Abstract
Morphological and molecular evidence strongly supported the monophyly of tribe Anemoneae DC.; however, phylogenetic relationships among genera of this tribe have still not been fully resolved. In this study, we sampled 120 specimens representing 82 taxa of tribe Anemoneae. One nuclear ribosomal internal transcribed spacer (nrITS) and six plastid markers (atpB-rbcL, matK, psbA-trnQ, rpoB-trnC, rbcL and rps16) were amplified and sequenced. Both Maximum likelihood and Bayesian inference methods were used to reconstruct phylogenies for this tribe. Individual datasets supported all traditional genera as monophyletic, except Anemone and Clematis that were polyphyletic and paraphyletic, respectively, and revealed that the seven single-gene datasets can be split into two groups, i.e. nrITS + atpB-rbcL and the remaining five plastid markers. The combined nrITS + atpB-rbcL dataset recovered monophyly of subtribes Anemoninae (i.e. Anemone s.l.) and Clematidinae (including Anemoclema), respectively. However, the concatenated plastid dataset showed that one group of subtribes Anemoninae (Hepatica and Anemone spp. from subgenus Anemonidium) close to the clade Clematis s.l. + Anemoclema. Our results strongly supported a close relationship between Anemoclema and Clematis s.l., which included Archiclematis and Naravelia. Non-monophyly of Anemone s.l. using the plastid dataset indicates to revise as two genera, new Anemone s.l. (including Pulsatilla, Barneoudia, Oreithales and Knowltonia), Hepatica (corresponding to Anemone subgenus Anemonidium).
Introduction
Tribe Anemoneae is a member of subfamily Ranunculoideae (Ranunculaceae) [1–4]. Traditionally, this tribe included three subtribes, i.e., Anemoninae, Clematidineae and Kingdoniae [1–3]. An overview of classifications for tribe Anemoneae is summarized in S1 Table. The subtribe Kingdoniae contains only one species, Kingdonia uniflora Balf. f. & W. W. Sm., which is characterized by one cordate-orbicular leaf, veins bifurcated and a short flower stalk with a small flower. Kingdonia uniflora grows at high elevations in western China [5]. Currently, morphological and molecular evidences show that K. uniflora should be excluded from tribe Anemoneae, even from Ranunculaceae [6], and it has been treated as an independent family Kingdoniaceae, or incorporated into family Circaeasteraceae since 2009 [4, 7]. Excluding K. uniflora, tribe Anemoneae was strongly supported as monophyletic in phylogenetic analyses [4, 8–11].
Traditionally, subtribe Clematidinae comprised three genera: Archiclematis (Tamura) Tamura, Clematis L., and Naravelia Adans. [1–3]. The largest genus Clematis has more than 300 species [12]. In some classification systems, this genus was treated as several genera on the basis of morphological, palynological, and anatomical data, e.g., Atragene L., Cheiropsis (DC.) Bercht. ex J. Presl, Clematopsis Bojer ex Hutch., Meclatis Spach, Viorna (Pers.) Rchb. [12]. In general, these ranks have been adopted as sections or subgenera under Clematis [12–16]. The flower of Archiclematis alternata (Kitam. & Tamura) Tamura (≡ Clematis alternata Kitam. & Tamura) resembles Clematis section Viorna (Reichb.) Prantl. [17], while this is the only species having alternate leaves in this subtribe. Wang and Li [12] treated Archiclematis as a section in Clematis. Naravelia is restricted to tropical Asia. In the full revision of Naravelia, Tamura [18] accepted seven species. Naravelia is distinguished from Clematis with the presence of petals and leaflet tendrils. According to molecular phylogenetic analyses [19, 20], Clematis is paraphyletic, including Naravelia and Archiclematis. However, the status of Naravelia need to be further confirmed because the studies [19, 20] included only two species without the generic type, i.e. N. eichleri Tamura and N. laurifolia Wall. ex Hook. f. & Thomson. Wang et al. [4] documented that Naravelia zeylanica L. is the sister to Clematis, though this study only included one Clematis species, C. ganpiniana (H. Lév. & Vaniot) Tamura.
Generally, subtribe Anemoninae consists of eight genera: Anemoclema (Franch.) W. T. Wang, Anemone L., Barneoudia C. Gray, Hepatica Miller, Knowltonia Salisb, Metanemone W. T. Wang, Oreithales Schldl., and Pulsatilla Mill. [1–3, 21, 22]. Among them, Anemoclema, Metanemone and Oreithales are monotypic (i.e., only one species). The genus Anemone contained more than 150 species, and it is distributed throughout the world. Molecular phylogenetic studies recognized that Hepatica, Pulsatilla and Knowltonia are nested within Anemone, and that they should be subsumed within Anemone [23–25]. Then, Hoot et al. [26] and Mayer et al. [25] revealed that two South American endemic genera Barneoudia and Oreithales should be also included in Anemone. Anemoclema contains a single species, A. glaucifolium (Franch.) W. T. Wang, endemic to the Hengduan Mountains in southwestern China [27]. Because of specific pinnatisect and penninerved leaves and spinulose pollen grains, Wang [28] proposed that Anemone sect. Anemoclema Franch. should be separated from Anemone as an independent genus. This treatment is widely adopted by Chinese researchers in Floras [21, 29, 30], checklists [31], and publications [27, 32, 33]. In contrast, non-Chinese taxonomists prefer treating this species as a monotypic section or subgenus in Anemone [1–3, 34–36]. However, Wang’s treatment is supported by results of karyotype and molecular phylogenies [32, 37, 38]. Furthermore, it has been documented that Anemoclema is close to Clematis, not to Anemone [4, 38, 39]. Therefore, Anemoclema has been transferred to subtribe Clematidinae [38], then subtribe Anemoninae includes Anemone s.l. and Metanemone.
To date, phylogenetic analyses of Anemone s.l. are mainly based on nuclear ribosomal internal transcribed spacers (nrITS) and plastid atpB-rcbL intergenic spacer, because the two regions show high rates of variable and parsimony-informative sites, and they are powerful to resolve phylogenies at the infrageneric level [24–26, 40, 41]. Monophyly of Anemone s.l. was strongly supported in these studies. However, the monophyly of Anemone s.l. was not resolved in other studies using other regions, but these were with limited samples [4, 39, 42]. In addition, phylogenetic relationship between subtribes Anemoninae and Clematidinae is inferred just using nrITS and atpB-rcbL datasets [24, 38, 41]. In this study, we extensively sampled Hepatica and Pulsatilla in subtribe Anemoninae, as well as Anemoclema and Naravelia in subtribe Clematidinae, and we sequenced nrITS, atpB-rbcL, and five additional plastid regions (matK, rbcL, psbA-trnQ, rpoB-trnC and rps16). For the atpB-rbcL region, we only used the intergenic spacer, so there is no overlapping with the rbcL gene. Based on comprehensive phylogenetic analyses, we sought to: (1) infer the phylogenetic relationships among genera within the two subtribes; (2) reevaluate the monophyly of Anemone s.l.; and (3) resolve the phylogenetic placement of Anemoclema and Naravelia.
Materials and methods
Plant samplings and ethics statement
We sampled nine of ten recognized genera in tribe Anemoneae (excluding Kingdonia). Metanemone was not sampled, because the single species M. ranunculoides has type material alone, and we were failed to collect in the field. In total, we sampled 122 accessions representing 77 species and five infraspecific taxa of tribe Anemoneae, including Anemoclema (1 species/6 individuals, 100% of total species, hereafter), Anemone (14/19, ~10%), Archiclematis (1/1, 100%), Barneoudia (3/3, 100%), Clematis (21/22, ~7%), Hepatica (9/21, ~90%), Knowltonia (5/5, 62.5%), Naravelia (6/10, 85.7%), Oreithales (1/2, 100%), and Pulsatilla (17/53, ~40%). Eleven species from five genera of Ranunculaceae (Adonis, Batrachium, Caltha, Halerpestes, and Ranunculus) were selected as outgroups. Silica-dried samples were collected from public land instead of protected areas in Southwestern and Western China; therefore, field permits were not required. Voucher specimens, geographic coordinates, and GenBank accessions are presented in S2 Table.
