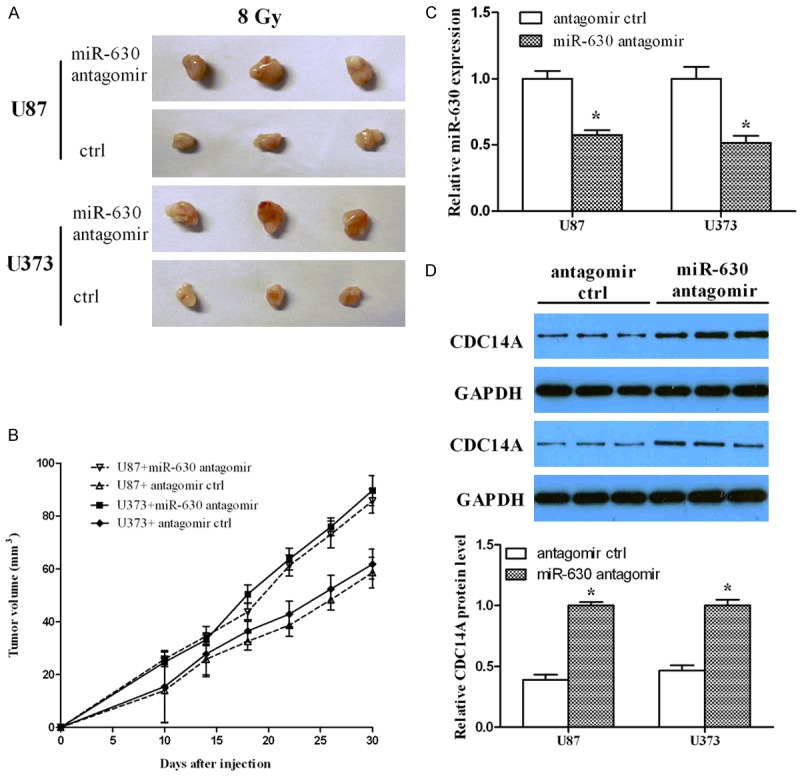
miR-630 inhibition enhanced radio-resistance of glioma cells in vivo. (A) Nude mice were subcutaneously injected with miR-630 antagomir or control, and radiation was delivered to tumors 10 days post-injection. The images showed the xenografts when the mice were killed on day 30. (B) The volume curve of xenografts treated as (A). (C) miR-630 expression was measured using qPCR in xenografts generated by IR and either miR-630 antagomir or control. (D) CDC14A expression was measured using Western blotting in xenografts generated by radiation and either miR-630 antagomir or control. *P<0.05.
Discussion
Although radio-resistance is a challenging obstacle for the effective therapy of glioma, it is still routinely used in glioma clinical treatment. Therefore, it is urgent to determine the underlying mechanism as to how glioma cells survive radiation. Accumulation evidence has demonstrated that aberrant miRNA expression is related to the cell response to radiation treatment in diverse cancers. The inhibition of miR-221 enhances the radio-sensitivity of colorectal carcinoma cells by targeting PTEN [26]. MiR-124 sensitizes colorectal cancer cells to radiation by downregulating PRRX1 [27]. The direct suppression of ARRB1 and TXN1 by miR-525-3p is required for cell survival under radiation [28]. These studies indicate that miRNAs may serve as therapeutic targets for modifying the efficiency of radiotherapy.
In the current study, we found that miR-630 was downregulated in glioma cells after radiation treatment. The in vitro studies showed that anti-miR-630 enhanced the survival fraction of glioma cells, increased the number of cells in S stage and colony-forming ability after radiation treatment. In contrast, miR-630 overexpression sensitized the glioma cells to radiation and suppressed cell proliferation. In addition, the in vivo study showed that anti-miR-630 could enhance the radio-resistance of xenografts bearing with glioma cells. These findings are in line with the previous study that miR-630 is downregulated in human colorectal cancer after radiation and miR-630 increases the radio-sensitivity by inducing cell death and apoptosis through targeting BCL2L2 and TP53RK [29]. In addition, a previous study has shown that the cell cycle regulators (such as p53 and p21) and cell cycle phase play an important role in determining radio-sensitivity; meanwhile, the cells in G2/M phase are most radio-sensitive, cells in G1 phase are less sensitive, and cells in S phase are least sensitive [30]. In our study, we also detected the roles of miR-630 on p53 and p21/WAF1 expression (Supplementary Figure 1) and found that miR-630 inhibition did not affect total p53 expression, but suppressed p53 phosphorylation. miR-630 inhibition also suppressed p21/WAF1 expression, while miR-630 overexpression resulted in opposite effects. Therefore, we conclude that miR-630 inhibition enhances the radio-resistance of glioma cells by influencing the cell cycle phase and cell cycle regulators. However, a previous study demonstrates that miR-630 is upregulated in radio-resistant cervical cancer cells; miR-630 overexpression increases the survival fraction of irradiated cells and inhibition of miR-630 reverses the cell radio-resistance [31]. The different roles of miR-630 in response to radiation in glioma cells and cervical cancer cells may be due to the different targets of miR-630.
