Abstract
APP/PS1 transgenic mice with Alzheimer disease (AD) are widely used as a reliable animal model in studies about behaviors, physiology, biochemistry and histomorphology of AD, but few studies have been conducted to investigate the role of lncRNAs in this model. In this study, lncRNA microarray was employed to detect the gene expression profile and lncRNA expression profile in the mouse brain. Then, bioinformatics was used to predict the differentially expressed genes related to AD (n=20). Among different lncRNAs (n=249), 99 were downregulated and 150 upregulated. Co-expression network was applied to analyze the co-expression of differential lncRNAs and different genes. In network, lncRNA Gm13498 and lncRNA 1700030L20Rik correlated with the most genes and their degrees were 6 and 5, respectively. Then, the function and signal transduction pathways related to the differentially co-expressed lncRNAs were analyzed with bioinformatics, and results showed that these lncRNAs were involved in the systemic development of neurons, intercellular communication, regulation of action potential of neurons, development and differentiation of oligodendrocytes, neurotransmitters transmission, and neuronal regeneration. Realtime PCR was employed to detect the expression of relevant lncRNAs and differentially expressed RNAs in 10 samples, and results were consistent with above findings from microarray.
Keywords: APP/PS1 transgenic mouse, Alzheimer’s disease, long non-coding RNA, gene microarray, RE1 protein silencing transcription factor
Introduction
The pathogenesis of Alzheimer’s disease (AD) is very complex and involving several nervous dysfunctions. β-amyloid protein (Aβ) deposition in the brain is a key event in the pathogenesis of AD [1,2]. The cleavage of amyloid precursor protein (APP) by endogenous beta-secretase may cause the binding between transmembrane segment and extracellular segment to form Aβ42 peptide [3,4]. Presenilin1 (PS1) has been found to be associated with AD. High expressions of APP and PS1 have been observed in AD [5,6]. Jackson’s laboratory prepared APP/PS1 transgenic mice via APPA and PS1 co-transfection. These mice at an age of 9-12 months developed behavioral, physiological, biochemical and histomorphological features similar to those in AD patients. To date, these transgenic mice have been widely used in studies about AD worldwide [7].
Long non-coding RNA (lncRNA) refers to non-protein coding transcripts longer than 200 nucleotides and has functions of dosage compensation, epigenetic modification and protein complex backbone [8-10]. LncRNA expression and/or dysfunction are closely related to human diseases. The abnormalities of lncRNAs in their sequence, spatial structure, expression and interaction with proteins have been found to play important roles in the pathogenesis of AD [11-13]. In the present study, lncRNA expression profile was investigated with Agilent Mouse lncRNA Microarray V2.0 in the hippocampus of APP/PS1 transgenic mice with AD and wide type C57 mice, and bioinformatics was employed for the analysis of differentially expressed genes. Furthermore, real time PCR was used to validate the expressions of relevant lncRNAs and genes in these mice.
Materials and methods
Ethics, consent and permissions
The Ethics Committee of Shanghai Tenth People’s Hospital approved this study, and international guidelines for animal welfare were followed.
Animal grouping and morphological identification
APP/PS1 transgenic AD mice and wide type C57 mice were purchased from the Animal Center of Nanjing, housed in separated cages (n=5 per cage) at 25°C, and given ad libitum access to water and food with 12 h/12 h light/dark cycle. The degenerative neuropathy was evaluated in 12 months-old animals by water maze test. Once behavioral changes were observed, AD mice (n=3) and wide type littermates were randomly selected for the identification by immunochemistry. The remaining mice were anesthetized, and then perfused with 4% paraformaldehyde. The brain was collected and fixed in 4% paraformaldehyde over night. After dehydration in 10%, 20% and 30% sucrose, brain tissues were embedded in OCT and cut into sections. Immunohistochemistry was performed for Aβ (abcam ab2539, CA, USA) and phosphorylated Tau (abcam ab52834, CA, USA).
Morris water maze test
The Morris water maze was 120 cm in diameter and 47 cm in height with a 9 cm platform. The water was 0.5 cm higher than the platform, and the water temperature was maintained at 25.0±0.5°C. Test was divided into hidden-platform acquisition training and probe trial testing. The hidden-platform acquisition training was employed to test the learning and memory of mice in the maze and conducted for 6 days. Before test, mice were allowed to swim freely for 2 min. The hidden platform was placed at the center of a quadrant. The location of this platform remained unchanged in the whole test. Mice were randomly placed in any quadrant with the head forward the wall. When the mouse reached the platform or stayed on the plat form for 60 s, the test was stopped. After the mouse reached the platform, it was allowed to stay on the platform for 10 s. If the mouse failed to find the platform within 60 s, the investigator guided the mouse to the hidden platform and stay on the platform for 10 s. Each mouse received training 4 times every day with an interval of 30 min between two trainings. The escape latency (time to reaching the platform) was recorded by reviewing the video. The probe trial testing was employed to evaluate the maintenance of memory. On day 7, the platform was removed, and the mouse was placed in any quadrant and then allowed to swim for 60 s in water. The duration of staying the target quadrant (TQ) was calculated within 60 s.
