Abstract
Autophagy, the lysosome-mediated self-degradation process, is implicated in survival during starvation in yeast, Dictyostelium and animals. In these eukaryotic taxa (collectively called Unikonts), autophagy is induced primarily through the Atg1/ULK1 complex in response to nutrient depletion. Autophagy has also been well-studied in non-unikont parasites, such as Trypanosoma and Plasmodium, and found important in their life-cycle transitions. However, how autophagy is induced in non-unikonts remains largely unrevealed. Using a bioinformatics approach, we examined the presence of Atg1 and of its complex in the genomes of 40 non-unikonts. We found that these genomes do not encode typical Atg1 proteins: BLAST and HMMER queries matched only with the kinase domain of Atg1, while other segments responsible for regulation and protein-binding were missing. Non-unikonts also lacked other components of the Atg1-inducing complex. Orthologs of an alternative autophagy inducer, Atg6 were found only in the half of the species, indicating that the other half may possess other inducing mechanisms. As key autophagy genes have differential expression patterns during life-cycle, we raise the possibility that autophagy in these protists is induced mainly at the post-transcriptional level. Understanding Atg1-independent autophagy induction mechanisms in these parasites may lead to novel pharmacological interventions, not affecting human Atg1-dependent autophagy.
Autophagy is a highly conserved self-degradation process of eukaryotic cells, and has found to be important in various cellular processes including stress-response, protein metabolism, differentiation and aging1. An increasing number of studies has provided data on the presence of autophagic structures (such as autophagosomes and autolysosomes) and the molecular mechanisms underlying autophagy in unicellular eukaryotes called protists2,3,4. Orthologs of various yeast autophagy-related genes (ATG) have been identified in these organisms5,6. Thus, autophagy may have emerged at very close to the eukaryote origin or even been coupled with the prokaryote-eukaryote transition7. Most of these studies focused on parasitic protists, such as Trypanosoma, Leishmania, Toxoplasma and Plasmodium species8. Autophagy in these organisms was found to be essential for the transition of parasitic life cycle stages, when the parasite changes its host organism9,10. These transitions require a rapid reorganization in the composition of the cytoplasmic compartments as pathogens adapt to a strikingly new environment. Despite its significance in their natural life cycle, how autophagy induction occurs in these protists remains to be elucidated.
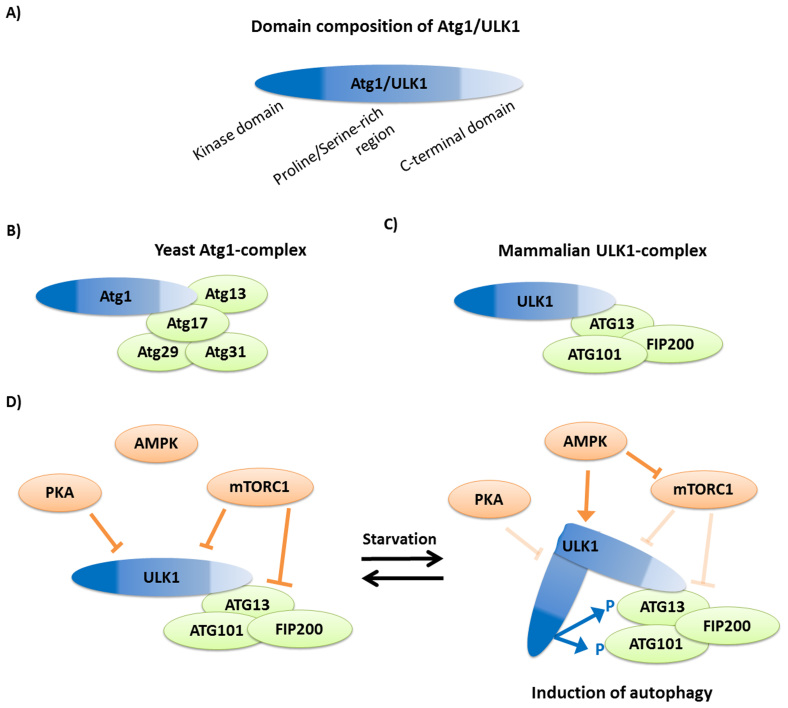
Induction of autophagy is tightly coupled to metabolic stress response in Metazoa (taxonomic group of animals) and in two intensively investigated unicellular model organisms, the yeast Saccharomyces cereviseae and the amoebozoan Dictyostelium discoideum11. All of these organisms belong to the taxonomy group Unikonts (which contains Animals, Fungi and Amoebozoans). In uni- and multicellular, starvation-induced activation of autophagy is mediated mainly by the TOR (target of rapamycin) and AMP (adenosine monophosphate) activated (AMPK) kinases that are highly conserved energy and nutrient sensors of eukaryotic cells12. In metazoans, as in yeast and Dictyostelium, autophagy is inhibited by the TOR kinase, which phosphorylates some components of the Atg1 autophagy induction complex12. This complex contains the Atg1, Atg13, Atg17, Atg29 and Atg31 proteins in yeast (Fig. 1a and 1b), and ULK1, ATG13, ATG101 and FIP200 in mammals (Fig. 1c)12. ULK1 is the mammalian ortholog of Atg1, while FLIP200 is the mammalian counterpart of Atg17. Nutrient deprivation leads to inhibition of TOR kinase and activation of AMPK, which enables ULK1/Atg1 and Atg13 to become partially dephoshorylated, resulting in the initiation of downstream autophagic events (autophagic membrane formation, autophagic membrane ubiquitination, and fusion with the lysosome) in yeast and in mammals (Fig. 1d)12.