DNA extraction, PCR and sequencing
Total genomic DNA was extracted from silica-dried leaves using modified CTAB buffer protocol. One nuclear (nrITS) and six plastid markers (atpB-rbcL, matK, rbcL, psbA-trnQ, rpoB-trnC and rps16) were amplified and sequenced. Primer information is given in S3 Table. Polymerase chain reaction (PCR) amplification for nrITS, matK, psbA-trnQ, rbcL and rps16 markers used the following protocol: one cycle 97°C for 3 min; then 33 cycles of 94°C for 50 s, 55°Cfor 50 s and 72°Cfor 60 s; and followed by 72°C for 5 min. In addition, the regions atpB-rbcL and rpoB-trnC were amplified using a different protocol: one cycle 80°C for 5 min; then 35 cycles of 95°C for 60 s, 50°Cfor 45 s and 65°C for 2 min; followed by 65°C for 3 min. PCR products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA, USA). Sequencing reactions were performed using the ABI Prism BigDye Terminator Kits (Applied Biosystems, Inc.) and followed the manufacturer’s protocol. Automated sequencing was performed on an ABI 3730xl DNA sequencer (Applied Biosystems).
Phylogenetic analyses
New sequences were assembled, aligned, and adjusted using Geneious 7.0 [43]. Aligned matrices of the seven DNA regions were firstly analyzed separately, then plastid matrices were concatenated using SequenceMatrix 1.7 [44]. The DNA matrix of seven DNA regions was deposit at Figshare (DOI: 10.6084/m9.figshare.4774753). No nucleotide positions were excluded from analyses. According to the topologies of single marker datasets, monophyly of Anemone s.l. was recovered in nrITS and atpB-rbcL datasets. Previous studies using the nrITS + atpB-rbcL dataset well resolved the monophyly of Anemone s.l., therefore, the two datasets were combined in this study. To combine the plastid datasets, we did two treatments: one has all six plastid regions (i.e. six-plastid-gene dataset), and the second has five plastid regions without atpB-rbcL (i.e. five-plastid-gene dataset). Topological incongruence among nrITS, atpB-rbcL, nrITS + atpB-rbcL and five plastid datasets was investigated using the approximately unbiased (AU) test [45] and the Shimodaira–Hasegawa (SH) test [46]. Topologies were constrained using Mesquite 3.2 [47]. The SH and AU tests were performed using PAUP 4.0 [48].
Maximum likelihood (ML) analyses were conducted using RAxML [49]. These analyses used the GTR substitution model with gamma-distributed rate heterogeneity among sites and the proportion of invariable sites estimated from the dataset. The multiple-gene datasets were partitioned by genes. Support values for the node and clade were estimated from 1000 bootstrap replicates. ML bootstrap support (BS) values ≥ 70% were considered well supported, and BS < 50 were seen as an indication of nonsupport. Bayesian inference (BI) analyses was performed using MrBayes 3.2.6 [50], with DNA substitution models selected for each gene partition by the Bayesian information criterion (BIC) using jModeltest 2.0 [51]. Markov Chain Monte Carlo (MCMC) analyses were run in MrBayes for 10,000,000 generations for each dataset. The BI analyses were started with a random tree and sampled one tree every 1000 generations. The first 20% of the trees were discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. Internodes with posterior probability values (PP) ≥ 0.95 were considered as statistically significant. The best-fit model of nucleotide substitution for the seven DNA regions is listed in Table 1.
Table 1. Summary information of seven DNA markers.
Including sequence characteristics and best-fit model of Bayesian information criterion (BIC) for Bayesian inference.
| Nuclear maker | Plastid marker | Combined dataset | |||||||
|---|---|---|---|---|---|---|---|---|---|
| nrITS | atpB-rbcL | matK | psbA-trnQ | rbcL | rpoB-trnC | rps16 | nrITS+atpB-rbcL | Plastid genes (no atpB-rbcL) | |
| No. of accessions/tribe Anemoneae | 118/107 | 112/101 | 89/80 | 85/80 | 84/73 | 89/78 | 65/54 | 129/118 | 107/96 |
| Aligned length (bp) | 854 | 1266 | 807 | 806 | 680 | 1538 | 1055 | 2120 | 4886 |
| Variable sites/ informative sites | |||||||||
| All samples | 404/325 | 460/285 | 338/188 | 310/195 | 93/68 | 599/409 | 346/230 | 864/610 | 1686/1090 |
| tribe Anemoneae | 321/256 | 352/200 | 146/89 | 251/151 | 49/31 | 310/174 | 181/102 | 456/673 | 937/545 |
| -lnL | 8430.3365 | 6435.4960 | 4043.4941 | 4107.4513 | 2000.6280 | 7281.6526 | 4444.5674 | — | — |
| K | 239 | 228 | 182 | 176 | 170 | 182 | 134 | — | — |
| BIC model | TIM2ef+I+G | TPM3uf+G | TPM1uf+G | TPM1uf+G | TPM1+I+G | TPM1uf+G | TPM1uf+G | — | — |
Results
Characteristics of DNA sequences
Sequence characteristics of the DNA regions and the concatenated datasets are summarized in Table 1. For the matrix of tribe Anemoneae, the proportions of both variable site and parsimony-informative site were highest for nrITS (variable: 37.59%, and parsimony-informative: 29.98%, hereafter), followed by psbA-trnQ (31.14% and 18.73%), atpB-rbcL (27.80% and 15.80%), rpoB-trnC (20.16% and 11.31%), matK (18.09% and 11.03%), rps16 (17.16% and 9.67%), and rbcL (7.21% and 4.56%). The best-fit BIC models for seven DNA regions were independent (Table 1), thus the BI analyses of the concatenated datasets were partitioned using a specific model for each DNA region.
Phylogenetic analyses of single DNA marker
Phylogenetic relationships among genera resulting from of the seven DNA markers analyzed separately using ML and BI methods are presented in S1 Fig. As for Barneoudia, Knowltonia, and Oreithales only nrITS and atpB-rbcL sequences were available from GenBank, the three genera were not included in phylogenetic analyses of the other five plastid datasets. In addition, all samples of Hepatica failed to amplify for the rps16 region.
Topologies of the seven datasets were divided into two types. The first type included nrITS and atpB-rbcL datasets, which supported the splitting of tribe Anemoneae into two clades, i.e. Clematis s.l. (including Archiclematis and Naravelia) + Anemoclema and Anemone s.l. (including Barneoudia, Hepatica, Knowltonia, Oreithales, and Pulsatilla). The clade Clematis s.l. + Anemoclema corresponds to a newly defined subtribe Clematidinae by Zhang et al. [38], and the clade Anemone s.l. corresponds to subtribe Anemoninae. The other type of dataset was the other five plastid regions. All five trees showed that Anemoclema was sister to Clematis s.l., while Anemone s.l. was paraphyletic. Overall, species of Anemone were divided into two clades in all seven trees, with one clade (Anemone I) close to Pulsatilla (not with atpB-rbcL), and another clade (Anemone II) close to Hepatica (but not with the nrITS and rps16 datasets). There is no species sharing between the two Anemone clades. In the clade Clematis s.l., six datasets of single marker, except matK dataset, strongly supported the monophyly of Naravelia.