LncRNA microarray assay
After Morris water maze test, animals were sacrificed, and the hippocampus was collected and grounded. Total RNA was extracted and processed for lncRNA microarray assay (AD: n=3; control: n=3).
Screening of differentially expressed lncRNAs and mRNAs
LncRNAs and mRNAs with fold change of >2 and with significant differences between them (P<0.05) were defined as differentially expressed genes.
Co-expression network analysis of lncRNAs and mRNAs
The regulation network of lncRNA regulatory genes was analyzed according to the passon correlation coefficient of genes and lncRNAs. The co-expression relationship between lncRNAs and genes could be used to establish the adjacency matrix (A=[aij], where aij refers to the weight of relationship between gene i and lncRNAj) between lncRNAs and genes. In the regulation network, genes were expressed as circles, lncRNA as triangles, and interactions as sides. The network center was expressed as the rank which refers to the contribution of a specific lncRNA to the surrounding genes or the contribution of a specific gene to the surrounding transcription factors. Core lncRNA has the highest rank in the network.
GO and pathway analysis
GO analysis was applied to analyze the main functions of the differential expression genes according to the Gene Ontology which is the key functional classification of NCBI. Generally, Fisher’s exact test and χ 2 test were used to classify the GO category, and the false discovery rate (FDR) was calculated to correct the P value. The smaller the FDR, the small the error in judging the P value. FDR was defined as FDR = 1-Nk/T, where Nk refers to the difference between P value of Fisher’s test and the P value of χ 2 test. P values for the GOs of all the different genes were calculated. Enrichment provides a measure of the significance of the function: as the enrichment increases, the corresponding function is more specific, which helps us to find those GOs with more concrete function description in the experiment. Within the significant category, the enrichment Re was given by: Re=(nf/n)/(Nf/N), where nf is the number of differential genes within the particular category, n is the total number of genes within the same category, Nf is the number of differential genes in the entire microarray, and N is the total number of genes in the microarray.
Similarly, Pathway analysis was used to find out the significant pathway of the differential genes according to KEGG, Biocarta and Reatome. Still, we turned to the Fisher’s exact test and χ 2 test to select the significant pathway, and the threshold of significance was defined by P-value and FDR. The enrichment Re was calculated like the equation above [14-16].
Real time PCR
Total RNA was extracted from the left hippocampus with 1 ml of Trizol reagent, and genomic DNA was digested with 1 μl of RQ1 DNase buffer. Then, 3 μg of total RNA was used for reverse transcription according to the manufacturer’s instructions (SuperScript II reverse transcriptase; Invitrogen). Real time PCR was conducted with TaKaRa SYBR Premix Ex Taq and 7500 real-time PCR instrument (Applied Biosystems) according to the manufacturer’s instructions. Primer 5.0 was used for primer design. Related primers are listed in Table 1.
Table 1.
Premars of related genes in realtime PCR
| Sequence (5’→3’) | Length | Tm | Location | ||
|---|---|---|---|---|---|
| APP | Forward Primer | TCCGAGAGGTGTGCTCTGAA | 20 | 62.4 | 860-879 |