Figure 1. The autophagy induction Atg1 complex in yeast and mammals.
(a) The protein domain composition of Atg1/ULK1. (b) In yeast, the complex contains Atg1, which directly binds to Atg13 and Atg17, while Atg29 and Atg31 are linked to the complex through Atg17. (c) The mammalian complex is formed by ULK1, the ortholog of Atg1, which interacts with ATG13. The two other members of the complex are ATG101 and FIP200. (d) In normal, nutrient rich conditions the mammalian TOR complex 1 (mTORC1) inhibits ULK1 and ATG13 by phosphorylation. This inhibition is enhanced by phosphorylation on ULK1 by another cellular nutrient sensor, the cAMP-dependent protein kinase A (PKA)51. We note that in yeast Tpk1, ortholog of the mammalian PKA, can also phosphorylate Atg1 but this reaction regulates only further steps of the autophagic process and not the induction52. Upon starvation nutrient limitation inhibits PKA and activates AMPK. Activated AMPK inhibits mTORC1 and activates ULK1 by direct phosphorylation53,54. Both reactions enable ULK1 to become active. ULK1 conformation is hypothesized to have an “open” and a “close” conformation19. Active ULK1 may take a close conformation, in which the middle proline/serine-rich regions move the kinase domain to the proximity of its possible targets, ATG13 and FIP200. Their phosphorylation triggers downstream events for autophagosome formation. Figures were made based on Ref. 12.
Atg1 function is thereby critical for autophagy induction: in yeast, the kinase activity of Atg1 is required for the disassembly of ATG proteins from the PAS (Preautophagosomal Structure) complex, a step that precedes autophagosome (double-membrane vesicle sequestering portions of cytosol and organelles) formation13. Interestingly, Atg1 also has a kinase-independent role in the assembly of PAS in yeast14,15. Accordingly, both Atg1 and ULK1 contain, in addition to an N-terminal kinase domain, a relatively long proline/serine-rich connecting region and a C-terminal regulatory domain (Fig. 1a)12. In mammals, analysis of the C-terminal region revealed its importance for the autophagosomal localization of ULK1 and for the regulation of ULK1 kinase activity13. We note that ULK1/Atg1 has roles in downstream autophagic processes, though its exact function is less known (for a review, see Ref. 12).
In non-unikont protists, there are only a limited number of observations on the role of starvation and TOR activity in autophagy induction. Most of these studies have been focused on parasitic species. For example, starvation in Trypanosoma cruzi leads to the formation of autophagosome-like structures that are positive for the autophagy marker Atg8 protein16. However, no molecular or cell biology evidence was provided to confirm that autophagy is indeed up-regulated in response to nutrient limitation in this organism. In another study, Trypanosoma brucei mutants defective for TOR kinase was shown to be able to form autophagosome-like structures, which, however, are not labelled with ATG817. Consistent with this finding, rapamycin treatment that inhibits TOR kinase and generally stimulates autophagy in Unikonts, failed to produce autophagic structures in wild-type T. brucei17. Conversely, a recent study from T. brucei showed that upon starvation, Atg8 localizes to punctate structures characteristic for autophagosomes, as confirmed by fluorescence and electron microscopy analysis18. In agreement with a previous study17, rapamycin treatment in T. brucei also failed to produce autophagic structures18. This study also suggested a life-cycle specific difference in autophagy induction18. In Toxoplasma gondii, rapamycin was found to induce autophagy, but the effective concentration of rapamycin was ten times higher than what is used in yeast or mammalian cells, indicating a potential non-specific effect19. Starvation-induced autophagy was detected in Giardia lamblia, however, rapamycin failed to induce autophagy in this species20. Recently, Trichomonas vaginalis was shown to be able to induce autophagy in response to glucose restriction, but the underlying mechanism was not studied21. In Plasmodium falciparum, PfAtg8 localizes to double membrane bound vesicles, and complements Atg8 in Atg8-deficient yeast cells22. Interestingly, in another study, Plasmodium berghei PbAtg8 was unable to complement yeast Atg823. It was recently reported that an in silico analysis find no member of the Atg1 autophagy induction complex in Plasmodium falciparum24. In this organism, starvation could not induce autophagy24. Thus, one can conclude that starvation may induce autophagy in some of these parasitic protists by a mechanism that is significantly different from those we learnt in yeast and in higher eukaryotes.