Phylogenetic analyses of nrITS +atpB-rbcL dataset
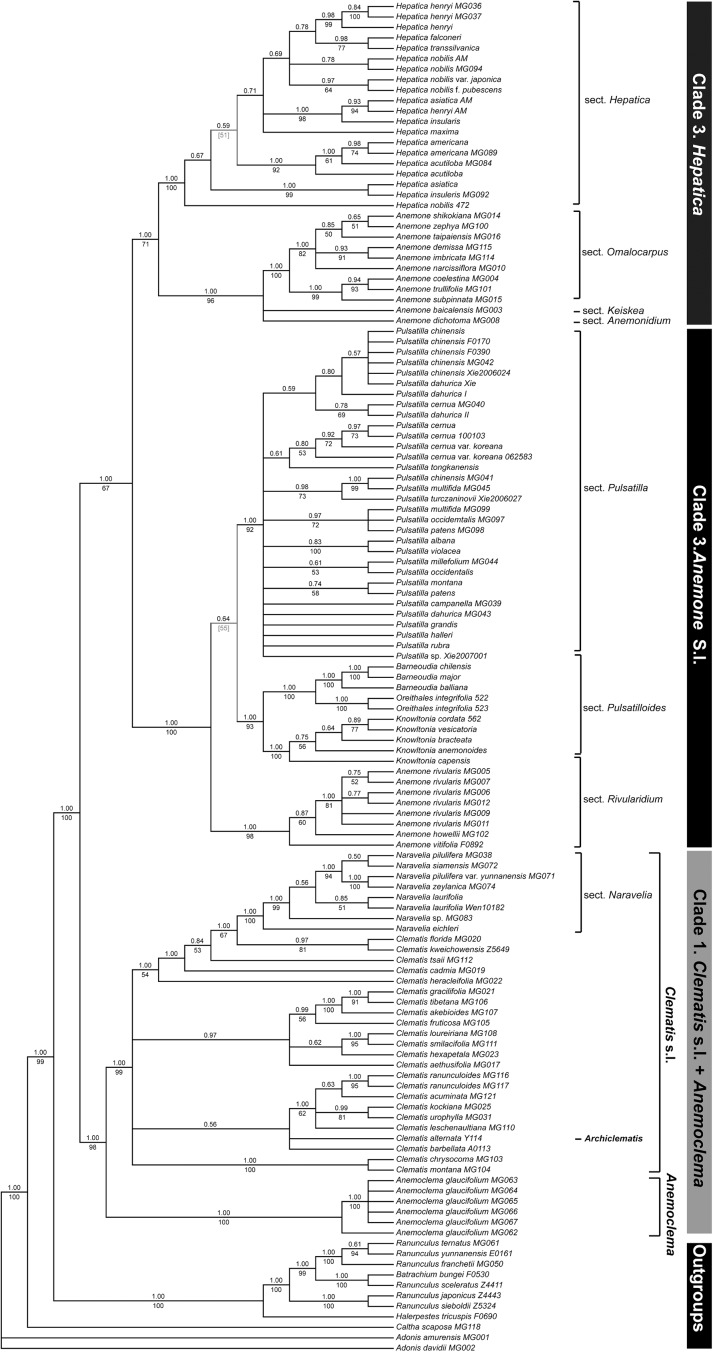
Topology of the combined nrITS and atpB-rbcL dataset is showed in Fig 1. Topological incongruence between ML and BI trees was found in two weakly resolved clades (Fig 1, S2 Fig). In the combined dataset analyses, Clematis s.l. + Anemoclema (subtribe Clematidinae, BS/PP = 98/1.00) and Anemone s.l. (subtribe Anemoninae, BS/PP = 67/1.00) were well supported as monophyletic. Three major clades were recognized (Fig 2): clade 1 corresponding to Clematis s.l. + Anemoclema; and clades 2 and 3 corresponding to two subgenera in Anemone s.l. [26]: subgenus Anemone and subgenus Anemonidium, respectively. Because subtribe Anemoninae was not supported as monophyletic by the plastid dataset (see below), we divided this subtribe into two clades to maintain consistent statements between two combined datasets.
Fig 1. Phylogenetic relationships within tribe Anemoneae based on the combination of nrITS and atpB-rbcL datasets.
The topology is that of the majority rule consensus of BI tree. Bootstrap values of ML are presented under branches, and posterior probability of BI above branches. Topological incongruence between ML and BI trees is indicated by colored nodes/branches, and topology of BI tree shows by dash lines with posterior probability in square bracket under branches.
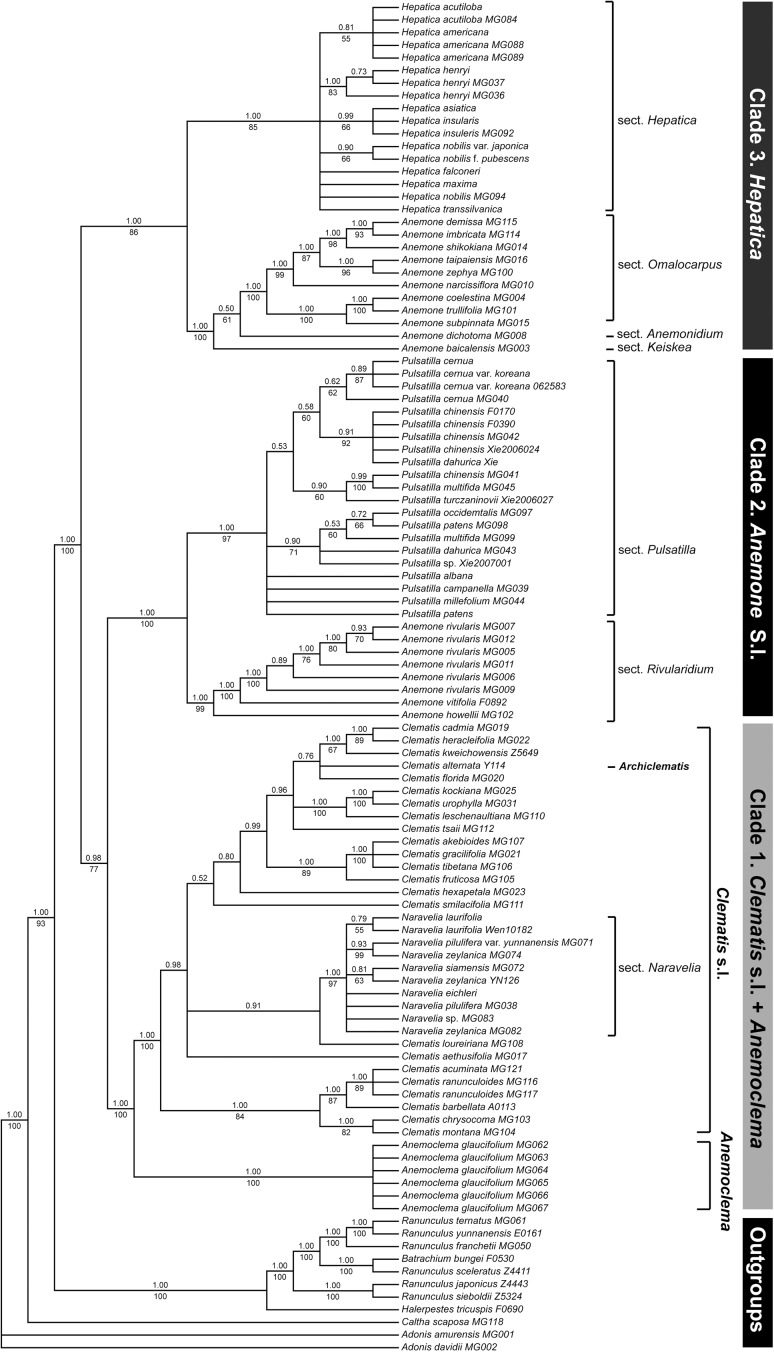
Fig 2. Phylogenetic relationships within tribe Anemoneae based on the combination of five-plastid-gene dataset.
The five plastid genes are matK, psbA-trnQ, rbcL, rpoB-trnC, and rps16. The topology is that of the majority rule consensus of ML tree. Bootstrap values of ML are presented above branches, and posterior probability of BI under branches. Topological incongruence between ML and BI trees is indicated by colored nodes/branches, and topology of BI tree shows by dashed lines with posterior probability in square bracket under branches.