| Reverse Primer | CCACATCCGCCGTAAAAGAATG | 22 | 61.8 | 974-953 | |
| PSEN1 | Forward Primer | TGCACCTTTGTCCTACTTCCA | 21 | 61 | 15-35 |
| Reverse Primer | GCTCAGGGTTGTCAAGTCTCTG | 22 | 62.5 | 145-124 | |
| REST | Forward Primer | GGCAGATGGCCGAATTGATG | 20 | 61.5 | 212-231 |
| Reverse Primer | CTTTGAGGTCAGCCGACTCT | 20 | 61.2 | 298-279 | |
| HHIP | Forward Primer | GAAGATGCTCTCGTTTAAGCTGC | 23 | 61.8 | 6-28 |
| Reverse Primer | CCACCACACAGGATCTCTCC | 20 | 61.3 | 212-193 | |
| CNTN2 | Forward Primer | TTGGACCCGTCTTTGAAGAGC | 21 | 62.3 | 110-130 |
| Reverse Primer | TACTGGGTTAGAGGCTAGGCA | 21 | 61.5 | 357-337 | |
| APOC2 | Forward Primer | ATGGGGTCTCGGTTCTTCCT | 20 | 62.2 | 2-21 |
| Reverse Primer | GTCTTCTGGTACAGGTCTTTGG | 22 | 60 | 176-155 | |
| GAPDH | Forward Primer | AGGTCGGTGTGAACGGATTTG | 21 | 62.6 | 8-28 |
| Reverse Primer | GGGGTCGTTGATGGCAACA | 19 | 62.6 | 102-84 | |
| GM13498 | Forward Primer | GACCCGTCAGGGACCAAAAC | 20 | 62.7 | 163-182 |
| Reverse Primer | AACGGTAAGGAATCACGATGTG | 22 | 60.4 | 349-328 | |
| 1700030L20RIK | Forward Primer | GGTGGCTGTTTTATGTCCCAA | 21 | 60.5 | 774-794 |
| Reverse Primer | CAACCACACCATTGTTGAGGA | 21 | 60.4 | 885-865 | |
| AK038159 | Forward Primer | CAGGTCTTCTTCAAACAACTGCT | 23 | 60.9 | 470-492 |
| Reverse Primer | TGCTTTCTCGGGAAGTCTGGA | 21 | 62.9 | 560-540 | |
| DQ113493 | Forward Primer | TGTCTGTGCGAGATGCAAC | 19 | 60.4 | 1514-1532 |
| Reverse Primer | CCATAGTGGGGTCATGCGAG | 20 | 62.1 | 1621-1602 | |
| U6 | Forward Primer | ACCCTGAGAAATACCCTCACAT | 22 | 60.2 | 140-161 |
| Reverse Primer | GACGACTGAGCCCCTGATG | 19 | 61.8 | 201-183 |
Statistical analysis
Quantitative data are expressed as mean ± standard deviation. The latency in Morris water maze test was compared by using t test, and the apoptosis index with chi square test. A value of P<0.05 was considered statistically significant.
Results
Identification of APP/PS1 transgenic AD mice
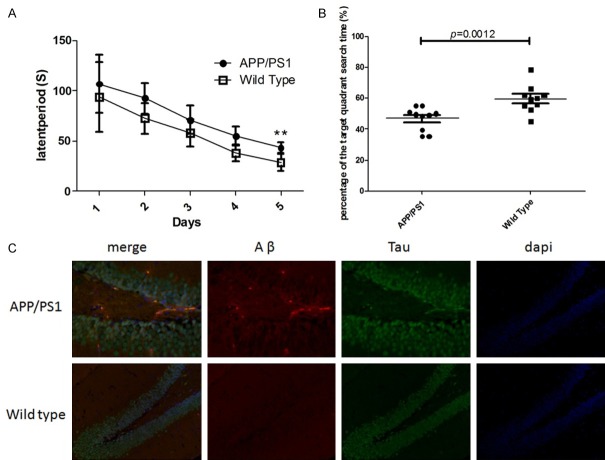
When the mice were 12 months old, these mice were subjected to Morris water maze test. As shown in Figure 1A and 1B, the escape latency was 28.6±8.3 s in wide type group and 43.6±5.4 s in APP/PS1 group, showing significant difference between two groups (P<0.01). It is suggested that the APP/PS1 transgenic mice had learning and memory impairment. In addition, the duration of staying in target quadrant was also markedly different between two groups (46.9±2.8 s in APP/PS1 group and 59.7±3.6 s wide type group). To further confirm the AD in APP/PS1 transgenic mice, 3 mice were randomly selected from each group, and pathological examination of the brain was performed. The brain was fixed in 4% paraformaldehyde, and the brain sections were subjected to immunohistochemistry for Aβ and phosphorylated Tau. As shown in Figure 1C, there was evident for Aβ deposition in the brain of APP/PS1 transgenic mice (red fluorescence), but it was not obvious in wide type Littermates. In addition, the change in phosphorylated Tau (green fluorescence) was similar to that of Aβ deposition. These indicated that the APP/PS1 transgenic mice aged 12 months might develop AD spontaneously.
Figure 1.
Identification of AD in APP/PS1 transgenic mice. A: The escape latency of APP/PS1 transgenic mice and wide type littermates in Morris water maze test; B: Duration of staying in the target quadrant; C: Immunofluorescence staining of Aβ and phosphorylated Tau in the brain. Data are shown as mean ± SD, **P<0.01 vs. wild type group.