There are some comprehensive studies on genomic searches for ATG genes among protists. Rigden et al. (2009)2 performed the first and influential research in this field. They examined 18 protist species and searched for homologs of yeast autophagy proteins. The presence or absence of autophagy proteins were discussed in details with great emphasis on the clear, bioinformatically proven differences in autophagy pathways between the examined species. They mentioned the possibility of differences in induction because of the difficulties in the identification of Atg1 orthologs, but did not go into detail. Kiel (2010)3 reviewed the available information on autophagy in non-yeast unicellular eukaryotes and focuses on kinetoplastids and Dyctiostelium. Duszenko et al. (2011) analyzed the presence or absence of autophagy proteins in protists4. The authors placed a particular emphasis on the evolutionary aspect and role of autophagy in the life cycle and pathogenicity of these parasitic species. Duszenko et al.4 examined the issue of autophagy induction but not focused on and discussed it in detail. Results from previous studies were obtained only by a single method (usually BLAST25 (BLAST, PSI-BLAST) or in some cases HMMER26,27). No previous study examined the difference in the autophagy inducing complex between unikont and non-unikont protists in detail.
This prompted us to examine whether the genomes of non-unikont parasites contain homologs of yeast and metazoan ATG genes implicated in starvation-induced activation of autophagy. We show, by using two different algorithm employing methods, that the examined non-unikont organisms lack the components of the Atg1 autophagy induction complex.
Results
Non-unikont protist species do not contain typical Atg1 proteins
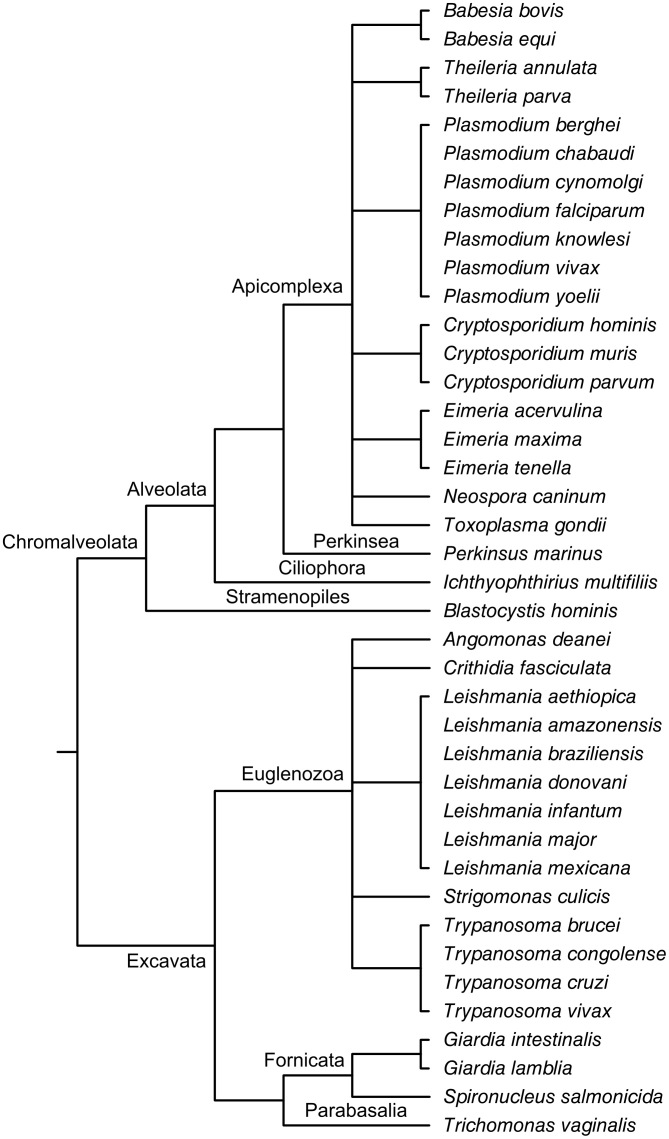
We searched for the presence of components of the Atg1 complex in 40 non-unikont parasitic protist species, including Leishmania, Plasmodium, Toxoplasma and Trypanosoma ones. The complete list of the examined species with their respective phylogenetic tree can be found in Fig. 2. We analyzed the genomes of these species by using BLASTp (protein-protein sequence homology search)25 and HMMER (profile Hidden Markov model)27,26. We used the sequences of autophagy-related proteins from Dictyostelium discoideum, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens as queries to find homologous proteins in the protist species examined. The detailed results of BLASTp and HMMER searches can be found in Fig. 3, Fig. 4 and Suppl. Table 1.
Figure 2. Phylogeny of the 40 examined non-unikont parasite species.
The cladogram were created according to the taxonomic classification of the species find at NCBI Taxonomy database55 and the Eukaryote tree of Tree of Life web project56,57.