Next, we validated that CDC14A was a direct target of miR-630. The luciferase assay showed that miR-630 reduced the luciferase intensity of CDC14A 3’UTR, but the inhibitory effect of miR-630 on CDC14A 3’UTR was abrogated when the binding sites between miR-630 and CDC14A were mutated. In addition, Western blotting assay showed that CDC14A protein level decreased when the cells were transfected with miR-630. CDC14A expression was increased after radiation (Supplementary Figure 1). Our study found that CDC14A overexpression could increase the survival fraction, the number of cells in S stage and colony number that were inhibited by miR-630 after radiation treatment. Importantly, the in vivo study also showed that CDC14A was upregulated in the xenografts tissues that had a lower miR-630 level. A previous study discovers that human CDC14A can interact with and dephosphorylates the ser315 site of the tumor suppressor p53 [32], suggesting that CDC14A may regulate the functions of p53, an inducer of cell cycle arrest. The localization of CDC14A in centrosome makes it closed to some cell cycle kinases and their substrates such as Cdk1 [33] and Cdk2 [34], indicating CDC14A may be involved in the regulation of cell cycle progression. Moreover, researchers have found that CDC14A expression in human tumor cell lines is inversely related to p53, and CDC14A may inactivate p53 and regulate Cdk/cyclin B [35]. All these results demonstrate that CDC14A may regulate cell cycle through the regulation of p53. In accord with the findings in the previous studies, our study showed that CDC14A silencing did not affect p53 expression, but increased p53 phosphorylation. CDC14A silencing also increased p21/WAF1 expression. However, miR-630 inhibition suppressed the above effects induced by CDC14A knockdown (Supplementary Figure 1).
In conclusion, our study indicates that miR-630 was reduced under radiation treatment and anti-miR-630 enhanced the radio-resistance of glioma cells by directly targeting CDC14A. The results suggest that miR-630 may serve as a therapeutic target for modifying the efficiency of radiotherapy in glioma patients.
Acknowledgements
The study was supported by the Tianjin Technology Program (No. 14ZCDZSY00037).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng J, Li P, Zhang Q, Yang Z, Fu S. A radiosensitivity gene signature in predicting glioma prognostic via EMT pathway. Oncotarget. 2014;5:4683–4693. doi: 10.18632/oncotarget.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen CL, Romner B. Current treatment of low grade astrocytoma: a review. Clin Neurol Neurosurg. 2013;115:1–8. doi: 10.1016/j.clineuro.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Zhang Y, Skalski M, Hayes J, Kefas B, Schiff D, Purow B, Parsons S, Lawler S, Abounader R. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 2014;74:1541–1553. doi: 10.1158/0008-5472.CAN-13-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma LN, Xu SL, Yang L, Wang QL, Dang WQ, Cui W, Yu SC, Ping YF, Cui YH, Kung HF, Qian C, Zhang X, Bian XW. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34:1407–1419. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 12.Guo M, Jiang Z, Zhang X, Lu D, Ha AD, Sun J, Du W, Wu Z, Hu L, Khadarian K, Shen J, Lin Z. miR-656 inhibits glioma tumorigenesis through repression of BMPR1A. Carcinogenesis. 2014;35:1698–1706. doi: 10.1093/carcin/bgu030. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23:997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 14.Guo P, Lan J, Ge J, Nie Q, Guo L, Qiu Y, Mao Q. MiR-26a enhances the radiosensitivity of glioblastoma multiforme cells through targeting of ataxia-telangiectasia mutated. Exp Cell Res. 2014;320:200–208. doi: 10.1016/j.yexcr.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, Kim JH, Yin J, Yoo H, Lee SH, Park JB. Silencing of MicroRNA-21 Confers Radio-Sensitivity through Inhibition of the PI3K/AKT Pathway and Enhancing Autophagy in Malignant Glioma Cell Lines. PLoS One. 2012;7:e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, Kroemer G. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 17.Cao JX, Li SY, An GS, Mao ZB, Jia HT, Ni JH. E2F1-regulated DROSHA promotes miR-630 biosynthesis in cisplatin-exposed cancer cells. Biochem Biophys Res Commun. 2014;450:470–475. doi: 10.1016/j.bbrc.2014.05.138. [DOI] [PubMed] [Google Scholar]