Differentially expressed lncRNAs and differentially expressed genes
The Mouse lncRNA Microarray v2.0 4×180 K microarray of Agilent G3 platform was used to detect the lncRNA expression profile in two groups, and a total of 249 differentially expressed lncRNAs were identified, of whom 99 had up-regulated expression and 150 displayed down-regulated expression. Then, a hotmap was constructed with these differentially expressed lncRNAs (Figure 2A). In addition, it was also found 209 had up-regulated expression and 242 displayed down-regulated expression (Figure 2A) among 451 differentially expressed genes. These lncRNAs and mRNAs could clearly separate one group from another. Then, the bioinformatics analysis was employed to confirm some important lncRNAs. Their information is shown in Table 2. In addition, the important mRNAs were also analyzed, and their information is shown in Table 3. To validate the changes in above lncRNAs and mRNAs expression, real time PCR was employed to detect their expression in addition samples (n=10). As shown in Figure 2C and 2D, the mRNA expression of Six3, Rest and Shh reduced significantly in AD mice as compared with wild type mice (expression ratio of AD mice to wild type mice: 0.04±0.03, 0.08±0.03 and 0.4±0.14, respectively; P<0.01 or 0.05), but the mRNA expression of Apoc2, Klf5 and Mef2c increased marked in AD mice as compared with wild type mice (ratio: 2.48±0.72, 2.63±0.31 and 2.83±0.58, respectively; P<0.01 or 0.05). In addition, real time PCR was also performed to detect the expression of lncRNA Gm13498, DQ113493, AK038159 and 1700030L20Rik, and results were similar to those from microarray (ratio: 2.87±0.8, 0.36±0.1, 0.24±0.03 and 2.75±0.96, respectively).
Figure 2.
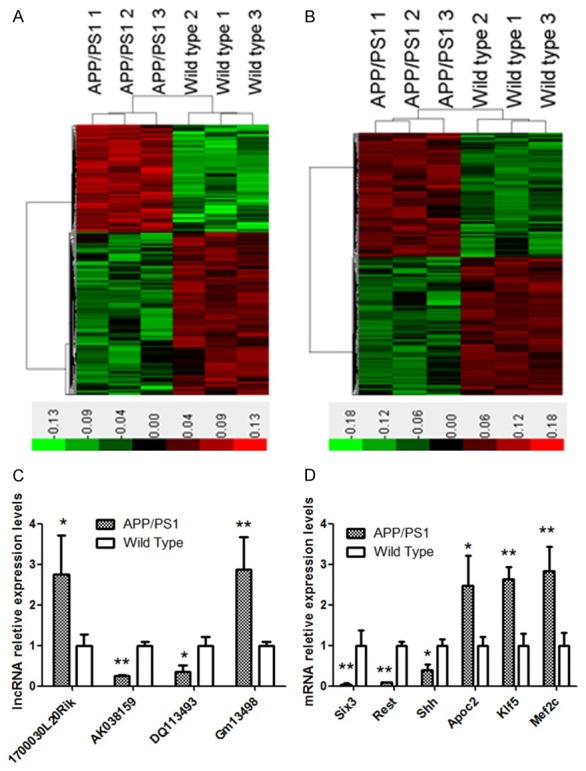
LncRNA and mRNA expression profiles of the hippocampus of APP/PS1 transgenic mice. A: Hotmap analysis of differentially expressed lncRNAs; B: Hotmap analysis of differentially expressed mRNA; C: Real time PCR for lncRNAs; D: Real time PCR for mRNA. Data are shown as mean ± SD, *P<0.05 vs. wild type group, **P<0.01 vs. wild type group, Student’s t-test.
Table 2.
Differentially expressed lncRNAs in the hippocampus between APP/PS1 transgenic mice and wild type mice
| LncRNA | P value | Fold change | Chr No. | Position | Start | End | Strand | Length |
|---|---|---|---|---|---|---|---|---|
| Gm13498 | 0.026709 | 1.107295 | 2 | 2qC1.1 | 50,909,684 | 50,911,846 | + | 2163 |
| 1700030L20Rik | 0.03086 | 1.088938 | 3 | 3qG3 | 136,435,270 | 136,449,349 | - | 985 |
| AK038159 | 0.000148 | -2.04013 | 9 | 9qA5.1 | 40,820,969 | 40,822,673 | + | 1705 |
| DQ113493 | 0.015561 | -1.00214 | 12 | 12qF1 | 114,357,761 | 114,358,218 | - | 458 |
Table 3.