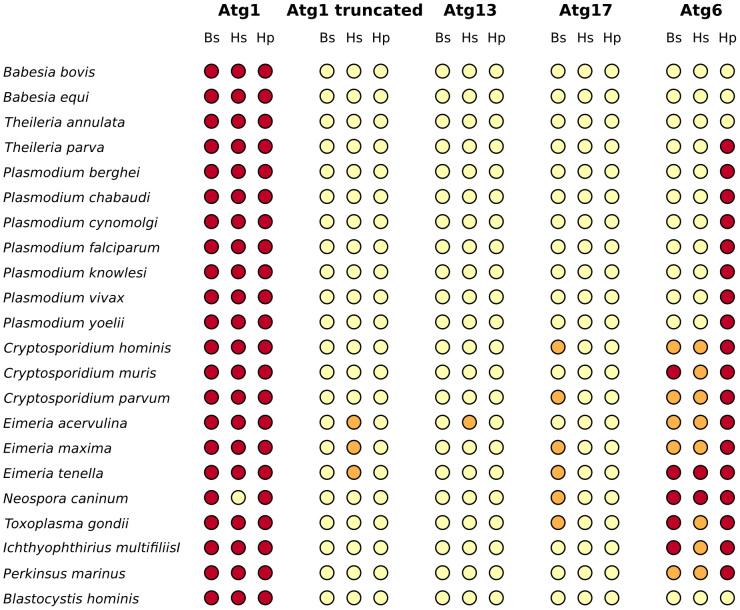
Figure 3. DOT-plot of BLAST and HMMER search results for the examined Atg1-complex members and Atg6 in the Chromalveolata group.
The figure shows which Chromalveolata species has a potential ortholog with the members of the Atg1-complex or Atg6 query sequences of five Unikonts. Target species – ordered according to their taxonomy, see Fig. 2 – are located in the rows and query proteins (Atg1, truncated Atg1, Atg13, Atg17 and Atg6) in the columns. For Atg6 and each protein of the Atg1-complex both BLAST and HMMER searches were applied using the Atg proteins of five unikont species (Dictyostelium discoideum, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens) as query sequences. Each colored dot indicates a significant hit (Expect values or E-values < 0.001) with a particular query sequence in the proteome of the target species. In columns named “Bs” and “Hs” you can find the results of BLAST and HMMER searches respectively. For these similarity searches the query sequences of five different unikont species were used one by one. Light yellow dots mean there were no significant hit using any of the query sequences. Orange dots mean there were some query sequences that gave a significant hit in a particular non-unikont species but not all five (or four in case of Atg17 where we did not find any orthologs in Caenorhabditis elegans). Dark red dots indicate that significant hits were found with all of the unikont query species. In columns named “Hp” the results of profile based HMMER searches can be found. For the profile search the protein sequences of the five unikont species were aligned by the Muscle algorithm58 implemented in SeaView software59. Target species that had a significant hit using the profile sequence as query indicated with dark red dots, and yellow dots stand for no hits were found searching with the profile. Based on the E-values alone, Atg1 orthologs are certainly present in every examined non-unikont protist species. However, our more detailed analysis suggests that these might be false positive results. (See the text for more details and the description of truncated Atg1 analysis, and Suppl. Table 1 for detailed results of BLAST and HMMER analyses).
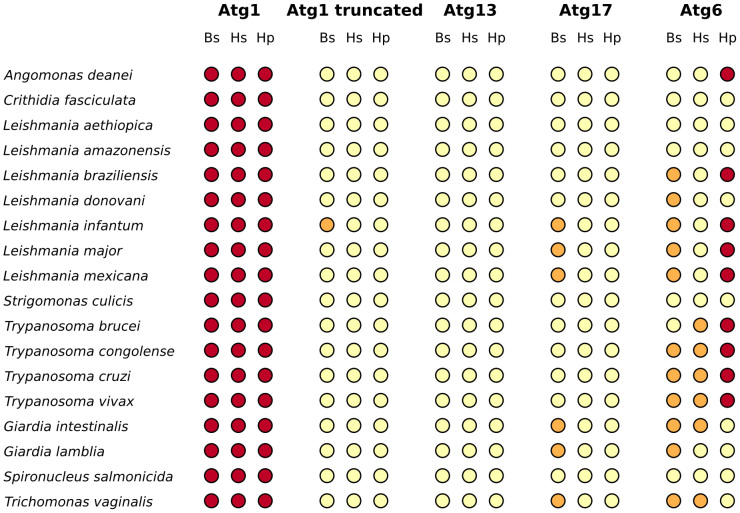
Figure 4. DOT-plot of BLAST and HMMER search results for the examined Atg1-complex members and Atg6 in the Excavata group.
The figure shows which Excavata species has a potential orthologous with the members of the Atg1-complex or Atg6 query sequences of five Unikonts. For more detailed description see the figure legend of Fig. 3.