- 18.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. Up-regulation of miR-630 in clear cell renal cell carcinoma is associated with lower overall survival. Int J Clin Exp Pathol. 2014;7:3318–3323. [PMC free article] [PubMed] [Google Scholar]
- 19.Chu D, Zhao Z, Li Y, Li J, Zheng J, Wang W, Zhao Q, Ji G. Increased microRNA-630 expression in gastric cancer is associated with poor overall survival. PLoS One. 2014;9:e90526. doi: 10.1371/journal.pone.0090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su J, Zhou H, Tao Y, Guo Z, Zhang S, Zhang Y, Huang Y, Tang Y, Hu R, Dong Q. HCdc14A is involved in cell cycle regulation of human brain vascular endothelial cells following injury induced by high glucose, free fatty acids and hypoxia. Cell Signal. 2015;27:47–60. doi: 10.1016/j.cellsig.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Sacristan MP, Ovejero S, Bueno A. Human Cdc14A becomes a cell cycle gene in controlling Cdk1 activity at the G(2)/M transition. Cell Cycle. 2011;10:387–391. doi: 10.4161/cc.10.3.14643. [DOI] [PubMed] [Google Scholar]
- 22.Paulsen MT, Starks AM, Derheimer FA, Hanasoge S, Li L, Dixon JE, Ljungman M. The p53-targeting human phosphatase hCdc14A interacts with the Cdk1/cyclin B complex and is differentially expressed in human cancers. Mol Cancer. 2006;5:25. doi: 10.1186/1476-4598-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Wang Y, Sheng K, Fei X, Guo Q, Larner J, Kong X, Qiu Y, Mi J. Serine/threonine protein phosphatase 6 modulates the radiation sensitivity of glioblastoma. Cell Death Dis. 2011;2:e241. doi: 10.1038/cddis.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Yaromina A, Krause M, Thames H, Rosner A, Hessel F, Grenman R, Zips D, Baumann M. Pre-treatment number of clonogenic cells and their radiosensitivity are major determinants of local tumour control after fractionated irradiation. Radiother Oncol. 2007;83:304–310. doi: 10.1016/j.radonc.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ, Li GX. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World J Gastroenterol. 2013;19:9307–9317. doi: 10.3748/wjg.v19.i48.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Zheng L, Huang J, Gao F, Lin X, He L, Li D, Li Z, Ding Y, Chen L. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9:e93917. doi: 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraemer A, Barjaktarovic Z, Sarioglu H, Winkler K, Eckardt-Schupp F, Tapio S, Atkinson MJ, Moertl S. Cell survival following radiation exposure requires miR-525-3p mediated suppression of ARRB1 and TXN1. PLoS One. 2013;8:e77484. doi: 10.1371/journal.pone.0077484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J, Li G. Novel Epigenetic CREB-miR-630 Signaling Axis Regulates Radiosensitivity in Colorectal Cancer. PLoS One. 2015;10:e0133870. doi: 10.1371/journal.pone.0133870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B, Chen J, Ren Z, Chen Y, Li J, Miao X, Song Y, Zhao T, Li Y, Shi Y, Ren D, Liu J. A specific miRNA signature promotes radioresistance of human cervical cancer cells. Cancer Cell Int. 2013;13:118. doi: 10.1186/1475-2867-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Ljungman M, Dixon JE. The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. J Biol Chem. 2000;275:2410–2414. doi: 10.1074/jbc.275.4.2410. [DOI] [PubMed] [Google Scholar]
- 33.Pockwinse SM, Krockmalnic G, Doxsey SJ, Nickerson J, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Penman S. Cell cycle independent interaction of CDC2 with the centrosome, which is associated with the nuclear matrix-intermediate filament scaffold. Proc Natl Acad Sci U S A. 1997;94:3022–3027. doi: 10.1073/pnas.94.7.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 35.Paulsen MT, Starks AM, Derheimer FA, Hanasoge S, Li L, Dixon JE, Ljungman M. The p53-targeting human phosphatase hCdc14A interacts with the Cdk1/cyclin B complex and is differentially expressed in human cancers. Mol Cancer. 2006;5:25. doi: 10.1186/1476-4598-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