In clade 1, both Clematis s.l. (BS/PP = 99/1.00) and Anemoclema (BS/PP = 100/1.00) are strongly supported as monophyletic. In Anemoclema, the Sichuan sample (MG062) was strongly supported as sister to the remaining Yunnan samples. The clade Clematis s.l. included Archiclematis and Naravelia. The backbone of the clade Clematis s.l. was poorly resolved. Four major groups were strongly supported by the BI analysis (PP > 0.95). The phylogenetic position of Archiclematis alternata (≡ C. alternata) was uncertain, as well as the position of C. barbellata Edgew. The monophyly of Naravelia (BS/PP = 100/1.00) was strongly supported, and the genus was sister to C. florida Thunb. + C. kweichowensis C. P'ei (BS/PP = 67/1.00). In the clade Naravelia, N. eichleri Tamura was sister to the remaining taxa, followed by an unknown species from Laos; N. pilulifera var. yunnanensis Y. Fei was close N. zeylanica (BS/PP = 100/1.00), but N. pilulifera Hance var. pilulifera was nested with N. siamensis Craib (PP = 0.50).
In clade subtribe Anemoninae (clades 2 + 3), four traditional genera (i.e., Barneoudia, Hepatica, Knowltonia, and Pulsatilla) were strongly supported as monophyletic, and Anemone spp. fell into two clades: Anomene II was close to Hepatica (BS/PP = 71/1.00); and Anomene I (sect. Rivularidium) was close to Pulsatilla in the ML analyses (BS = 51, S2 Fig), while it was close to the clade Pulsatilla + Knowltonia–Barneoudia (sect. Pulsatilloides) in the BI analysis (PP = 0.59, S2 Fig). In clades Hepatica and Pulsatilla, morphology-based species were not resolved as monophyletic yet. Anemone section Omalocarpus DC. was recovered as monophyletic in the clade Anemone II.
Additional ML analyses excluding samples of Barneoudia, Knowltonia, and Oreithales recovered three major clades (S3 Fig). In comparison with the full dataset, there is little difference in support values of the resolved clades. For example, BS value for monophyly of Anemone s.l. was 59 (vs. 67), that of Anemone II in clade 3 was 95 (vs. 96), and that of Clematis s.l. in clade 1 was 100 (vs. 99).
Phylogenetic analyses of the five-plastid-gene dataset (without atpB-rbcL)
Phylogenetic trees of the five-plastid-gene dataset are shown in Fig 2. Topologies were consistent in both BI and ML analyses (S4 Fig). Three strongly supported clades were recognized in tribe Anemoneae, and clades were numbered following the nrITS + atpB-rbcL dataset. The topology resulting from this dataset was different from that of the nrITS + atpB-rbcL dataset in that clade 2 was nested with clade 1, Clematis s.l. + Anemoclema (BS/PP = 77/0.98). The monophyly of subtribe Anemoninae was rejected by the plastid dataset.
Three traditional genera (Hepatica, Naravelia and Pulsatilla) were strongly supported as monophyletic, and all six samples of Anemoclema formed one clade. Clematis, including Naravelia, was paraphyletic; and Anemone was polyphyletic, separated into two subclades, Anemone I in clade 2 and Anemone II in clade 3. Clade 3 was sister to clades 1 + 3 (BS/PP = 77/0.98). Clade 3 included two subclades, Hepatica (BS/PP = 93/1.00) and Anemone II (BS/PP = 100/1.00). Within Hepatica, H. henryi (BS/PP = 83/1.00) and H. nobilis (BS/PP = 66/0.90) were monophyletic, respectively. In the clade Anemone II, A. section Omalocarpus was recovered as monophyletic. Subsequently, clade 2 divided into two subclades, Anemone I and Pulsatilla, and phylogenetic resolution in the clade Pulsatilla was poor, and some of the species appeared to non-monophyletic. In clade 1, Anemoclema was sister to Clematis s.l. The clade C. montana Buch.-Ham. ex DC.–C. acuminata DC. (BS/PP = 84/1.00) was sister to the remaining Clematis (including Archiclematis) and Naravelia. Clematis loureiroana DC. was resolved as sister to Naravelia (PP = 0.91). Interspecific relationship in Naravelia was not resolved. Clematis smilacifolia Wall. and C. hexapetala Pall. was sister to the remaining Clematis (BI = 0.96), then they formed three well or strongly supported clades, C. fruticosa Turcz.–akebioides (Maxim.) H.J. Veitch (BS/PP = 89/1.00), C. leschenaultiana–C. kockiana C.K. Schneid. (BS/PP = 100/1.00), and C. kweichowensis–C. cadmia Buch.-Ham. ex Hook. f. & Thomson (BS/PP = 67/1.00).
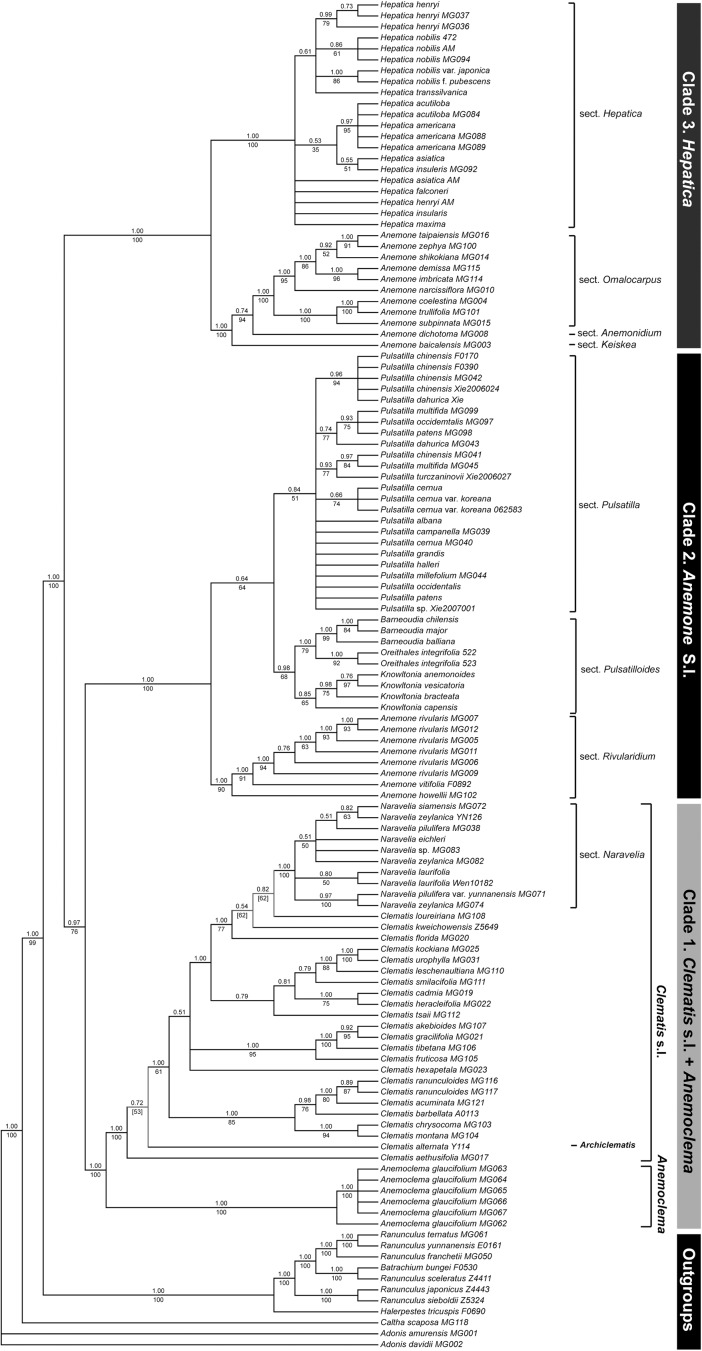
Phylogenetic analyses of the six-plastid-gene dataset
Topology of the six-plastid-gene dataset (Fig 3) recovered the same relationship of three major clades using five-plastid-gene dataset. However, two weakly incongruent clades between BI and ML trees were found in the clade Clematis s.l.: ML tree supported the clade C. alternata + C. aethusifolia Turcz. (BS = 53) and the clade C. florida + C. kweichowensis + C. loureiriana DC. (BS = 62), however, both were rejected in the BI tree (S5 Fig).