Differentially expressed mRNAs in hippocampus of APP/PS1 transgenic mice with Alzheimer disease
| Gene symbol | Genbank accession | P value | Fold change | Description |
|---|---|---|---|---|
| Res0t | NM_011263 | 6.82E-06 | -3.54924 | RE1-silencing transcription factor |
| Shh | NM_009170 | 0.018668 | -2.6385 | Sonic hedgehog |
| Six3 | NM_011381 | 0.022093 | -2.49683 | Sine oculis-related homeobox 3 |
| Slitrk6 | NM_175499 | 0.024647 | -2.00858 | SLIT and NTRK-like family 6 |
| Ret | NM_001080780 | 0.028781 | -1.89211 | Ret proto-oncogene |
| Hhip | NM_020259 | 0.005445 | -1.65801 | Hedgehog-interacting protein |
| Gjc3 | NM_080450 | 0.0347360 | -1.46313 | Gap junction protein |
| Fos | NM_010234 | 0.034798 | -1.44827 | FBJ osteosarcoma oncogene |
| Foxp2 | AK164319 | 0.006651 | -1.31022 | Forkhead box P2 |
| Insc | NM_173767 | 0.040053 | -1.30909 | Inscuteable homolog |
| Cntn2 | NM_177129 | 0.045533 | -1.0722 | Contactin 2 |
| Drd2 | NM_010077 | 0.01017 | -1.06875 | Dopamine receptor D2 |
| Klf5 | NM_009769 | 0.012230 | 1.123472 | Kruppel-like factor 5 |
| Efnb2 | NM_010111 | 0.018935 | 1.149162 | Ephrin B2 |
| Apoc2 | NM_001277944 | 0.026465 | 1.168242 | Apolipoprotein C-II |
| Rbp4 | NM_001159487 | 0.011893 | 1.310413 | Retinol binding protein 4 |
| Cwh43 | NM_181323 | 0.031735 | 1.356818 | Cell wall biogenesis 43 C-terminal homolog |
| Plxnd1 | NM_026376 | 0.012214 | 1.462215 | Plexin D1 |
| Figf | NM_010216 | 0.019358 | 1.635167 | C-fos induced growth factor |
| Mef2c | NM_025282 | 0.031811 | 1.696655 | Myocyte enhancer factor 2C |
Coexpression analysis of lncRNAs and AD related genes
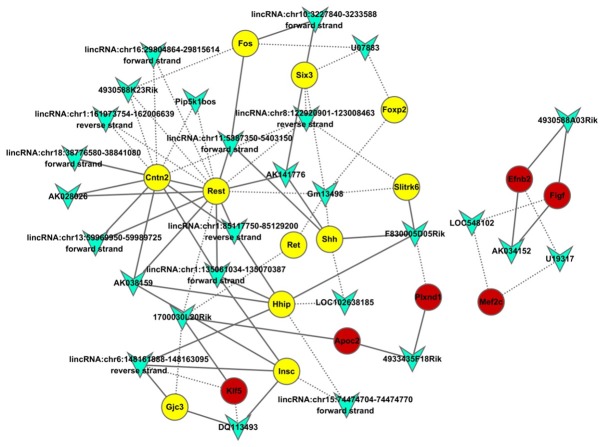
According to the passon correlation coefficient between lncRNAs and AD related genes, lncRNAs with a coefficient of > ±0.95 were used to construct regulation network of lncRNA regulatory genes. As shown in Figure 3, the number of genes coexpressed with REST gene was the largest, the degree was 15 and they were the most important genes in the network. In the network, lncRNA Gm13498 and lncRNA 1700030L20Rik had the widest relation with other genes and the degree was 6 and 5, respectively. In the figure, triangle referred to lncRNA, circle to AD related genes, yellow to down-regulation in wide type mice, red to up-regulation in wide type mice, the solid line to the positive relationship and the dotted line to the negative relationship.
Figure 3.
Co-expression analysis of differentially expressed lncRNAs and differentially expressed mRNA. The passon correlation coefficient between differentially expressed lncRNAs and differentially expressed mRNA was calculated to construct the co-expression network of differentially expressed lncRNAs and differentially expressed mRNA. The circles represent differentially expressed mRNA (yellow: down-regulated; red: up-regulated). Polygons represent differentially expressed lncRNAs. Connection line: co-expression between differentially expressed lncRNAs and differentially expressed mRNA. Solid line: positive correlation; dotted line: negative correlation.