Using the complete Atg1 sequence as query, we found sequence similarities only for the kinase domain of Atg1 in the species examined (Suppl. Table 1). Truncated Atg1 sequences lacking the kinase domain as query however yielded no sequence similarity in non-unikont protists (Suppl. Table 1). (As a positive and a negative control for our analysis, we searched the genome of Schizosaccharomyces pombe and Escherichia coli, respectively.) Thus, parts of the Atg1 kinase that are responsible for regulation and protein binding appear to be missing from the non-unikont protist species we studied. Consequently, yeast Atg1 has many kinase homologs in kinase domain in these protists but they lack further homology. Thus, these kinase orthologs cannot be considered as putative Atg1-like proteins. This suggests that these homologs are not able to respond to TOR and AMPK activities, and facilitate PAS assembly. It would be interesting to learn whether these kinase domain-containing proteins are implicated in the autophagic process. A future genetic analysis can certainly address this important issue.
BLAST and HMMER analyses gave almost completely identical results with both full and truncated Atg1/ULK1 sequences (Fig. 3 and Fig. 4). The DOT-plot28 representation of the results convincingly shows that full Atg1/ULK1 query sequences have homologs in all protists examined, while truncated Atg1/ULK1 query sequences do not. The very few exceptions can be explained by random sequence similarities (Suppl. Table 2).
Non-unikont protists also lack other components of the Atg1 autophagy-inducing complex
We also searched non-unikont protist genomes for homologs of yeast proteins that participate in the composition of the Atg1 complex, including Atg13, Atg17, Atg29 and Atg31. Consistent with the absence of Atg1, we found no appreciable sequence similarity for the other members of the autophagy inducing complex (Fig. 3, Fig. 4 and Suppl. Table 1). We found sequence matches above the cut-off value only in very few cases (Suppl. Table 2). Note that in these rare cases the coverage of the query and target sequences was very low, typically below 20%. We conclude that the genomes of non-unikont parasites we examined here do not encode components of the Atg1 autophagy induction complex. This raises the intriguing possibility that autophagy in these unicellular organisms is induced in a way that is independent of the Atg1 complex.
BLAST and HMMER results supported each other with almost no contradiction (Fig. 3 and Fig. 4). The only exception is the human FIP200, whose query sequence gave noticeably better sequence similarities than any other sequences examined (Suppl. Table 2). It is interesting, since FIP200 considered as non-orthologous to yeast Atg17, though FIP200 is the mammalian counterpart of yeast Atg17 with mostly similar function29.
Atg6/Beclin 1, an alternative autophagy inducer, is missing from non-unikont protists
In metazoans, Atg6/Beclin 1 protein can also induce autophagy by integrating stress signals such as hypoxia, endoplasmic reticulum (ER) stress and infection, largely independently of Atg11. In mammals a direct phosphorylation interaction between ULK1/Atg1 and Beclin 1 was recently found, strengthening further the importance of Beclin 1 in autophagy induction30. As Beclin 1 could serve as an alternative autophagy inducer in non-unikont protists, we searched for and could identify possible Beclin 1 orthologs in the half of these species (Suppl. Table 1). We conclude that in these organisms autophagy could be induced by Beclin 1orthologs. However, further experimental analysis is needed to provide specific function for Beclin 1 in autophagy induction in these species.
Interestingly, for the other half of the protists examined, we found no Beclin 1 homolog (Suppl. Table 1). The lack of this potential post-translational regulatory mechanism raises the possibility that in these organisms autophagy might be regulated predominantly at the transcriptional or post-transcriptional level, with an analogous mechanism to the one recently described in humans31.
Although for Atg6 the picture was not as clear as in the case of Atg1-complex members, the BLAST and HMMER analyses provided very similar results (Fig. 3 and Fig. 4). Potential Atg6 orthologs typically either uniformly present or absent in a certain genus. Interestingly, the pattern of the absence of Atg6 homologs did not follow any taxonomical or biological rule, indicating various effects and potentially different induction mechanisms these organisms faced and developed during their evolution.
Discussion
Here we report that non-unikont parasitic protists do not contain components of the Atg1 complex (Atg1, Atg13, Atg17, Atg29 and Atg31) having an important role in autophagy induction in yeast and metazoan species (Fig. 3, Fig. 4 and Suppl. Table 1). Thus, in these unicellular eukaryotes autophagy might be activated by an Atg1 complex-independent mechanism. We also showed that Atg6/Beclin 1, an alternative autophagy inducer, can be involved in autophagy induction only in less than half of the examined 40 non-unikont parasites (Fig. 3, Fig. 4). However, in the other half of the examined species, we detected no Atg6/Beclin 1 homologs in the genomes. Starvation response may induce autophagy in these protists, but currently there is no clear experimental observation that could unambiguously prove the existence of such regulatory relationship. The molecular mechanisms underlying starvation-induced up-regulation of autophagy in these organisms remain to be elucidated. Our results are strengthened by two different algorithm applying methods (BLAST and HMMER), which gave consistent results.