Fig 3. Phylogenetic relationships within tribe Anemoneae the combination of six-plastid-gene dataset.
The six plastid genes are atpB-rbcL, matK, psbA-trnQ, rbcL, rpoB-trnC, and rps16. The topology is that of the majority rule consensus of ML tree. Bootstrap values of ML are presented above branches, and posterior probability of BI under branches. Topological incongruence between ML and BI trees is indicated by colored nodes/branches, and topology of BI tree shows by dash lines with posterior probability in square bracket under branches.
Clade 1 and clade 2 were well supported as sister (BS/PP = 76/0.97). In clade 1, Anenoclema was sister to Clematis s.l. Then, C. alternata and C. aethusifolia were sister to remaining Clematis spp. (BS/PP = 61/1.00), followed the clade C. montana–C. ranunculoides Franch. (BS/PP = 85/1.00). The clades C. fruticosa–akebioides (BS/PP = 89/1.00) and C. leschenaultiana–C. kockiana (BS/PP = 100/1.00) were recovered as monophyletic. Clematis florida, C. kweichowensis and C. loureiriana were sister to Naravelia (BS/PP = 77/1.00). In clade 2, three clades were the same to those in nrITS + atpB-rbcL dataset. The clade Pulsatilla was weakly supported (BS/PP = 51/0.84). The clade 3 was strongly supported by both analyses (BS/PP = 100/1.00), as well as two subclades (BS/PP = 100/1.00). In clade Anemone II, sect. Omalocarpus was recovered as monophyletic. In clade Hepatica, three of four samples from H. henryi formed a clade (BS/PP = 79/0.99), five samples of H. nobilis split as two groups, and two samples of H. acutiloba and three samples of H. america were sisters (BS/PP = 95/0.97).
Additional ML analyses excluding samples of Barneoudia, Knowltonia, and Oreithales recovered three major clades (S3 Fig). In comparison with the full dataset, there is little difference in support values of the resolved clades. For example, BS value for clades 1 + 2 was 78 (vs. 76), that of the clade Naravelia in clade 1 was 99 (vs. 100). One exception was that monophyly of Pulsatilla was strongly supported (BS = 95 vs. BS/PP = 51/0.84).
Topological comparisons and dataset combinations
The SH and UA tests for constrained relationships using nrITS, atpB-rbcL, nrITS + atpB-rbcL and five-plastid-gene datasets are presented in Table 2. We only found that the unconstrained topology of the five-plastid dataset showed significant difference in both SH and AU tests when compared with the constraint nrITS topology, and in AU test when compared with the constrained atpB-rbcL topology. For combined analyses, the atpB-rbcL dataset was more suitable for concatenating with nrITS than the five-plastid-gene dataset, and nrITS dataset and the five-plastid-gene dataset should be analyzed separately.
Table 2. Summary of the Shimodaira-Hasegawa (SH) and the approximately unbiased (AU) tests.
P values were less than 0.05 in boldface. Log likelihood scores for the unconstrained analysis are given, as well as the difference in log likelihood scores between the unconstrained and the constraint topologies (∂).
| Ln likelihood | ∂ | SH | AU | |
|---|---|---|---|---|
| nrITS analyses compared with constraint clades from atpB-rbcL and five-plastid genes analyses | ||||
| Unconstrained nrITS analysis | 9064.41715 | |||
| atpB-rbcL: ((A,B),((C,D),(E,(F,G))))* | 9072.49361 | 8.07647 | 0.2888 | 0.2248 |
| Plastid: ((C,D),((A,B),(F,(E,G)))) | 9074.76080 | 10.34366 | 0.2696 | 0.0785 |
| atpB-rbcL analyses compared with constraint clades from nrITS and five-plastid gene analyses | ||||
| Unconstrained atpB-rbcL analysis | 6927.72870 | |||
| nrITS: ((A,B),(C,(D,(F,(E,G)))))) | 6934.32995 | 6.60125 | 0.38310 | 0.2220 |
| Plastid I: ((C,D),((A,B),(E,(F,G)))) | 6931.46488 | 3.73618 | 0.53430 | 0.2301 |
| Plastid II: ((C,D),((A,B),(F,(E,G)))) | 6929.99927 | 2.27058 | 0.64490 | 0.5139 |
| nrITS + atpB-rbcL analyses compared with constraint clades from five-plastid gene analyses | ||||
| Unconstrained nrITS + atpB-rbcL analysis | 16844.75792 | |||
| Plastid I: ((C,D),((A,B),(E,(F,G)))) | 16862.85264 | 18.09472 | 0.2150 | 0.1165 |
| Plastid II: ((C,D),((A,B),(F,(E,G)))) | 16861.05424 | 16.29632 | 0.1137 | 0.0588 |
| Five-plastid-gene analyses compared with constraint clades from nrITS and atpB-rbcL analyses | ||||
| Unconstrained five-plastid-gene analysis | 23900.91280 | |||
| nrITS: ((A,B),(C,(D,(E,G)))) | 23911.26222 | 80.32224 | 0.0001 | 0.0000 |
| atpB-rbcL: ((A,B),((C,D),(E,G))) | 23981.23504 | 10.34943 | 0.3485 | 0.0296 |
*Notes: A, Anemoclema; B. Clematis s.l.; C. Hepatica; D, Anemone II; E. Anemone I; F, (Knowltonia, (Barneoudia, Oreithales)); G, Pulsatilla.
Discussion
Phylogenetic incongruence among datasets
Monophyly of tribe Anemoneae was strongly supported by seven single marker datasets (S1 Fig). Within tribe Anemoneae, five major groups were recognized in all seven datasets, six major groups in the six datasets (except rps16 dataset), and nine major groups in both nrITS and atpB-rbcL datasets. Species of Barneoudia, Knowltonia and Oreithales were absent from the psbA-trnQ, rbcL rpoB-trnC and rps16 datasets, and Hepatica from the rps16 dataset because we failed to generate sequences from the samples, or there was no sequence in GenBank. For the five datasets, the remaining major groups were well supported as monophyletic. Overall, phylogenetic resolution of the backbone was poor using the single marker datasets (S1 Fig), and relationships among groups were incongruent. Based on the similarity of topologies, and the SH and AU tests, the seven datasets tended to split in two groups: one group included nrITS and atpB-rbcL, and the other group included the remaining five plastid datasets. We confirmed that taxa sampling had no effect on backbone relationships obtained with either the nrITS or atpB-rbcL datasets, because clades Clematis + Anemoclema and Anemone s.l. were also supported when Barneoudia, Knowltonia and Oreithales were excluded (S3 Fig). Generally, the conflicting topologies in plants are found between nuclear and plastid datasets [52–56]. In tribe Anemoneae, the topologies based on the nrITS and atpB-rbcL datasets were consistent [26, 41, 57]. However, topological incongruence was found between the five-plastid-dataset and atpB-rbcL suggested that plastid genes may be evolved independently in tribe Anenomeae. In a large-scale analysis, Zeng et al. [58] have documented that topologies showed differences between the single copy region genes and inverted repeat region genes, because genes in the inverted repeated region are more conservative than those in the single copy region. Meanwhile, the coding genes are more conservative than the non-coding genes. In this study, six plastid genes were not powerful enough to clarify this question. Based on published plastomes of Ranunculaceae, at least two large rearrangements (rps4 CDS and trnH tRNA- rps16 CDS) were found tribe Anenomeae, which has been detected using restriction enzymes [59]. As more and more chloroplast genomes are published [60], comparative analyses of whole chloroplast genomes may help to understand the evolutionary history of plastid genes.
Compared to the single marker datasets, phylogenetic resolution was significantly improved when the nrITS dataset was combined with the atpB-rbcL dataset, and five plastid datasets were concatenated. Meanwhile, phylogenetic conflicts between the two combined datasets became significant (AU test: P = 0.0588). In the topology, monophyly of subtribe Anemoninae was well supported by the nrITS + atpB-rbcL dataset; whereas subtribe Anemoninae was paraphyletic using the plastid dataset. In addition, support values for the clades 1 + 2 were not increased yet when the atpB-rbcL dataset was combined with the other five plastid datasets. The AU test indicated that the atpB-rbcL and the five-plastid gene datasets were tended to analyze separately.
Phylogenetic placement of Anemoclema and Naravelia
Anemoclema is upgraded as an independent genus primarily based morphological characters [28]. The flowers of Anemoclema glaucifolium resemble to Anemone, and its persistent styles with hairs to Pulsatilla [28]. Therefore, Anemoclema should belong to Anemone s.l or subtribe Anemoninae. However, preliminary phylogenetic analyses show that Anemoclema is the sister to Clematis + Naravelia, while Anemone and Pulsatilla form another clade [4]. Due to the study of Wang et al. [4] focusing on resolving the relationships of Ranunculales, Anemoclema and the other three genera (Anemone, Clematis and Pulsatilla) only included one sample/species. Subsequently, Zhang et al. [38] sampled multiple species of Anemone, Clematis, and Pulsatilla, and three individuals of Anemoclema, and they sequenced the nrITS and atpB-rbcL regions. Their results strongly support the transfer of Anemoclema to subtribe Clematidinae. In this study, we sampled six individuals of Anemoclema representing its whole distribution regions in southwestern China, and 18 taxa of Pulsatilla, and sequenced nrITS and six plastid regions. Phylogenetic analyses revealed that seven single marker datasets and three combined datasets all recovered the clade Anemoclema + Clematis s.l. Therefore, Anemoclema is clearly excluded from Anemone s.l. or subtribe Anemoninae as a distinctive genus that is sister to Clematis s.l.
Morphological delimitation of the genus Clematis is very controversial, several small genera have been proposed [12]. Of these genera, Naravelia is widely accepted as an independent genus [2, 3, 18, 21, 29, 61], although it is subsumed within Clematis s.l. by some taxonomists [14, 22, 62]. Naravelia is separated from Clematis as an independent genus by having narrow and long petals and leaflet tendrils. Traditionally, Clematis section Atragene (L.) DC. is supposed to have petals. However, floral development has shown that petals in Clematis macropetala are initiated from stamen primordia, and then antherless filaments expand to petal-like staminodia [63]. Therefore, we suggested that the “petals” of Naravelia may be the narrow and long staminodia.
Miikeda at al. [19] firstly revealed that Naravelia was nested with Clematis, then N. laurifolia and N. eichleri formed a clade. Subsequent studies [20, 24, 37, 39] confirmed the result of Miikeda at al. [19] because they used same/similar dataset of Naravelia from GenBank, or sequenced the same species. Based on our extensive sampling of Naravelia, we recovered the monophyly of Naravelia (including N. eichleri), which should be treated as a subgenus or section. Naravelia eichleri was originally placed in Naravelia by Tamura [18] based on fruiting and imperfect specimens, then Tamura [64] himself transferred it to Clematis after he collected fertile specimens without petals and leaflet tendrils. However, the sequenced sample of N. eichleri was collected by Tamura from Thailand [19]. In the present study, we demonstrated that N. eichleri was included the Naravelia group. The nrITS + atpB-rbcL dataset strongly supported N. eichleri as sister to remaining species of Naravelia, indicating that species with petal-like staminodia and leaflet tendrils may be derived from an ancient without staminodia and leaflet tendrils only once.
Generic delimitation in subtribe Anemoninae
According to molecular phylogenies [25, 26, 41, 65], Barneoudia, Hepatica, Knowltonia, Oreithales, and Pulsatilla were suggested to subsumed with Anemone. When Anemoclema has transferred to subtribe Clematidinae [38], current subtribe Anemoninae includes Anemone s.l. and Metanemone. To date, the only species of Metanemone, M. ranunculoides W. T. Wang, was collected only one time from the type locality in Weixi County, northwestern Yunnan. There is no sample of Metanemone included in any phylogenetic analyses, so the systematic placement of this genus remains unclear.
Anemone s.l. has been suggested to include Barneoudia, Hepatica, Knowltonia, Oreithales, and Pulsatilla, because this group is strongly supported as monophyletic by the combined nrITS and atpB-rbcL dataset [25, 26, 41, 65]. Our phylogenetic analyses also recovered the monophyly of Anemone s.l. using nrITS + atpB-rbcL dataset. Based on 26S rDNA and other three plastid markers (matK, rbcL, trnL-F), however, Wang et al. [4] revealed that the clade Pulsatilla + Anemone was nested with Clematis s.l., and that Hepatica was the sister to them. This conflicting result might be caused by limited sampling from tribe Anemoneae [26]. Nevertheless, the concatenated plastid dataset with extensive sampling of this tribe also revealed the paraphyly of Anenome s.l. in this study. Therefore, Barneoudia, Knowltonia, Oreithales, and Pulsatilla in clade 2 are strongly supported to subsume with Anemone s.l. [26], whereas Hepatica and Anemone II in clade 3 tends to be treated as an independent genus, i.e. Hepatica. The clade 3 corresponds to subgenus Anemonidium (Spach) Juz. [23, 26], which is characterized by a chromosome number equal to 7; achenes are globose (usually wider than long) and nearly glabrous (or with short, straight hairs) with thick walls; and each head may yield no more than 50 achenes.
Recommendations for reclassification of tribe Anemoneae
Morphologically, two subtribes have been recognized in tribe Anemoneae [1, 21]. Subtribe Anemoninae is characterized by erect herbs with basal leaves and imbricate sepals, and subtribe Clematidinae by lianas with opposite leaves (except Archiclematis alternata) and valvate sepals. However, Anemoclema, an Anemoninae-type genus, tends to transfer to subtribe Clematidinae [38]. When this treatment was adopted, diagnostic characters between subtribes Anemoninae and Clematidinae became confused. Moreover, the concatenated plastid datasets have demonstrated that subtribe Anemoninae is paraphyletic. Therefore, the subtribe rank in this tribe becomes inapplicable, and it should be abolished in future classifications.
Clematis s.l. is strongly supported as monophyletic in all phylogenetic analyses [19, 20, 24]. Therefore, Archiclematis and Naravelia must be subsumed with Clematis [20, 22]. Because phylogenetic resolution within Clematis s.l. is poor, morphology-based infrageneric classifications are not supported [19, 20]. Phylogenetic placements of Archiclematis and Naravelia are not resolved; however, monophyly of Naravelia is strongly supported. According to previous morphological classification, we suggested that Archiclematis and Naravelia should be conservatively retained as sections in Clematis [14, 66, 67].
Phylogenetic conflicts between nrITS + atpB-rbcL and the concatenated plastid datasets for Anemone s.l. provide new clues to redefine generic boundaries in this group. Phylogenetic clustering integrating morphological delimitations tend to split Anemone s.l. into two genera. Subgenus Anemone, defined by Hoot et al. [23, 26], corresponds to the new Anemone s.l., including Barneoudia, Knowltonia, Oreithales, and Pulsatilla. This genus includes four sections: Anemone, Rivularisium, Pulsatilla, and Pusatilloides [23, 26]. The subgenus Anemoniudium (Spach) Juz. needs to be separated as an independent genus, Hepatica. In the new genus Hepetica, four sections were recognized, Hepatica Spreng, Anemonidium Spach, Keiska Tamura, and Omalocarpus DC. [23, 26].
Conclusions
Monophyly of tribe Anemoneae has been demonstrated by several studies [4, 8–11]. However, phylogenetic relationship among genera was not full resolved, due to limited DNA markers were used, and/or incomplete genera samplings were analyzed. In this study, we included nine of ten recognized genera in tribe Anemoneae (only Metanemone was not sampled) and used one nuclear and six plastid markers to reconstruct a comprehensive phylogeny of tribe Anemoneae. Based on evaluation of topological incongruence, seven DNA markers were classified as two groups, nrITS and atpB-rbcL, and the remaining five plastid genes. The combined datasets resolved tribe Anemoneae as three major clades: clade 1 included Anemoclema and Clematis s.l. (including Archiclematis and Naravelia), clades 2 and 3 corresponded to Anemone subgenus Anemone (including Barneoudia, Knowltonia, Oreithales, and Pulsatilla), and subgenus Anemonidium (including Hepatica), respectively. The nrITS + atpB-rbcL supported the monophyletic of Anomone s.l. (including clades 2 and 3). However, the five-plastid-gene dataset made subgenus Anemone (clade 2) sister to the clade Anemoclema + Clematis s.l. (clade 1). Our results strongly supported to subsume Archiclematis and Naravelia within Clematis s.l., and to retain Anemoclema as an independent genus. For the genus Anemone s.l., all analyses supported to include Barneoudia, Knowltonia, Oreithales, and Pulsatilla in this genus. However, the five-plastid-gene dataset tended to retain Hepatica as a separated genus, corresponding to Anemone subgenus Anemonidium. Therefore, the updated tribe Anemoneae consists of four revised genera, Anemoclema, Anemone s.l., Clematis s.l. and Hepatica, and an unresolved genus, Metanemone.
Supporting information
(XLSX)
Note: TBD, accession number of new sequences to be determined by GenBank.
(XLSX)
(DOC)
Topology shows the majority rule consensus of ML tree. Topological incongruence between ML and BI trees are indicated by colored nodes/branches and posterior probability in square bracket under branches.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful to the Germplasm Bank of Wild Species for providing requested DNA samples; Jun He, Hua-Jie He, Jie Liu, Chun-Lei Xiang for theirs assistance in the field or providing DNA samples; to Zhen-Shan He, Jing Yang, Ji-Xiong Yang, Wen-Bin Yuan, Chun-Xia Zeng and Zhi-Rong Zhang for their assistance in molecular experiments; to Lily Zeng for her English editing in early version; to Curator of herbaria of Kunming Institute of Botany, Chinese Academy of Sciences and Field Museum of Natural History for allowing us to access specimens; to Carl. S. Keener and Claude W. dePamphilis for their valuable discussions and suggestions; and to two anonymous reviewers for their valuable comments and suggestions.
Data Availability
Data are available from Figshare (DOI: 10.6084/m9.figshare.4774753).
Funding Statement
This study was supported by Natural Science Foundation of China (31200158) and Natural Science Foundation of Zhejiang Province (LQ12C02002).
References
- 1.Tamura M. A new classification of the family Ranunculaceae 2. Acta Phytotaxon Geobot. 1991;42:177–87. [Google Scholar]
- 2.Tamura M. Ranunculaceae In: Kubitzki K, Rohwer JG, Bittrich V, editors. The families and genera of vasular plants. Vol. II Flowering plant. Dicotyledons. Berlin etc.: Springer-Verlag; 1993. p. 563–383. [Google Scholar]
- 3.Tamura M. Ranunculaceae In: Hiepko P, editor. Die Naturlichen Pflanzenfamilien,. 17 (4). Berlin: Duncker & Humblot; 1995. p. 223–555. [Google Scholar]
- 4.Wang W, Lu A-M, Ren Y, Endress ME, Chen Z-D. Phylogeny and classification of Ranunculales: Evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Syst. 2009;11:81–110. [Google Scholar]
- 5.Hu Z-H, Li K-M, Lee CL. Distribution and general morphology in Kingdonia uniflora. Acta Bot Sin. 1964;12:351–8. [Google Scholar]
- 6.Ren Y, Li Z-j, Chang H-l, Lei Y-j, Lu A-m. Floral development of Kingdonia (Ranunculaceae s. l., Ranunculales). Plant Syst Evol. 2004;247:145–53. [Google Scholar]
- 7.Bremer B, Bremer K, Chase MW, Fay MF, Reveal JL, Soltis DE, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–21. [Google Scholar]
- 8.Hoot SB. Phylogeny of the Ranunculaceae based on epidermal microcharacters and macromorphology. Syst Bot. 1991;16:741–55. [Google Scholar]
- 9.Hoot SB. Phylogeny of the Ranunculaceae based on preliminary atpB, rbcL and 18S nuclear ribosomal DNA sequence data In: Jensen U, Kadereit JW, editors. Systematics and evolution of the Ranunculiflorae. Plant Systematics and Evolution Supplement 9. Vienna: Springer; 1995. p. 241–51. [Google Scholar]
- 10.Johansson JT, Jansen RK. Chloroplast DNA variation and phylogeny of the Ranunculaceae. Plant Syst Evol. 1993;187:29–49. [Google Scholar]
- 11.Ro KE, Keener CS, McPheron BA. Molecular phylogenetic study of the Ranunculaceae: utility of the nuclear 26S ribosomal DNA in inferring intrafamilial relationships. Mol Phylogenet Evol. 1997;8:117–27. 10.1006/mpev.1997.0413 [DOI] [PubMed] [Google Scholar]
- 12.Wang WT, Li LQ. A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotaxon Sin. 2005;43:431–88. [Google Scholar]
- 13.Tamura M. A classification of genus Clematis. Acta Phytotaxon Geobot. 1987;38:33–44. [Google Scholar]
- 14.Johnson M. Slaktet Klematis. Sodertalje: Bengt Sundstrom; 1997. [Google Scholar]
- 15.Grey-Wilson C. Clematis: the genus. London: B T Batsford; 2000. [Google Scholar]
- 16.Wang WT. A revision of Clematis sect. Cheiropsis (Ranunculaceae). Acta Phytotaxon Sin. 2002;40:193–241. [Google Scholar]
- 17.Yang TYA, Moore DM. A revision of the Viorna group of species (section Viorna sensu Prantl) in the genus Clematis (Ranunculaceae). Syst Geogr PI. 1999;68:281–303. [Google Scholar]
- 18.Tamura M. A revision of genus Naravelia. Acta Phytotaxon Geobot. 1986;37:106–10. [Google Scholar]
- 19.Miikeda O, Kita K, Handa T, Yukawa T. Phylogenetic relationships of Clematis (Ranunculaceae) based on chloroplast and nuclear DNA sequences. Bot J Linn Soc. 2006;152:153–68. [Google Scholar]
- 20.Xie L, Wen J, Li L-Q. Phylogenetic analyses of Clematis (Ranunculaceae) based on sequences of nuclear ribosomal ITS and three plastid regions. Syst Bot. 2011;36:907–21. [Google Scholar]
- 21.Wang W-T, Chang M-C, Fang M-Y, Ling P-P, Ting C-T, Wang S-H, et al. Ranunculaceae subfam. Ranunculoideae. In: Wang W-T, editor. Flora Reipublicae Popularis Sinicae. 281980. p. 1–345.