Functional prediction of differentially expressed lncRNAs
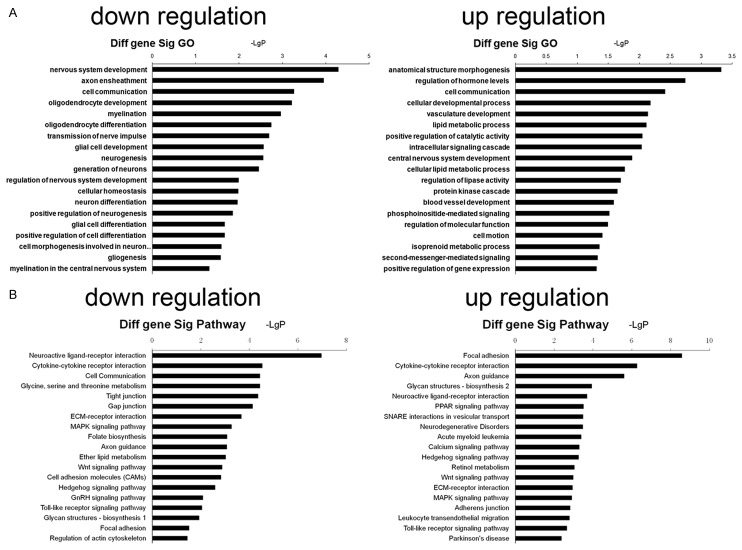
LncRNAs belong to non-coding RNA SIMILAR TO miRNA. They may affect the expression or activity of proteins via regulating target genes to regulate the biological processes. However, the ways in which lncRNA regulates target genes are different from that of miRNA, and several ways have been identified for lncRNA regulating target genes. Of note, the lncRNA and its target genes may form the co-expression relationship. Thus, the functions and pathways of genes that are co-expressed with differentially expressed lncRNA are helpful for the investigation of the potential functions and pathways of these lncRNA. The co-expressed genes were subjected to GO and pathway analysis (Figure 4). Results showed that the GO of genes with downregulation mainly related to the nervous system development, axon ensheathment, cell communication, oligodendrocyte development, myelination, oligodendrocyte differentiation, transmission of nerve impulse, glial cell development, neurogenesis, generation of neurons, regulation of nervous system development, cellular homeostasis, neuron differentiation, positive regulation of neurogenesis, and glial cell differentiation, and GO of genes with upregulation were mainly associated with cell communication, vasculature development, lipid metabolic process and others. In addition, the pathways related to genes with down-regulated expression included neuroactive ligand-receptor interaction, cytokine-cytokine receptor interaction, cell communication, glycine, serine and threonine metabolism, gap junction, ECM-receptor interaction, MAPK signaling pathway, axon guidance, Wnt signaling pathway and regulation of actin cytoskeleton, and those of genes with up-regulated expression mainly included focal adhesion, cytokine-cytokine receptor interaction, calcium signaling pathway, MAPK signaling pathway, ECM-receptor interaction, and MAPK signaling pathway.
Figure 4.
Function and pathway analysis of co-expressed genes. Differentially expressed genes related to differentially expressed lncRNAs were subjected to GO and pathway analysis, and the potential GO and pathways affected by differentially expressed lncRNAs were predicted. A: GO analysis of differentially expressed genes co-expressed with differentially expressed lncRNAs (left: GO of genes with down-regulated expression; right: GO of genes with up-regulated expression); B: Pathway analysis of differentially expressed genes co-expressed with differentially expressed lncRNAs (left: pathway of genes with down-regulated expression; right: pathway of genes with up-regulated expression).
Discussion
In the present study, APP/PS1 transgenic mice with AD and wide type CD57 mice matched in age were used, and Morris water maze test was conducted to examine the learning and memory of these mice, aiming to evaluate the presence of degenerative neurological diseases. Results showed that the latency to finding the platform was 28.6±8.3 s in wide type mice and 43.6±5.4 s in AD mice, suggesting the compromised memory in AD mice. It was suggested that APP/PS1 mice with AD spontaneously develop AD at the age of 12 months, which was consistent with previously reported [17,18].
Bioinformatics analysis was employed to find AD related genes among the differentially expressed genes. These genes were mainly related to the neuronal development, intercellular communication, neuronal action potential, development and differentiation of oligodendrocytes, transduction of neurotransmitters, neuronal regeneration and lipid metabolism, which have been found to be closely related to the progression of AD. There is evidence showing that Rest gene plays a negative regulatory role in the gene expression during the neuronal development and can regulate the expressions of some neurons related cytokines and affect the differentiation of neuronal stem cells [19-21]. Shh gene encodes Shh peptide and can activate Shh signaling pathway, exerting neuroprotective effect via downstream transcription factor Gli to promote synaptic regeneration and reconstruction and recovery of neurofunction [22]. Apoc2 is a membrane of ApoE family. Some studies showed that Apoc2 mutation was highly related to AD, and Apoc2 could affect the AD progression via oxidative stress pathway [23]. By using GWAS, Blennow et al found that genes including Mef2c played important roles in the progression of AD [24].