It was already observed in numerous studies that there are differences in the presence of autophagy proteins among different organisms. Key autophagy proteins, including the major inductors of autophagy, were found to be present universally in metazoans, Arabidopsis and 21 fungal species with only few exceptions32. However, there is difference between fungi and human or fungi and Arabidopsis in case of proteins important for specific autophagy-related processes32. Several comprehensive bioinformatics studies were carried out on protozoan genomes, and discussed the presence of ATG genes and the differences in presence among species5,8,33,34,35. Lack of some ATG genes were demonstrated2,20, including the Atg12 ubiquitin-like conjugation system2,5,6,34 or Atg172. Remarkably, many of these studies showed the presence of ATG1 gene in non-unikont parasites2,3,4,19. The major differences between these studies and our current report are that (1) they used only the complete sequence of Atg1 orthologs as a query and did not examine other parts of the proteins (Fig. 2), (2) these studies did not focus especially on autophagy induction.
An extensive and profound study mentioned the possibility of different induction pathways, but showed that most protozoan genome contains orthologs of Atg12. For example, in two bioinformatics studies on Trypanosoma, the authors claimed their uncertainty that the many potential orthologous sequences of Atg1 could be real Atg1 proteins at all5,6. In another study with Toxoplasma gondii, amino acid deprivation induced mitophagy (selective elimination of mitochondria) was detected, and Atg proteins (including putative TgAtg1) were identified by bioinformatic analysis36. However, authors had no experimental proof regarding the function of putative TgAtg1, but had difficulties with electronmicroscopic detection of autophagosomes due to the lack of adequate autophagy markers36. Very recently, an in silico analysis found no member of the Atg1 autophagy induction complex in Plasmodium falciparum24. This finding is consistent with our observation that non-unikont protists do not contain the Atg1 complex. These results do contradict with earlier results of several comprehensive analysis, but point out the importance of analysing the domain structure of potential Atg1 orthologs, and examining the full induction complex. Altogether, these findings indicate an Atg1 complex-independent mechanism of autophagy induction in the 40 non-unikont parasites examined.
A recent study raises the possibility of another, TOR-related autophagy induction mechanism: a comprehensive analysis showed that in Trypanosoma brucei, TOR4, a specific TOR form, is down-regulated upon low cellular energy levels (at high AMP:ATP ratio)37. Moreover, TOR4, but not TOR1, was capable of regulating developmental switch between the life-cycle stages of T. brucei37. However, no molecular or cell biology analysis has yet been confirmed the role of TOR4 in autophagy regulation. As TOR4 is specific to Trypanosoma and Leishmania species38, and we found that these species lack Atg1, one may speculate that a further experimental analysis may connect TOR4 and its putative substrates to starvation induced autophagy. As an alternative autophagy inducing mechanism, it has recently been suggested that in Plasmodium falciparum the process may be regulated by PfAtg8 availability through cleavage from lipid association by PfAtg422. Experimental testing of this mechanism could show that such an alternative autophagy inducing process does exist, explaining the remarkable difference we found in this report.
In higher eukaryotes, several transcription factors, such as TFEB31, FoxO339, NRF240, p5341 and E2F142, are known to activate autophagy by regulating the expression of key ATG genes in a context-dependent manner. Similarly, in Trypanosoma, Leishmania, Plasmodium and Toxoplasma species several ATG genes display a differential expression pattern, according to the EuPathDB web resource43. For example, in T. brucei some ATG genes (e.g., ATG4, ATG5, ATG7, ATG16) are constitutively expressed, while others (e.g., ATG3, ATG8, ATG10 and ATG12) are expressed specifically in distinct stages of the life cycle44. Interestingly, these components have essential roles in different phases of the autophagic process. Note that in the mentioned kinetoplastid parasites, there is no regulation of transcription initiation, thus, we should not expect a precise transcriptional mechanism (i.e., transcription factors) responsible for autophagy regulation as seen in higher Eukaryotes. However, based on concordant expression patterns in the above listed parasites, we may speculate that either a post-transcriptional mechanism or other genetic program potentially modulate autophagic activity by differentially expressing certain ATG genes. We also assume that the factors that regulate the expression of ATG genes during life cycles may also act as sensors for metabolic stress signals. Further experimental studies may uncover components and possible mechanisms of autophagy regulation at the transcriptional level.
We note that our present study relies on the current knowledge of autophagy regulation and functional annotations of genes from these parasitic protozoan genomes. As new results will be published, we may shed deeper insight into how induction of autophagy occurs in non-unikont protists.