- 22.Takhtajan A. Flowering plants. Berlin: Springer; 2009. [Google Scholar]
- 23.Hoot SB, Reznicek AA, Palmer JD. Phylogenetic relationships in Anemone (Ranunculaceae) based on morphology and chloroplast DNA. Syst Bot. 1994;19:169–200. [Google Scholar]
- 24.Barniske A-M. Mutational dynamics and phylogenetic utility of plastid introns and spacers in early branching eudicots: Technische Universiät Dresden; 2009. [Google Scholar]
- 25.Meyer KM, Hoot SB, Arroyo MTK. Phylogenetic affinities of south American Anemone (Ranunculaceae), including the endemic segregate genera, Barneoudia and Oreithales. Int J Pl Sci. 2010;171:323–31. [Google Scholar]
- 26.Hoot SB, Meyer KM, Manning JC. Phylogeny and reclassification of Anemone (Ranunculaceae), with an emphasis on Austral species. Syst Bot. 2012;37:139–52. [Google Scholar]
- 27.Guan M-M, Ma R, Gong X. Conservation genetics of an endemic plant, Anemoclema glaucifolium, in the Jinsha River Valley. Plant Diver Resource. 2013; 35:555–62. [Google Scholar]
- 28.Wang W-T. Duo genera nova Rannuculacearum Sinensium. Acta Phytotaxon Sin. 1964;9:103–7. [Google Scholar]
- 29.Wang W-T, Fu D-Z, Li L-Q, Bartholomew B, Brach AR, Dutton BE, et al. Ranunculaceae In: Wu C-Y, Raven P, editors. Flora of China. 6 Beijing & St. Louis: Sicence Press & Missouri Botanical Garden; 2001. p. 133–438. [Google Scholar]
- 30.Wang WT. Ranunculaceae Flora Yunnanica, Volume 11 Beijing: Science Press; 2001. p. 208–53. [Google Scholar]
- 31.Wu ZY, Lu AM, Tang YC, Chen ZD, Li DZ. The families and genera of angiosperms in China: a comprehensive analysis. Beijing: Science Press; 2003. [Google Scholar]
- 32.Zhang G-L, Gong X. The karyotype analysis of Anemoclema glaucifolium and Heteroplexis microcephala both endemic to China. Acta Bot Yunnan. 2002;24:765–8. [Google Scholar]
- 33.Jiang N, Guan KY. Lectotypification of Anemoclema glaucifolium (Ranunculaceae), endemic to southwestern China. Phytotaxa. 2012;57:56–8. [Google Scholar]
- 34.Franchet A. Plantas Yunnanenses: A. Cl. J. M. Delavay collectas. Bull Soc Bot France. 1886;33:358–467. [Google Scholar]
- 35.Ehrendorfer F. Evolutionary trends and patterns in the Anemoninae In: Jensen U, Kadereit JW, editors. Systematics and Evolution of the Ranunculiflorae. Plant Systematics and Evolution Supplement 9. Suppl. 9. Vienna: Springer; 1995. p. 283–93. [Google Scholar]
- 36.Ziman SN, Bulakh EV, Kadota Y, Keener CS. Modern view on the taxonomy of the genus Anemone L. sensu stricto (Ranunculaceae). J Jap Bot. 2008;83:127–55. [Google Scholar]
- 37.Jiang N. Molecular phylogeny and repoductive biology of Clematis L. (Ranunculaceae) [Ph.D thesis]. Kunming: Kunming Institute of Botany, Graudate University of Chinese Academy of Sciences; 2010.
- 38.Zhang Y, Kong HH, Yang QE. Phylogenetic relationships and taxonomic status of the monotypic Chinese genus Anemoclema (Ranunculaceae). Plant Syst Evol. 2015;301:1335–44. [Google Scholar]
- 39.Lehtonen S, Christenhusz MJM, Falck D. Sensitive phylogenetics of Clematis and its position in Ranunculaceae. Bot J Linn Soc. 2016;182:825–67. [Google Scholar]
- 40.Ehrendorfer F, Samuel R. Contributions to a molecular phylogeny and systematics of Anemone and related genera (Ranunculaceae-Anemoninae). Acta Phytotaxon Sin. 2001;39:293–308. [Google Scholar]
- 41.Zhang Y, Hong Y, Ren C, Tang M, Hoot SB, Yang QE. Palynology, cytology, and molecular systematics of Anemone section Begoniifolia (Ranunculaceae). Plant Syst Evol. 2015;301:411–24. [Google Scholar]
- 42.Pfosser M, Sun B-Y, Stuessy TF, Jang C-G, Guo Y-P, Taejin K, et al. Phylogeny of Hepatica (Ranunculaceae) and origin of Hepatica maxima Nakai endemic to Ullung Island, Korea. STAPFIA. 2011;95:16–27. [Google Scholar]
- 43.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–80. [DOI] [PubMed] [Google Scholar]
- 45.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. 10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- 46.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. 1999;16:1114. [Google Scholar]
- 47.Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.2 2017 [10 Feb, 2017]. Available from: http://mesquiteproject.wikispaces.com/home.
- 48.Swofford D . PAUP*. Phylogenetic analysis using parsimony (* and other methods). version 4. Sunderland, Massachusetts, USA: Sinauer Associates; 2003. [Google Scholar]
- 49.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–71. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 50.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. [DOI] [PubMed] [Google Scholar]
- 51.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Meth. 2012;9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raamsdonk LWDV, Smiech MP, Sandbrink JM. Introgression explains incongruence between nuclear and chloroplast DNA-based phylogenies in Allium section Cepa. Bot J Linn Soc. 1997;123:91–108. [Google Scholar]
- 53.Nishimoto Y, Ohnishi O, Hasegawa M. Topological incongruence between nuclear and chloroplast DNA trees suggesting hybridization in the urophyllum group of the genus Fagopyrum (Polygonaceae). Genes Genet Syst. 2003;78:139–53. [DOI] [PubMed] [Google Scholar]
- 54.Pelser PB, Kennedy AH, Tepe EJ, Shidler JB, Nordenstam B, Kadereit JW, et al. Patterns and causes of incongruence between plastid and nuclear Senecioneae (Asteraceae) phylogenies. Am J Bot. 2010;97:856–73. 10.3732/ajb.0900287 [DOI] [PubMed] [Google Scholar]
- 55.Zhang YX, Zeng CX, Li DZ. Complex evolution in Arundinarieae (Poaceae: Bambusoideae): Incongruence between plastid and nuclear GBSSI gene phylogenies. Mol Phylogenet Evol. 2012;63:777–97. 10.1016/j.ympev.2012.02.023 [DOI] [PubMed] [Google Scholar]
- 56.Yu W-B, Huang P-H, Li D-Z, Wang H. Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS ONE. 2013;8:e74828 10.1371/journal.pone.0074828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mlinarec J, Satovic Z, Malenica N, Ivancic-Bace I, Besendorfer V. Evolution of the tetraploid Anemone multifida (2n = 32) and hexaploid A. baldensis (2n = 48) (Ranunculaceae) was accompanied by rDNA loci loss and intergenomic translocation: evidence for their common genome origin. Ann Bot. 2012;110:703–12. 10.1093/aob/mcs128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng LP, Zhang Q, Sun RR, Kong HZ, Zhang N, Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 2014;5:4956 10.1038/ncomms5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoot SB, Palmer JD. Structural rearrangements, including parallel Inversions, within the chloroplast genome of Anemone and related genera. J Mol Evol. 1994;38:274–81. [DOI] [PubMed] [Google Scholar]
- 60.Tonti-Filippini J, Nevill PG, Dixon K, Small I. What can we do with 1000 plastid genomes? Plant J. 2017;89: [DOI] [PubMed] [Google Scholar]
- 61.De Candolle A. Regni vegetabilis Systema naturale: Sive ordines, genera et species, Vol. I Argentorati et Londini: Treuttel et Würtz; 1818. [Google Scholar]
- 62.Kuntze O. Monographie der Gattung Clematis. Verh Bot Vereins Prov. 1885;26:83–202. [Google Scholar]
- 63.Ren Y, Chang HL, Endress PK. Floral development in Anemoneae (Ranunculaceae). Bot J Linn Soc. 2010;162:77–100. [Google Scholar]
- 64.Tamura M. Synoptic Flora of the Ranunculaceae in Thailand. Thai Forest Bull. 1997;25:63–80. [Google Scholar]
- 65.Schuettpelz E, Hoot SB, Samuel R, Ehrendorfer F. Multiple origins of Southern Hemisphere Anemone (Ranunculaceae) based on plastid and nuclear sequence data. Plant Syst Evol. 2002;231:143–51. [Google Scholar]
- 66.Prantl K. Clematis. beträge zur morphologie und systematik der Ranunculaceen. Bot Jahrb Syst. 1888;9:325–73. [Google Scholar]
- 67.Tamura M. Notes on Clematis of Eastern Asia 3. Acta Phytotaxon Geobot. 1956;16:79–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Note: TBD, accession number of new sequences to be determined by GenBank.
(XLSX)
(DOC)
Topology shows the majority rule consensus of ML tree. Topological incongruence between ML and BI trees are indicated by colored nodes/branches and posterior probability in square bracket under branches.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
Data are available from Figshare (DOI: 10.6084/m9.figshare.4774753).