LncRNAs refer to non-encoding RNA longer than 200 nucleotides. Previously, lncRNA were regarded garbage fragments in the genome. In recent years, studies reveal that lncRNA are not garbage fragments in cells, but play crucial roles in the biological evolution, embryonic development, cell metabolism, cell differentiation and tumorogenesis [13,25]. The lncRNA expression is temporally and spatially specific. In different tissues and stages of development, the expression of lncRNAs varies significantly. The abundance of lncRNA is the highest and the type of lncRNA is the largest in the brain [26]. LncRNA is involved in several neuron related processes such as neuronal differentiation, brain development, synaptic plasticity and neurodegenerative diseases [27]. In addition, lncRNA may act on AD via different ways. For example, BACE1-AS is an lncRNA as a result of transcription of BACE1 antisense strand and shows a high expression in the AD. This lncRNA cannot bind to the mRNA BACE1 to inhibit its activity, but blocks the binding site of miR-485-5p on the BACE1 mRNA, which hinders the regulation of miRNA on BACE1 [28]. lncRNA17A is mapped to the 3rd intron region of G protein-coupled receptor 51 (GRP51, GAGA B2) gene and can regulate the variable slicing of GRP51 to influence the GABA B signaling pathway, increase the transformation of Aβ42 and elevate its toxicity [29]. lncRNA BC200 selectively localizes in the synapses between neurons and is crucial for the maintenance of synaptic plasticity. Studies have shown that lncRNA BC200 localized around the nucleus in the brain of AD patients, and this spatial ectopia caused the loss of function in the maintenance of synaptic plasticity [30]. In this study, lncRNA microarray assay showed 249 differentially expressed lncRNAs, of which 99 showed down-regulation and 150 had up-regulation. Coexpression analysis of these lncRNAs and AD related genes revealed 4 important lncRNAs: Gm13498, DQ113493, AK038159 and 1700030L20Rik. Further qPCR confirmed that the expressions of these lncRNAs were consistent with those from microarray assay. This indicated that these lncRNAs played regulatory roles in the progression of AD and provided clues for further investigation of target molecules in the pathogenesis of AD.
The study of Lu et al [31] showed the Rest expression reduced significantly in AD and its intracellular localization was also crucial. In healthy old mice, Rest is highly expressed, and fluorescence staining shows Rest is mainly expressed in the cytoplasm and nucleus evenly. However, in AD, Rest first disappears in the nucleus, then its expression reduces gradually, and finally the neuroprotection of Rest loses. In the coexpression network of lncRNAs and mRNAs, lncRNA Gm13498 and lncRNA 1700030L20Rik had the widest relationship with other genes and were negatively related to Rest. It is indicated that lncRNA Gm13498 and lncRNA 1700030L20Rik may bind to Rest protein to block its translocation into the nucleus, resulting in the loss of neuroprotective effect of Rest and the reduction in Rest expression. However, more studies are required to confirm these findings.
Acknowledgements
The study was designed and conducted with funds made available by Shanghai Health and Family Planning Commission Projects (201540063) and National Natural Science Foundation of China (81371212).
Disclosure of conflict of interest
None.
References
- 1.Malkki H. Alzheimer disease: possible prion-like transmission of AD-like pathology in humans. Nat Rev Neurol. 2015;11:612. doi: 10.1038/nrneurol.2015.190. [DOI] [PubMed] [Google Scholar]
- 2.Birks JS, Chong LY, Grimley Evans J. Rivastigmine for Alzheimer’s disease. Cochrane Database Syst Rev. 2015;9:CD001191. doi: 10.1002/14651858.CD001191.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbas Bukhari SN, Jantan I. Synthetic curcumin analogs as inhibitors of beta-amyloid peptide aggregation: potential therapeutic and diagnostic agents for Alzheimer’s disease. Mini Rev Med Chem. 2015;15:1110–1121. doi: 10.2174/138955751513150923101841. [DOI] [PubMed] [Google Scholar]
- 4.Jhoo JH, Kim HC, Nabeshima T, Yamada K, Shin EJ, Jhoo WK, Kim W, Kang KS, Jo SA, Woo JI. Beta-amyloid (1-42)-induced learning and memory deficits in mice: involvement of oxidative burdens in the hippocampus and cerebral cortex. Behav Brain Res. 2004;155:185–196. doi: 10.1016/j.bbr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Georgakopoulos A, Xu J, Xu C, Mauger G, Barthet G, Robakis NK. Presenilin1/gamma-secretase promotes the EphB2-induced phosphorylation of ephrinB2 by regulating phosphoprotein associated with glycosphingolipid-enriched microdomains/Csk binding protein. Faseb J. 2011;25:3594–3604. doi: 10.1096/fj.11-187856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo M, Marcello N, Curcio SA, Colao R, Geracitano S, Bernardi L, Anfossi M, Puccio G, Frangipane F, Clodomiro A, Mirabelli M, Vasso F, Smirne N, Muraca G, Di Lorenzo R, Maletta R, Ghidoni E, Bugiani O, Tagliavini F, Giaccone G, Bruni AC. A novel pathogenic PSEN1 mutation in a family with Alzheimer’s disease: phenotypical and neuropathological features. J Alzheimers Dis. 2011;25:425–431. doi: 10.3233/JAD-2011-110185. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Tian T, Qin S, Li W, Zhang X, Wang X, Gao Y, Huang G. Folic acid deficiency enhances abeta accumulation in APP/PS1 mice brain and decreases amyloid-associated miRNAs expression. J Nutr Biochem. 2015;26:1502–1508. doi: 10.1016/j.jnutbio.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Cooper C, Vincett D, Yan Y, Hamedani MK, Myal Y, Leygue E. Steroid receptor RNA activator bi-faceted genetic system: heads or tails? Biochimie. 2011;93:1973–1980. doi: 10.1016/j.biochi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtale G, Citarella F. Dynamic nature of noncoding RNA regulation of adaptive immune response. Int J Mol Sci. 2013;14:17347–17377. doi: 10.3390/ijms140917347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan L, Yu JT, Hu N, Tan L. Non-coding RNAs in Alzheimer’s disease. Mol Neurobiol. 2013;47:382–393. doi: 10.1007/s12035-012-8359-5. [DOI] [PubMed] [Google Scholar]
- 12.Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Cathcart P, Lucchesi W, Ottaviani S, De Giorgio A, Krell J, Stebbing J, Castellano L. Noncoding RNAs and the control of signalling via nuclear receptor regulation in health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29:529–543. doi: 10.1016/j.beem.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabasi AL, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 16.Schlitt T, Palin K, Rung J, Dietmann S, Lappe M, Ukkonen E, Brazma A. From gene networks to gene function. Genome Res. 2003;13:2568–2576. doi: 10.1101/gr.1111403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pradier L, Carpentier N, Delalonde L, Clavel N, Bock MD, Buee L, Mercken L, Tocque B, Czech C. Mapping the APP/presenilin (PS) binding domains: the hydrophilic N-terminus of PS2 is sufficient for interaction with APP and can displace APP/PS1 interaction. Neurobiol Dis. 1999;6:43–55. doi: 10.1006/nbdi.1998.0212. [DOI] [PubMed] [Google Scholar]
- 18.Weitzner DS, Engler-Chiurazzi EB, Kotilinek LA, Ashe KH, Reed MN. Morris water maze test: optimization for mouse strain and testing environment. J Vis Exp. 2015:e52706. doi: 10.3791/52706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li K, Jiang Q, Xu A, Liu G. REST rs3796529 variant does not confer susceptibility to Alzheimer’s disease. Ann Neurol. 2015;78:835–836. doi: 10.1002/ana.24503. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Tanila H, Puolivali J, Kadish I, van Groen T. Gender differences in the amount and deposition of amyloidbeta in APPswe and PS1 double transgenic mice. Neurobiol Dis. 2003;14:318–327. doi: 10.1016/j.nbd.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Yankner BA. REST and Alzheimer disease. Ann Neurol. 2015;78:499. doi: 10.1002/ana.24420. [DOI] [PubMed] [Google Scholar]
- 22.He P, Staufenbiel M, Li R, Shen Y. Deficiency of patched 1-induced Gli1 signal transduction results in astrogenesis in Swedish mutated APP transgenic mice. Hum Mol Genet. 2014;23:6512–6527. doi: 10.1093/hmg/ddu370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malkki H. Alzheimer disease: APOE*epsilon4-associated increase in AD risk linked to phospholipid dysregulation. Nat Rev Neurol. 2015;11:610. doi: 10.1038/nrneurol.2015.180. [DOI] [PubMed] [Google Scholar]
- 24.Blennow K, Mattsson N, Scholl M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol Sci. 2015;36:297–309. doi: 10.1016/j.tips.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Lisitsyn NA, Chernyi AA, Karpov VL, Beresten SF. [A role of long noncoding RNAs in carcinogenesis] . Mol Biol (Mosk) 2015;49:561–570. doi: 10.7868/S0026898415040102. [DOI] [PubMed] [Google Scholar]
- 26.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 2016;1859:16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Guennewig B, Cooper AA. The central role of noncoding RNA in the brain. Int Rev Neurobiol. 2014;116:153–194. doi: 10.1016/B978-0-12-801105-8.00007-2. [DOI] [PubMed] [Google Scholar]
- 28.Kang MJ, Abdelmohsen K, Hutchison ER, Mitchell SJ, Grammatikakis I, Guo R, Noh JH, Martindale JL, Yang X, Lee EK, Faghihi MA, Wahlestedt C, Troncoso JC, Pletnikova O, Perrone-Bizzozero N, Resnick SM, de Cabo R, Mattson MP, Gorospe M. HuD regulates coding and noncoding RNA to induce APP-->Abeta processing. Cell Rep. 2014;7:1401–1409. doi: 10.1016/j.celrep.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E, Russo C, Florio T, Dieci G, Cancedda R, Pagano A. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]