Together – based on extensive in silico analysis of 40 non-unikont parasitic protist genomes – metabolic stress response appears to induce autophagy in a way independent of the Atg1-complex. Further studies should clarify the function of TOR in autophagy induction in these species, as well as the possibility that autophagy is induced via an Atg1 complex-independent way. The lack of the Atg1 complex may raise the possibility that autophagy induction occurs at the transcriptional or/and post-translational levels in these organisms. We pointed out that differential expression of ATG genes may be involved in the regulation of autophagy during starvation. In addition, post-translational modification of Atg6/Beclin 1 can be involved in autophagy induction in some non-unikont parasites. Potential identification of Atg1-independent autophagy induction pathways in these protists may lead to novel parasite-specific pharmacological interventions. Without affecting Atg1-dependent autophagy up-regulation in humans, modification of the parasite-specific autophagy induction may have important therapeutic applications.
Methods
BLAST25 searches were run to identify autophagy-related proteins in non-unikont protist species. BLAST (Basic Local Alignment Searching Tool) is a service of NCBI and generally used for searching for sequence similarities. BLAST uses heuristic algorithm and performs local alignments. We used BLAST Expect value (E-value) to measure significance: The smaller the E-value, the more significant the alignment. According to similar studies2, we set the E-value cut-off to 0.001 in every BLAST searches to identify homolog sequences. Apart from the significance level, we used the default settings. We also used HMMER to strengthen our results. HMMER is also used to search for similar protein sequences27,26. HMMER applies probabilistic methods, profile hidden Markov models. For the HMMER searches UniProtKB protein sequence database45 were used with default settings.
The sequences of the respective proteins of Dictyostelium discoideum, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens were applied as query sequences. We acquired the yeast sequences from the Saccharomyces Genome Database46, the D. discoideum sequences from dictyBASE47, the C. elegans sequences from Wormbase48, the D. melanogaster sequences from Flybase49 and the human sequences from Ensemble database50, respectively. We selected the sequences of ATG1/ULK1-complex member proteins, since this complex is, according to our current knowledge, the main inductor of autophagy in Unikonts. The main components of Atg1/ULK1-complex are Atg1/ULK1, Atg13, Atg17/FIP200 (and in yeast Atg29, Atg31).
We searched the genomes of 40 non-unikont parasite species. All the non-unikont parasites species that had completely sequenced genome (“Genome representation: full” according to the status of NCBI Genome database in December 2013; http://www.ncbi.nlm.nih.gov/sites/genome) were included to the study, excluding those genomes, that had no significant kinase-domain similarities with Saccharomyces cerevisiae Atg1 protein query-sequence. The list of the 40 species can be found in Fig. 2.
Supplementary Material
Supplementary Dataset
Acknowledgments
We are grateful for the technical help of Dávid Fazekas. We thank members of the Vellai lab, the NetBiol group and the LINK-Group for helpful discussions. This work was supported by the European Union and the European Social Fund [TAMOP-4.2.2/B-10/1-2010-0013] and the Hungarian Scientific Research Fund [OTKA K83314, K75334, NK78012]. TK is a grantee of the János Bolyai Scholarship of the Hungarian Academy of Sciences, and is supported by a fellowship in computational biology at The Genome Analysis Centre, in partnership with the Institute of Food Research, and strategically supported by BBSRC.
Footnotes
The supporters had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no competing financial interests.
Author Contributions T.V. initiated the project. L.F.-N. and T.K. conceived the research. L.F.-N. performed data analysis. All (E.A., P.C., L.F.-N., T.K., T.V.) authors contributed to biological interpretation of the results. L.F-N. and T.K. wrote the main manuscript text. E.A., L.F.-N. and T.K. prepared figures. All authors reviewed the manuscript.
References
- He C. & Klionsky D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D. J., Michels P. A. & Ginger M. L. Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 5, 784–794 (2009). [DOI] [PubMed] [Google Scholar]
- Kiel J. A. K. W. Autophagy in unicellular eukaryotes. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 365, 819–830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M. et al. Autophagy in protists. Autophagy 7, 127–158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D. J., Herman M., Gillies S. & Michels P. A. M. Implications of a genomic search for autophagy-related genes in trypanosomatids. Biochem. Soc. Trans. 33, 972–974 (2005). [DOI] [PubMed] [Google Scholar]
- Herman M., Gillies S., Michels P. A. & Rigden D. J. Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy 2, 107–118 (2006). [DOI] [PubMed] [Google Scholar]
- Hughes T. & Rusten T. E. Origin and evolution of self-consumption: autophagy. Adv. Exp. Med. Biol. 607, 111–118 (2007). [DOI] [PubMed] [Google Scholar]
- Brennand A. et al. Autophagy in parasitic protists: unique features and drug targets. Mol. Biochem. Parasitol. 177, 83–99 (2011). [DOI] [PubMed] [Google Scholar]
- Besteiro S., Williams R. A. M., Morrison L. S., Coombs G. H. & Mottram J. C. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 281, 11384–11396 (2006). [DOI] [PubMed] [Google Scholar]
- Herman M., Pérez-Morga D., Schtickzelle N. & Michels P. A. M. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy 4, 294–308 (2008). [DOI] [PubMed] [Google Scholar]
- Yang Z. & Klionsky D. J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335, 1–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Ro S.-H., Cao J., Otto N. M. & Kim D.-H. mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. Y. W., Longatti A., McKnight N. C. & Tooze S. A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29, 157–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J. & Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 668–681 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T. & Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19, 2039–2050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez V. E. et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 283, 3454–3464 (2008). [DOI] [PubMed] [Google Scholar]
- Barquilla A. & Navarro M. Trypanosome TOR as a major regulator of cell growth and autophagy. Autophagy 5, 256–258 (2009). [DOI] [PubMed] [Google Scholar]
- Li F.-J. et al. A role of autophagy in Trypanosoma brucei cell death. Cell. Microbiol. 14, 1242–1256 (2012). [DOI] [PubMed] [Google Scholar]
- Besteiro S., Brooks C. F., Striepen B. & Dubremetz J.-F. Autophagy protein Atg3 is essential for maintaining mitochondrial integrity and for normal intracellular development of Toxoplasma gondii tachyzoites. PLoS Pathog. 7, e1002416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi S., Oniku A. E., Topping K., Mamhoud Z. N. & Paget T. A. Programmed cell death in Giardia. Parasitology 139, 894–903 (2012). [DOI] [PubMed] [Google Scholar]
- Huang K.-Y. et al. Adaptive responses to glucose restriction enhance cell survival, antioxidant capability, and autophagy of the protozoan parasite Trichomonas vaginalis. Biochim. Biophys. Acta 1840, 53–64 (2014). [DOI] [PubMed] [Google Scholar]
- Tomlins A. M. et al. Plasmodium falciparum ATG8 implicated in both autophagy and apicoplast formation. Autophagy 9, 1540–1552 (2013). [DOI] [PubMed] [Google Scholar]
- Coppens I. Metamorphoses of malaria: the role of autophagy in parasite differentiation. Essays Biochem. 51, 127–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes S. et al. The multifunctional autophagy pathway in the human malaria parasite, Plasmodium falciparum. Autophagy 10, 80–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. A new generation of homology search tools based on probabilistic inference. Genome Inform 23, 205–211 (2009). [PubMed] [Google Scholar]
- Field H. I., Coulson R. M. R. & Field M. C. An automated graphics tool for comparative genomics: the Coulson plot generator. BMC Bioinformatics 14, 141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko V. Y. & Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat. Cell Biol. 15, 727–728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M. & Kiel J. A. K. W. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3, 106–116 (2007). [DOI] [PubMed] [Google Scholar]
- Sinai A. P. & Roepe P. D. Autophagy in Apicomplexa: a life sustaining death mechanism? Trends Parasitol. 28, 358–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand A., Rico E. & Michels P. A. M. Autophagy in trypanosomatids. Cells 1, 346–371 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickel N. et al. Features of autophagic cell death in Plasmodium liver-stage parasites. Autophagy 9, 568–580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D., Walton J. L., Roepe P. D. & Sinai A. P. Autophagy is a cell death mechanism in Toxoplasma gondii. Cell. Microbiol. 14, 589–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A. et al. Third target of rapamycin complex negatively regulates development of quiescence in Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 109, 14399–14404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquilla A., Crespo J. L. & Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc. Natl. Acad. Sci. U.S.A. 105, 14579–14584 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007). [DOI] [PubMed] [Google Scholar]
- Jain A. et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576–22591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M. C. et al. Autophagy regulation by p53. Curr. Opin. Cell Biol. 22, 181–185 (2010). [DOI] [PubMed] [Google Scholar]
- Polager S., Ofir M. & Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 27, 4860–4864 (2008). [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C. et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 35, D427–D430 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani S. et al. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 10, 427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42, D191–D198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. et al. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 41, D676–D683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. W. et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38, D463–467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J. & FlyBase Consortium. FlyBase 101--the basics of navigating FlyBase. Nucleic Acids Res. 40, D706–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. et al. Ensembl 2014. Nucleic Acids Res. 42, D749–D755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan J. S., Yeh Y.-Y., Ramachandran V., Deminoff S. J. & Herman P. K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U.S.A. 106, 17049–17054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J. & Herman P. K. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 102, 13933–13938 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Park S., Takahashi Y. & Wang H.-G. The association of AMPK with ULK1 regulates autophagy. PLoS ONE 5, e15394 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison D. R., Schulz K.-S. & Wayne P. Maddison. The Tree of Life Web Project. Zootaxa 1668, 19–40 (2007). [Google Scholar]
- Keeling P., Leander B. S. & Simpson A. Eukaryotes. Eukaryotes. Eukaryota, Organisms with nucleated cells. Version 28 October 2009. http://tolweb.org/Eukaryotes/3/2009.10.28 in The Tree of Life Web Project (2009) Date of access: 12/12/2010.
- Edgar R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S. & Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset