Abstract
Thioester-containing proteins (TEPs) form an ancient and diverse family of secreted proteins that play central roles in the innate immune response. Two families of TEPs, complement factors and α2-macroglobulins, have been known and studied in vertebrates for many years, but only in the last decade have crystal structures become available. In the same period, the presence of two additional classes of TEPs has been revealed in arthropods. In this review, we discuss the common structural features TEPs and how this knowledge can be applied to the many arthropod TEPs of unknown function. TEPs perform a wide variety of functions that are driven by different quaternary structures and protein–protein interactions between a common set of folded domains. A common theme is regulated conformational change triggered by proteolysis. Structure-function analysis of the diverse arthropod TEPs may identify not just new mechanisms in innate immunity but also interfaces between immunity, development and cell death.
Keywords: Innate immunity, Infectious disease, Complement system, Crystallography
Introduction
Thioester-containing proteins (TEPs) are large (>100 kDa), secreted glycoproteins found in both deuteromes and protostomes. Two classes of TEPs are well known. Complement factors are monomeric and deposit on surfaces when activated (Müller-Eberhard 1975). In contrast, α2-macroglobulins (A2Ms) are typically multimeric, pan-protease suicide inhibitors that encapsulate targets once cleaved in a protease-sensitive “bait region” (Barrett and Starkey 1973). These discrete functions are unified by the role of an internal β-cysteinyl-γ-glutamyl thioester bond that mediates covalent attachment of TEPs to substrates (Janatova et al. 1980; Law et al. 1980; Tack et al. 1980). Both complement and A2M play key roles in innate immune responses. Complement deposition on pathogen surfaces causes enhanced phagocytosis (opsonization), recruitment of phagocytes to sites of infection (chemotaxis), and direct lysis, while A2Ms inactivate and clear protease virulence factors. Besides their immune functions, complement and A2M play important roles in homeostasis of immune responses and host serum proteases (Ricklin et al. 2010; Rehman et al. 2013).
Complement activity was identified in other vertebrates and invertebrates prior to discovery of the thioester mechanism of binding (Day et al. 1970; Jensen et al. 1981). Shortly after the discovery of A2Ms, they were identified in both vertebrates and invertebrates, notably the horseshoe crab Limulus polyphemus (Starkey and Barrett 1982b, a; Starkey et al. 1982; Quigley and Armstrong 1983, 1985; Armstrong and Quigley 1987). Today, TEPs identified as complement factors and A2Ms have been identified across animal phyla (chordates, arthropods, echinoderms, coelenterates, molluscs), and A2Ms have been identified in bacteria. However, complement and A2M have been lost in certain lineages, most notably in the protostomes. While complement factors have been identified in crustaceans and arachnids, insects have two novel classes of TEP: (1) insect TEP (iTEP) and (2) macroglobulin/complement-related (MCR).
Dramatic progress in the past decade has provided a plethora of structural data to guide the analysis of TEPs with unknown function (Table 1). The complement system as well as the A2Ms have recently been reviewed (Armstrong 2006; Ricklin et al. 2010), including discussion of their structural features (Forneris et al. 2012; Gros et al. 2008; Lea and Johnson 2012; Rehman et al. 2013). In this review, we discuss the common chemistry and architecture of TEPs based on the present knowledge of complement factors, A2Ms, and the one iTEP of known structure, Anopheles gambiae thioester-containing protein 1 (AgTEP1). We then discuss the known or predicted features of arthropod TEPs, especially the complement-like mechanism of AgTEP1. The roles of quaternary structure and conformational change in TEP function are major obstacles to predicting function from primary sequence data. Structure-function studies are therefore required to understand the molecular properties of iTEPs, which are likely to be a source of novel biochemical diversity within this ancient protein family.
Table 1.
Crystal structures of thioester-containing proteins
| TEP structure description | PDB |
|---|---|
| Complement, pre-activation | |
| C3 (human) (Janssen et al. 2005) C3 (bovine) (Fredslund et al. 2006) C4 (Kidmose et al. 2012) C5 (Fredslund et al. 2008) C5-SSL7 (Laursen et al. 2010) C5-CVF (Laursen et al. 2010) |
2A73 2B39 4FXK,4FXG 3CU7 3KLS,3KM9 3PVM,3PRX |
| Complement, post-activation | |
| C3b (Janssen et al. 2006) C3b-CR1g (Wiesmann et al. 2006) C3b-fH (Wu et al. 2009) C3bBb (Forneris et al. 2010) C3bBbD (Forneris et al. 2010) C3c (Janssen et al. 2005) C3c-CR1g (Wiesmann et al. 2006) C3d (Nagar et al. 1998) C3d-CR2 (Szakonyi et al. 2001; van den Elsen and Isenman 2011) C5b6 (Hadders et al. 2012) |
2I07 2ICF 2WII 2XWJ 2XWB 2A74 2ICF 1C3D 1GHQ,3OED 4A5W |
| A2M structures | |
| A2M MG2 (Doan and Gettins 2007) A2M RBD (human) (Jenner et al. 1998) A1M RBD (rat) (Xiao et al. 2000) A2M(MeNH2) (Marrero et al. 2012) |
2P9R 1AYO 1EDY 4ACQ |
| iTEP structures | |
| AgTEP1*R1 (Baxter et al. 2007) AgTEP1*S1 (Le et al. 2012) |
4D94 4LNV |
The architecture and chemistry of TEPs
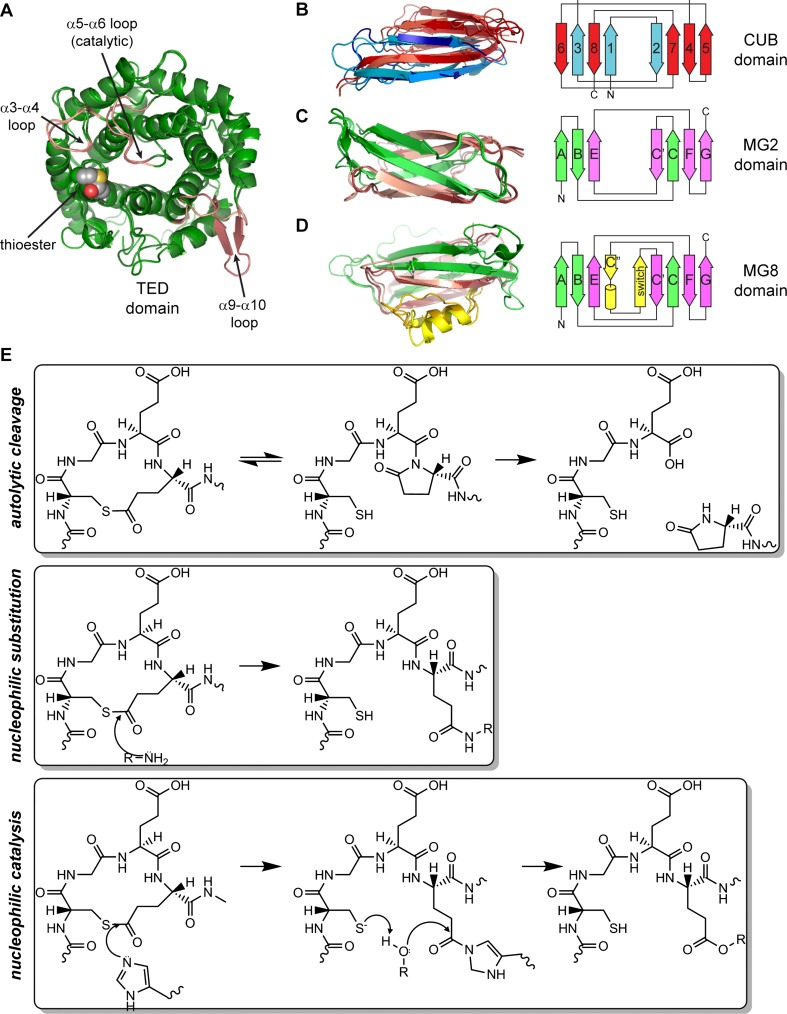
TEPs are modular proteins that contain three types of domains. The thioester domain (TED) is a 12-helix α-α barrel with the inner helical N-termini forming a concave face (Fig. 1a) (Nagar et al. 1998). The TED is inserted within the TEP CUB domain, which contains eight β-strands in two antiparallel β-sheets (Fig. 1b) (Bork and Beckmann 1993; Romero et al. 1997; Varela et al. 1997). The TEP CUB starts at the third strand of the canonical fold, so the N- and C-terminal β-strands occupy adjacent positions in a single β-sheet. The CUB in turn inserts between a series of macroglobulin domains (MG), seven β-strands arranged in two antiparallel β-sheets with a jelly-roll topology (Fig. 1c). MG topology is similar to the fibronectin-III (fnIII) fold (Leahy et al. 1992), found in numerous protein families such as integrins and transglutaminases.
Fig. 1.
a Thioester domain (TED), thioester bond in VDW spheres, α3-a4, α5-α6 (catalytic) and α9-α10 (β-hairpin) loops shown in pink. b-d Topology and ribbon diagram of the three β-sheet domains of complement C3, α2-macroglobulin (A2M) and AgTEP1. b CUB domain, strand 1 corresponds to strand 3 of the canonical CUB fold. c MG2, β-strands labeled according to fnIII, with A-B-E forming one sheet, and C-C’-F-G the other. d MG8, β-α-β insertion highlighted in yellow. e Chemical reactions of the thioester loop CGEQ: autolytic cleavage, nucleophilic substitution and nucleophilic catalysis. PDB IDs (Table 1): a 2A73, 4D94, b 2WII, 4ACQ, 4D94, c 2A74, 2P9R, 4D94, d 2A73, 1AYO, 4D94
All TEPs have eight MG domains (MG1–8). The MG8 domain, known as the receptor-binding domain (RBD) in A2Ms, contains an additional β-α-β motif within the C-C’ loop that packs against strand E in the first sheet and strand C of the second sheet (Fig. 1d). Complement factors have two more domains: the anaphylatoxin (ANA) and C345C domain. ANAs are small (74–77aa) 4-helix bundles stabilized by three disulfide bonds (Morikis et al. 2005; Klos et al. 2013), while the C345C domain is a ~150aa α/β domain with a netrin fold (Bányai and Patthy 1999; Ishii et al. 1992) appended to the C-terminus of the MG8 domain by a 2-disulfide anchor (ANK) motif.
The key feature of thioester-containing proteins is an internal β-cysteinyl-γ-glutamyl thioester bond (Janatova et al. 1980; Law et al. 1980; Tack et al. 1980). The thioester is contained in a specific sequence motif, Cys-Gly-Glu-Gln (CGEQ), located in the loop before the first inner helix (α2) of the TED (Fig. 1a). Thioester bonds are labile to hydrolysis or nucleophilic attack by amine and hydroxyl functional groups. In a synthetic model peptide, and the protein under denaturing conditions, substitution by the peptide nitrogen to form a lactam is preferred (Khan and Erickson 1981; 1982; Khan et al. 1986) resulting in autolytic cleavage of the peptide bond (Fig. 1e) (Sim and Sim 1981). Autolysis of the peptide chain is prohibited by folding of the TED in the native state. However, the thioester is still susceptible to hydrolysis if exposed to the solvent. Hydrolysis is prevented by sequestration of the thioester in a protein interface between the TED and MG8 domains. Regulated proteolysis within a separate protease-sensitive region in both complement factors and A2Ms causes a large conformational change that disrupts the TED-MG8 domain interface, thereby exposing the thioester bond.
Hydrolysis is an example of general nucleophilic attack on the thioester (Fig. 1e). Small amines such as methylamine (MeNH2) can access the TED-MG8 interface to react with the thioester. Primary amines or alcohols are better nucleophiles than water. Hence, activation of TEPs in proximity to a protein or cell surface leads to covalent attachment of the TEP to the protein or surface, respectively. Within the complement system, proteolytic activation is tightly regulated by recognition of non-self to avoid an auto-immune response. In contrast, A2M activation is unregulated. Cleavage in the bait region leads to protease sequestration by covalent attachment to, and/or entrapment of, the activating protease (Barrett and Starkey 1973; Crews et al. 1987; Feldman et al. 1985). A2Ms not only sequester pathogenic proteases but also physiological proteases, helping sense and maintaining a homeostatic level of protease activity within the serum (Chu et al. 1994; Rehman et al. 2013).
In some TEPs, thioester reactivity is enhanced by nucleophilic catalysis, specifically by a residue in the catalytic loop conserved as histidine in many complement factors. In an elegant series of studies, Dodds and Law demonstrated that the histidine acts as nucleophilic catalyst, accelerating the reaction of complement factors with hydroxyl nucleophiles, including hydrolysis (Law et al. 1984; Dodds and Law 1988, 1990; Sepp et al. 1993; Dodds et al. 1996; Law and Dodds 1997). For A2Ms, the histidine is usually replaced by acidic or neutral residues, nucleophilic substitution is uncatalyzed, and the preference for amine nucleophiles is enhanced. Differences in reactivity and selectivity between different complement factors, however, suggest that other residues in the catalytic and surrounding loops also affect the thioester’s reactivity for a given substrate (Dodds and Law 1990).
Inter-domain interactions and conformational change
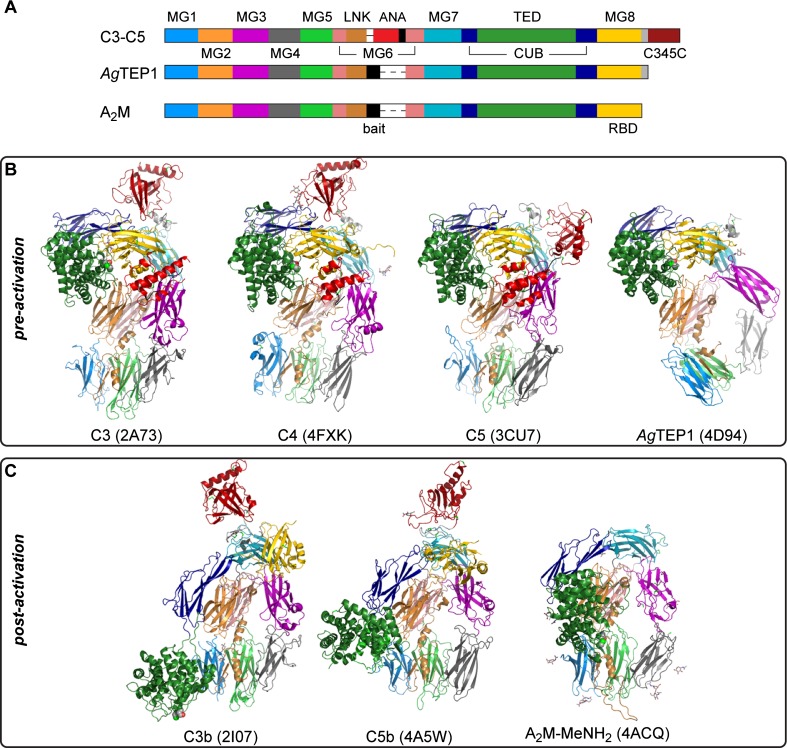
The TED, CUB, and MG domains of TEPs adopt a common quaternary structure (Fig. 2a) that is crucial to their function. Conformational changes accompanying activation expose both the thioester bond and so-called ‘cryptic’ binding sites for surface receptors and serum proteins, which mediate downstream signaling and effector mechanisms of TEP activation. Structures have been determined for the pre-activation state (Fig. 2b) of the three vertebrate complement factors C3–C5 and AgTEP1 (Janssen et al. 2005; Fredslund et al. 2006, 2008; Baxter et al. 2007; Laursen et al. 2010), and the post-activation state (Fig. 2c) of C3 and C5 (C3b, C5b), and MeNH2-treated A2M (A2M-MeNH2) (Janssen et al. 2006; Wiesmann et al. 2006; Hadders et al. 2012; Marrero et al. 2012). Distortion of the quaternary structure, such as the pre-activation state of AgTEP1 and disorder of RBD in the post-activation state of A2M, is discussed below. Some TEPs, such as C5, do not actually possess a thioester bond but have the same quaternary structure. Thus, proteolytic regulation of conformational change is a function independent of covalent binding to substrates.
Fig. 2.
a Schematic diagram of TEP domain structure. The furin-sensitive cleavage site before the ANA domain in complement is called the ‘protease-sensitive’ region for iTEPs and the ‘bait region’ for A2M. The MG8 domain in complement and iTEPs is called the RBD for A2M. b Crystal structures of TEPs in pre-activation states: C3, C4, C5, and AgTEP1. c Crystal structures of TEPs in post-activation states: C3b, C5b, and A2M-MeNH2. Domains in (b, c) are colored according to their position in (a)
In all TEP structures, the first six MG domains form a right-handed super-helix called the β-ring, in which the A/B/E β-sheet of MG1 and MG2 pack against the C/C’/F/G or MG5 and MG6, respectively. The LNK extends down the side of MG2-MG6 and MG1-MG5, donating a fourth β-strand to the MG1 A/B/E sheet. The protease-sensitive region spans the void created by β-ring, between the end of the LNK and the resumption of the MG6 fold. Thus, proteolysis splits TEPs into N- and C-terminal chains—the α and β chain, respectively—that remain associated as one molecule. Disulfide bonds connect the α and β chains in many TEPs but they are not required; the AgTEP1 structure, for instance, has no intra-chain disulfide bonds. Finally, multimeric A2Ms have intermolecular contacts, but the existence of monomeric A2Ms implies such structural features are not required for A2M function so they are not discussed further.
The TED-MG8 interface protects the thioester bond prior to activation (Fig. 2b). The interface is stabilized by domain interactions with both the α and β chains. The TED inserts into the CUB β3-β4 loop. CUB in turn inserts between MG7 and MG8, contacting TED and MG8 to form a ‘superdomain’(Fredslund et al. 2006) superimposable in C3–C5 and AgTEP1. MG7 contacts both CUB and MG8 and the whole α chain sits on top of the β-ring, the MG2-MG6 dimer contacting the TED-CUB-MG8 superdomain while MG3 contacts MG7. There is currently no A2M crystal structure with an intact thioester bond, but flexible modeling of SAXS and EM data for tetrameric human A2M and monomeric E. coli A2M (ECAM) support a similar domain arrangement to that of complement and AgTEP1 (Marrero et al. 2012; Neves et al. 2012).
TEPs undergo a large-scale conformational change upon activation (Fig. 2c). The TED-MG8 interface separates and the TED moves 50–100 Å to a position at the base of the β-ring. Disruption of the TED–MG8 interface is a key feature in activation, as hydrolysis and aminolysis of the thioester have similar structural and functional consequences as proteolysis (basal complement activity due to hydrolysis is known as ‘tick-over’) (Pangburn and Müller-Eberhard 1980; Isenman et al. 1981). During activation, MG7 and MG8 rotate around the central axis of the β-ring; in A2M-MeNH2, the RBD separates from MG7, becoming flexible relative to the remaining structure (Marrero et al. 2012). The β-ring adopts a similar prolate conformation in all post-activation TEP structures, suggesting that it represents a stable conformation for these domains.
The ANA domain plays a key structural role in complement activation. Complement factors contain a furin-sensitive site at the start of the protease-sensitive region and are cleaved at this position prior to secretion. The ANA, placed between this cleavage site and the resumption of the MG6 domain, acts as a molecular wedge between the MG3 and MG8 domains. A specific protease complex, or convertase, cleaves a scissile bond at the C-terminus of the ANA domain, which dissociates, destabilizing the MG8 domain and thereby causing activation. A similar structural trigger presumably exists in the bait region of A2Ms. In AgTEP1, MG3 is repositioned to stabilize the MG8 domain in the absence of the ANA by formation of a triangular MG3-MG7-MG8 interface (Fig. 2b), distorting the MG1-MG6 super-helix relative to complement factors.
Structures of complement factors bound to regulatory molecules or receptors (Table 1) reveal a variety of cryptic binding sites generated by TEP activation (Gros et al. 2008; Lea and Johnson 2012; Forneris et al. 2012). Regulatory factors are not conserved between vertebrate and arthropod lineages, so neither may be the cryptic binding sites involved in TEP regulation. Nevertheless the importance of cryptic binding sites to the function of TEPs is underlined by the fact that pathogens such as S. aureus produce specific factors that bind C3b and occlude the binding site for factor B, inhibiting complement-mediated immune responses (Rooijakkers et al. 2009; Garcia et al. 2010). Conversely, cobra venom contains a specific factor that mimics a C3b cryptic binding site for factor B to activate the complement system, stimulating vasodilation (Janssen et al. 2009; Krishnan et al. 2009; Laursen et al. 2010). Thus, manipulation of TEPs by pathogens, parasites and predators is widespread, and can be a tool to identify significant physiological interactions between TEPs with other immune factors.
Structure-based prediction and analysis for insect TEPs
Phylogenetic analysis of arthropod TEPs
One may think the function of arthropod TEPs could be predicted by phylogenetic analysis in comparison to vertebrate complement factors and A2Ms. This is not necessarily true, as TEP function is dictated by quaternary structure that may be poorly correlated with primary sequence. The evolutionary history of arthropods is complex with multiple terrestrial colonizations by the subfamilies of arachnida and pancrustacea (Grimaldi 2010b, a). There is no singular molecular tree for arthropods (Edgecombe 2010), and immune genes like TEPs are subject to species-specific expansion (Christophides et al. 2002), So while mosquitoes (Anopheles, Aedes, Culex) have over a dozen TEP genes, Drosophila melanogaster has five and bees (Apis mellifera, Bombus impatiens) have only three.
Only a few arthropod TEPs have been functionally characterized and only one, AgTEP1, is of known structure. Despite its structural and functional homology to complement C3 (Baxter et al. 2007; Levashina et al. 2001), AgTEP1 clusters with arthropod TEPs, separate from both complement factors and A2Ms (Blandin and Levashina 2004; Bou Aoun et al. 2011; Mone et al. 2010) and even from TEPs of other Diptera such as Drosophila (Bou Aoun et al. 2011). Such lack of phylogeny led to a hypothesis that mosquitoes separately evolved a complement-like system in an instance of convergent evolution (Jacob 1977; Waterhouse et al. 2007). If so, Drosophila and other insects may have separately evolved either complement-like or A2M-like TEPs, making functional assignment by phylogeny difficult.
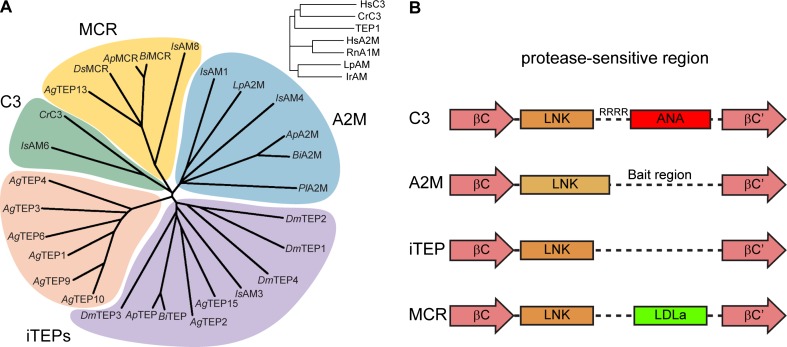
The common ancestor of AgTEP1 and complement factors, however, was probably a complement-like protein if not a complement factor. In a multiple sequence alignment with complement C3 from humans and the horshoe crab Carcinoscorpius rotundicauda (Zhu et al. 2005), rat A1M and A2Ms from humans, the horseshoe crab L. polyphemus (tetrameric), and the hard tick Ixodes ricinus (Buresova et al. 2009), AgTEP1 clusters with complement factors (Fig. 3, inset). Both complement factors and iTEPs are found in the crayfish Pacifastacus leniusculus, the spider Hasarius adansoni, and ticks have members of all TEP classes (Kopacek et al. 2000; Buresova et al. 2009, 2011; Kopacek et al. 2012). Thus, the common ancestors of insects and other arthropods possessed both complement factors and A2Ms.
Fig. 3.
a Phylogenetic analysis of selected arthropod TEPs via Clustal Omega alignment of the MG3-ANK region. Crustacea: C. rotundica C3; L. polyphenus A2M; P. lacificas A2M. Arachnida: I. scapularis AM1, AM3, AM4, AM6, AM8 (Buresova et al. 2011). Hymenoptera: A. mellifera TEP (GB45417), A2M (GB42455), MCR (GB484204); B. impatiens TEP (BIMP24442), A2M (BIMP19175), MCR (BIMP19195). Diptera: D. melanogaster TEP1–4, MCR; A. gambiae TEP1–4, TEP6, TEP9–10, TEP12, MCR (TEP13), TEP14). Inset, A. gambiae TEP1 clusters with human and crustacean complement factors rather than A2Ms. b Schematic diagram of the protease-sensitive region for the four clades of TEPs found in arthropods. The LNK region of C3 and iTEPs are structurally homologous whereas the A2M LNK region is extended, terminating with a disulfide bond. A2Ms and iTEPs contain an unstructured region of broad protease-sensitivity while C3 and MCR have small, disulfide-rich domains between the LNK region and resumption of the MG6 fold
From the known structures of TEPs, the region starting from the conserved MG2-MG3 linker FXVXE(F/Y)VL to the end of the MG8 domain may be useful for classification of TEPs. The MG3-MG4 domain adopts distinct conformations in AgTEP1 and C3 and is a dimerization interface in A2M. The LNK and protease-sensitive region (from a conserved aspartic acid in MG6 βC to a conserved tryptophan in βC’) contains the 6-cysteine ANA domain in complement factors, a conserved disulfide followed by the bait-region in A2M, and neither extra domains nor disulfides in AgTEP1. Finally, AgTEP1 and complement factors share the 4-cysteine ANK motif terminating the MG8 domain whereas A2Ms terminate with only a single cysteine.
Aligning the MG3-ANK regions of selected arthropod TEPs from Crustacea (L polyphemus A2M, C. rotundicauda C3, P. leniusculus A2M), Arachnida (I. scapularis), Hymenoptera (A. mellifera, B. impatiens), and Diptera (D. melanogaster, A. gambiae), four clades of arthropod TEPs are identified (Fig. 3): complement (C3), iTEPs, A2Ms, and MCRs. True complement factors are only in crustaceans and arachnids, but all arthropods have complement or iTEPs. suggesting that a complement-like system of TEP-mediated opsonization is broadly conserved in arthropods. The hymenoptera represent a minimum TEP repertoire for insects: an iTEP, an A2M, and an MCR. A subset of A. gambiae iTEPs exist as a species-specific expansion, while a separate, low-branched clade combines other Anopheles TEPs with Drosophila, the hymenopteran TEP, and Ixodes IsAM3. An open question is whether this broad class are also complement-like or if some have A2M-like or unique/hybrid properties, since a homolog of known A2Ms has not been identified in the Diptera.
Functional analysis of A. gambiae TEP1
The malaria vector A. gambiae has multiple iTEPs, but AgTEP1 has been most extensively studied. AgTEP1 is a key factor in the immune response of mosquitoes to malaria, binding to Plasmodium ookinetes in the basal lamina of the midgut epithelium and targeting them for lysis (Blandin et al. 2004; Fraiture et al. 2009; Povelones et al. 2009, 2011; Baxter et al. 2010; Le et al. 2012). Unlike complement C3 or A2M, AgTEP1 is secreted as a full-length protein but is cleaved within the protease-sensitive region to produce a two-chain molecule AgTEP1cut. Both full-length AgTEP1 and AgTEP1cut are present in the hemolymph (Blandin et al. 2004; Fraiture et al. 2009). AgTEP1 has two classes of alleles, AgTEP1*S and AgTEP1*R, found in A. gambiae strains that are susceptible (S) or refractory (R) to Plasmodium infection, respectively (Blandin et al. 2004, 2009; Molina-Cruz et al. 2012; White et al. 2011). AgTEP1*S and R alleles are >90 % identical in sequence.
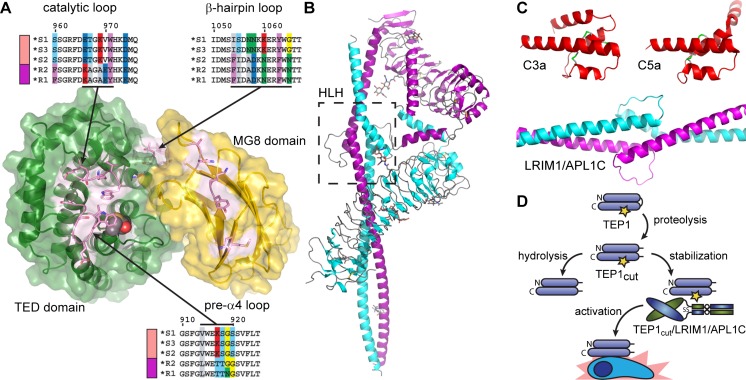
Variation between AgTEP1 alleles is mostly confined to the TED, CUB, or MG8 domains, and especially the α3-α4 loop, the catalytic loop (α3-α4) (Fig. 4a). Thus, the distinct phenotypes between S and R mosquitoes may be directly related to the reactivity of the AgTEP1 thioester. Based upon the effect of variation in the catalytic loop in C4 allotypes (Law and Dodds 1997), substitutions in the catalytic and pre-α4 loops can be expected to influence the reactivity of the thioester bond. Indeed, AgTEP1*S and AgTEP1*R alleles have significantly different rates of hydrolysis after cleavage in the protease-sensitive region (Le et al. 2012).
Fig. 4.
a Exploded view of the AgTEP1 TED-MG8 interface, highlighting distinct allotypes for the catalytic loop and the α3-α4 loop. b The crystal structure of the heterodimeric complex of LRIM1/APL1C, the helix-loop-helix (HLH) motif indicated by the boxed region. c Scale comparison of C3a, C5a and LRIM/APL1C HLH motif. d Current model of AgTEP1 regulation by LRIM1/APL1C. Short and long rounded rectangles represent the N- and C-terminal chains of TEP1, joined by a curved line representing the protease-sensitive, which is constitutively cleaved in the insect hemolymph. A yellow star indicates an intact thioester bond
Like complement, cryptic binding sites revealed by activation are expected to play an important role of AgTEP1. Unfortunately, most known complement system proteins are not conserved in insects, but some predictions can be made based upon existing complement structures. For instance, complement-control protein (CCP) domains are found in the Anopheles genome, including C-type lectin selectin-like 1 and 2 (CTLSE1, CTLSE2) with significant homology to human CR2 (>24 % identity, BLAST E < 10−5), but no phenotype has been assigned. Low-density lipoprotein receptor 1 (LRP1), the known receptor for A2M, has been identified as a hemocyte receptor for activated AgTEP1 (Moita et al. 2005). LRP1 is a multidomain protein with repeats of small disulfide-rich domains called low-density lipoprotein class A and B (LDLa, LDLb), which are also found in other complement proteins such as C6, C8, and Factor I. Actual binding of these, or any other protein with AgTEP1, is yet to be proven. Notably, the β-hairpin within the TED α9-α10 loop, which is a cryptic binding site for C6 on C5b (Hadders et al. 2012), is a site of hypervariation between AgTEP1 alleles (Baxter et al. 2007; Blandin et al. 2009). Hence, allotypes of AgTEP1 may differ vary not only in their binding to pathogens but also in the effector pathways regulated by their activation.
In the absence of homologs to vertebrate complement components, new molecular mechanisms of TEP regulation have been discovered. AgTEP1 is regulated by the leucine-rich repeat (LRR) proteins LRIM1 and APL1C, two members of a structurally distinct and mosquito-specific LRR protein family (Fig. 4b) (Osta et al. 2004; Riehle et al. 2006; Waterhouse et al. 2010). AgTEP1cut requires the heterodimer of LRIM1 and APL1C for stability in vitro and in vivo (Fraiture et al. 2009; Povelones et al. 2009, 2011; Baxter et al. 2010; Le et al. 2012). The crystal structure of LRIM1/APL1C (Fig. 4b) revealed a heterodimeric complex formed via their C-terminal coiled-coil domains including an interposed helix-loop-helix motif. The helix-loop-helix motif is an unusual structural feature for an extracellular complex. In comparison to the C3a and C5a fragments (Fig. 4c), the dimensions of the loop are similar to that of the disulfide-bridged helices but the hand of the parallel helices is reversed. Deletion of the helix-loop-helix abrogates LRIM1/APL1C binding to AgTEP1 (Povelones et al. 2011). The helix-loop-helix may act as a structural homolog of the ANA, inserting between the MG3 and MG8 domains to stabilize a reactive intermediate of AgTEP1cut, that otherwise rapidly hydrolyzes in the fluid phase.
AgTEP1cut slowly precipitates due to the hydrolysis of the thioester bond, but is stabilized by the presence of LRIM1/APL1C (Fig. 4d) (Baxter et al. 2010; Le et al. 2012). This stabilization strongly suggests that a ternary complex is formed between AgTEP1cut and LRIM1/APL1C, though efforts in this laboratory have thus far failed to determine its structure. It may be that the ternary complex itself is of a weak or transitory nature, and exists in equilibrium with free AgTEP1cut. The AgTEP1cut/LRIM1/APL1C is presumed to be recruited to the site of infection where dissociation or degradation of LRIM1/APL1C leads to activation of AgTEP1 in direct proximity to the pathogen surface.
Much about the AgTEP1 complement-like system remains to be discovered. APL1A and APL1B, closely related proteins to APL1C, direct the AgTEP1 response to different pathogens (Mitri et al. 2009), but the mechanism is unknown. CLIPs, an arthropod-specific protease family (Jiang and Kanost 2000; Jang et al. 2008), are reportedly involved in amplifying AgTEP1 activation following initial activation on a pathogen surface (Povelones et al. 2013), again by mechanisms unknown. Activation of NOS signaling within the midgut epithelia during their traversal by Plasmodium parasites is important for later recognition (Oliveira et al. 2012). Finally, the P. falciparum protein P47 was recently reported to be responsible for parasite evasion of the AgTEP1 response (Molina-Cruz et al. 2013), again by mechanisms unknown.
Functional analysis of other iTEPs
The homolog of AgTEP1 in Aedes aegyptii has been implicated in the immune response to flaviviral infection (Cheng et al. 2011). Drosophila melanogaster has four TEPs, DmTEP1–4, with an intact thioester bond. DmTEP1, DmTEP2, and DmTEP4, are upregulated on immune challenge (Lagueux et al. 2000). In a cell-based RNAi assay, DmTEP2 knockdown impaired phagocytosis of E. coli, while DmTEP3 knockdown impairs the phagocytosis of Staphylococcus aureus, suggesting they function as opsonins (Stroschein-Stevenson et al. 2006). DmTEP2 is also upregulated in S2 cells infected with the alphavirus Sindbis (Mudiganti et al. 2010). Yet, while AgTEP1 knockdown in vivo exhibits a strong phenotype for Plasmodium infection, TEP1–4-deficient flies are neither more susceptible to bacterial nor to fungal infection (Bou Aoun et al. 2011). It has been suggested the lack of a phenotype for DmTEPs is a function of the infection model tested. If so, other classes of pathogens (e.g., kinetoplastids, parasitoid wasps), or natural routes of infection, involving invasion of the gut or barrier epithelia, may reveal an in vivo phenotype.
Transcriptional activation of TEPs upon infection has also been reported for tetse flies (Weiss et al. 2011) and honey bees (Erler et al. 2011), but no functional analysis has been reported. Despite the large body of work on lepidopteran immunity, especially the tobacco hornworm Manduca sexta (Jiang et al. 2010), there are almost no reports on lepidopteran TEPs. Two TEP genes were reported in the M. sexta immunotranscriptome, transcript levels were low and changes following mixed-microbe injection small (Gunaratna and Jiang 2013). A similar result has been reported for TEPs (described as ‘macroglobulins’) in the silkworm Bombyx mori (Zhao et al. 2012). Hence, it is yet to be determined if iTEPs constitute a broadly conserved complement-like system in insects. If so, their mechanism of regulation is bound to be distinct from AgTEP1, since the LRIM1/APL1 family of LRR proteins are only found in mosquitoes (Waterhouse et al. 2010).
The hard tick Ixodes ricinus has nine TEP genes: three genuine complement factors, three A2Ms, one iTEP (IrTEP, also IrAM3), and two putative MCRs (Buresova et al. 2011). IrTEP knockdown reduced the phagocytosis of injected E. coli, but not C. indologenes, while knockdown of complement factor IrC3-3 reduced phagocytosis of both bacteria. It remains to be determined if ticks actually operate a simultaneous complement and complement-like system, or if IrTEP has a specific or unique role in the immune response to infection.
Macroglobulin/complement-related (MCR)
The MCR class of arthropod TEPs, named according to its annotation in the D. melanogaster genome, is worth special mention. While iTEPs vary greatly in number across insect genomes, a single copy of MCR is highly conserved across the hexapoda. Genetic knockout of MCR is larval lethal in Drosophila, but RNAi knockdown of MCR was found to impair the phagocytosis of C. albicans in Drosophila S2 cells (Stroschein-Stevenson et al. 2006). As for DmTEP1–4, however, no in vivo phenotype was observed for MCR against the fungal pathogen B. bassiania (Bou Aoun et al. 2011).
Unique structural features are apparent in the sequence of MCR in comparison to other TEPs. The thioester bond itself is mutated in almost all MCRs. MCRs are larger than most iTEPs, with an N-terminal extension prior to the start of the MG1 domain and an expanded MG3 domain. MCRs have a 6-cysteine domain within the protease-sensitive region, but, unlike complement factors, it belongs to the LDLa fold. Finally, MCRs contain a C-terminal predicted transmembrane helix following the ANK region, indicating that they are bound cell surface factors. Indeed, ISH staining of Drosophila larvae identified MCR associated with imaginal discs, the progenitors of adult organs (Bou Aoun et al. 2011).
An essential function of DmMCR has recently been discovered (Hall et al. 2014; Bätz et al. 2014). MCR is a key component of the septate junction (SJ) between cells in polarized epithelia, co-localizing with other known SJ proteins, Coracle (Cor) and Neuroglian (Nrg). Loss of MCR results in mislocalization of SJ components and permeability of barrier epithelia. In hemocytes, MCR is localized to intracellular vesicles. A hypothetical function for MCR in immunity may be the coagulation of hemocytes and encapsulation of foreign bodies too large to be phagocytosed. Intriguingly, MCR is also strongly expressed in stage 1 of the germarium and in polar follicle cells. Oogenesis in Drosophila requires the formation of specific cellular junctions through which nurse cells deliver cytoplasm to the developing oocyte. The same nurse cells undergo non-apoptotic programmed cell death during late oogenesis, the mechanism of which is unclear (Jenkins et al. 2013). A potential role of MCR in either of these processes remains to be explored.
Conclusion
Thirty years after the discovery of the intramolecular thioester bond, and almost a decade following the first crystal structure of complement factor C3, the general structure and function of TEPs can be understood at a high level of detail. The diversity of specific functions adopted by TEPs, however, arise from distinct conformations and interactions at the quaternary structural level, which are both more subtle and less easy to predict. This same diversity is part of fundamental inter-cellular decision-making processes: the recognition of self versus non-self and the choice between preservation and destruction
The complement system, a subject of study for over a century, continues to be a source of new cellular functions, such as the recent discovery of an intracellular role in T cell homeostasis (Liszewski et al. 2013). The invertebrate TEPs, in contrast, are a virtually unexplored world of novel functions and mechanisms of innate immunity. Further structural and functional studies of iTEPs and MCRs may reveal not only just new mechanisms for TEPs in innate immunity but also new interfaces between immunity, development, and cell death. The lack of conservation between arthropod and vertebrate TEPs may also be leveraged to develop insect-specific adjuvants or immune-suppressants for future application in agriculture and control of infectious diseases transmitted by insect vectors.
Acknowledgments
The authors express their thanks to members of the Baxter Laboratory and their collaborators for helpful discussions over the years.
Compliance with Ethical Standards
ᅟ
Funding
The authors received no funding for this work.
Conflict of interest
Marni Williams declares that she has no conflict of interest. Richard Baxter declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human or animal subjects performed by the any of the authors.
References
- Armstrong PB. Proteases and protease inhibitors: a balance of activities in host-pathogen interaction. Immunobiology. 2006;211(4):263–281. doi: 10.1016/j.imbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Armstrong PB, Quigley JP. Limulus a2-macroglobulin. First evidence in an invertebrate for a protein containing an internal thiol ester bond. Biochem J. 1987;248(3):703–707. doi: 10.1042/bj2480703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai L, Patthy L. The NTR module: domains of netrins, secreted frizzled related proteins, and type I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 1999;8(8):1636–1642. doi: 10.1110/ps.8.8.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Starkey PM. The interaction of alpha 2-macroglobulin with proteinases. Characteristics and specificity of the reaction, and a hypothesis concerning its molecular mechanism. Biochem J. 1973;133(4):709–724. doi: 10.1042/bj1330709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bätz T, Forster D, Luschnig S. The transmembrane protein Macroglobulin complement-related is essential for septate junction formation and epithelial barrier function in Drosophila. Development. 2014;141(4):899–908. doi: 10.1242/dev.102160. [DOI] [PubMed] [Google Scholar]
- Baxter RHG, Chang C-I, Chelliah Y, Blandin S, Levashina EA, Deisenhofer J. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci USA. 2007;104(28):11615–11620. doi: 10.1073/pnas.0704967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter RHG, Steinert S, Chelliah Y, Volohonsky G, Levashina EA, Deisenhofer J. A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae. Proc Natl Acad Sci USA. 2010;107(39):16817–16822. doi: 10.1073/pnas.1010575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandin S, Levashina EA. Thioester-containing proteins and insect immunity. Mol Immunol. 2004;40(12):903–908. doi: 10.1016/j.molimm.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Blandin S, Shiao S-H, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116(5):661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, Levashina EA, Steinmetz LM. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science. 2009;326(5949):147–150. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231(2):539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N. Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J Innate Immun. 2011;3(1):52–64. doi: 10.1159/000321554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buresova V, Hajdusek O, Franta Z, Sojka D, Kopacek P. IrAM-An a2-macroglobulin from the hard tick Ixodes ricinus: characterization and function in phagocytosis of a potential pathogen Chryseobacterium indologenes. Dev Comp Immunol. 2009;33(4):489–498. doi: 10.1016/j.dci.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Buresova V, Hajdusek O, Franta Z, Loosova G, Grunclova L, Levashina EA, Kopacek P. Functional genomics of tick thioester-containing proteins reveal the ancient origin of the complement system. J Innate Immun. 2011;3(6):623–630. doi: 10.1159/000328851. [DOI] [PubMed] [Google Scholar]
- Cheng G, Liu L, Wang P, Zhang Y, Zhao YO, Colpitts TM, Feitosa F, Anderson JF, Fikrig E. An in vivo transfection approach elucidates a role for Aedes aegypti thioester-containing proteins in flaviviral infection. PLoS ONE. 2011;6(7):e22786. doi: 10.1371/journal.pone.0022786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller H-M, Osta MA, Paskewitz SM, Reichhart J-M, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298(5591):159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Chu CT, Howard GC, Misra UK, Pizzo SV. a2-macroglobulin: a sensor for proteolysis. Ann NY Acad Sci. 1994;737:291–307. doi: 10.1111/j.1749-6632.1994.tb44319.x. [DOI] [PubMed] [Google Scholar]
- Crews BC, James MW, Beth AH, Gettins P, Cunningham LW. In support of the trap hypothesis. Chymotrypsin is not rigidly held in its complex with human a2-macroglobulin. Biochemistry. 1987;26(19):5963–5967. doi: 10.1021/bi00393a003. [DOI] [PubMed] [Google Scholar]
- Day NK, Gewurz H, Johannsen R, Finstad J, Good RA. Complement and complement-like activity in lower vertebrates and invertebrates. J Exp Med. 1970;132(5):941–950. doi: 10.1084/jem.132.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan N, Gettins PGW. Human a2-macroglobulin is composed of multiple domains, as predicted by homology with complement component C3. Biochem J. 2007;407(1):23–30. doi: 10.1042/BJ20070764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds AW, Law SKA. Structural basis of the binding specificity of the thioester-containing proteins, C4, C3 and a2-macroglobulin. Complement. 1988;5(2):89–97. doi: 10.1159/000463039. [DOI] [PubMed] [Google Scholar]
- Dodds AW, Law SKA. The complement component C4 of mammals. Biochem J. 1990;265(2):495–502. doi: 10.1042/bj2650495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds AW, Ren X-D, Willis AC, Law SK. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379(6561):177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- Edgecombe GD. Arthropod phylogeny: an overview from the perspectives of morphology, molecular data and the fossil record. Arthropod Struct Dev. 2010;39(2–3):74–87. doi: 10.1016/j.asd.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Erler S, Popp M, Lattorff HMG. Dynamics of immune system gene expression upon bacterial challenge and wounding in a social insect (Bombus terrestris) PLoS ONE. 2011;6(3):e18126. doi: 10.1371/journal.pone.0018126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman SR, Gonias SL, Pizzo SV. Model of a2-macroglobulin structure and function. Proc Natl Acad Sci USA. 1985;82(17):5700–5704. doi: 10.1073/pnas.82.17.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330(6012):1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Wu J, Gros P. The modular serine proteases of the complement cascade. Curr Opin Struct Biol. 2012;22(3):333–341. doi: 10.1016/j.sbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Fraiture M, Baxter RHG, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, Hoffmann JA, Blandin S, Levashina EA. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. 2009;5(3):273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Fredslund F, Jenner L, Husted LB, Nyborg J, Andersen GR, Sottrup-Jensen L. The structure of bovine complement component 3 reveals the basis for thioester function. J Mol Biol. 2006;361(1):115–127. doi: 10.1016/j.jmb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Fredslund F, Laursen NS, Roversi P, Jenner L, Oliveira CL, Pedersen JS, Nunn MA, Lea SM, Discipio R, Sottrup-Jensen L, Andersen GR. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat Immunol. 2008;9(7):753–760. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- Garcia BL, Ramyar KX, Tzekou A, Ricklin D, McWhorter WJ, Lambris JD, Geisbrecht BV. Molecular basis for complement recognition and inhibition determined by crystallographic studies of the staphylococcal complement inhibitor (SCIN) bound to C3c and C3b. J Mol Biol. 2010;402(1):17–29. doi: 10.1016/j.jmb.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi DA. 400 million years on six legs: on the origin and early evolution of Hexapoda. Arthropod Struct Dev. 2010;39(2–3):191–203. doi: 10.1016/j.asd.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Grimaldi DA. Fossil record and phylogeny of the Arthropoda: introduction. Arthropod Struct Dev. 2010;39(2–3):72–73. doi: 10.1016/j.asd.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Gros P, Milder FJ, Janssen BJC. Complement driven by conformational changes. Nat Rev Immunol. 2008;8(1):48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- Gunaratna RT, Jiang H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev Comp Immunol. 2013;39(4):388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadders MA, Bubeck D, Roversi P, Hakobyan S, Forneris F, Morgan BP, Pangburn MK, Llorca O, Lea SM, Gros P. Assembly and regulation of the membrane attack complex based on structures of C5b6 and sC5b9. Cell Rep. 2012;1(3):200–207. doi: 10.1016/j.celrep.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S, Bone C, Oshima K, Zhang L, McGraw M, Lucas B, Fehon RG, Ward RE. Macroglobulin complement-related encodes a protein required for septate junction organization and paracellular barrier function in Drosophila. Development. 2014;141(4):889–898. doi: 10.1242/dev.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenman DE, Kells DIC, Cooper NR, Müller-Eberhard HJ, Pangburn MK. Nucleophilic modification of human complement protein C3: correlation of conformational changes with acquisition of C3b-like functional properties. Biochemistry. 1981;20(15):4458–4467. doi: 10.1021/bi00518a034. [DOI] [PubMed] [Google Scholar]
- Ishii N, Wadsworth WG, Stern BD, Culotti JG, Hedgecock EM. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron. 1992;9(5):873–881. doi: 10.1016/0896-6273(92)90240-e. [DOI] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196(4295):1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Janatova J, Lorenz PE, Schechter AN, Prahl JW, Tack BF. Third component of human complement: appearance of a sulfhydryl group following chemical or enzymatic inactivation. Biochemistry. 1980;19(19):4471–4478. [PubMed] [Google Scholar]
- Jang I-H, Nam H-J, Lee W-J. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 2008;41(2):102–107. doi: 10.5483/bmbrep.2008.41.2.102. [DOI] [PubMed] [Google Scholar]
- Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437(7058):505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444(7116):213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Janssen BJC, Gomes L, Koning RI, Svergun DI, Koster AJ, Fritzinger DC, Vogel C-W, Gros P. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28(16):2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol. 2013;23(11):567–574. doi: 10.1016/j.tcb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Husted LB, Thirup S, Sottrup-Jensen L, Nyborg J. Crystal structure of the receptor-binding domain of a2-macroglobulin. Structure. 1998;6(5):595–604. doi: 10.1016/s0969-2126(98)00061-6. [DOI] [PubMed] [Google Scholar]
- Jensen JA, Festa E, Smith DS, Cayer M. The complement system of the nurse shark: hemolytic and comparative characteristics. Science. 1981;214(4520):566–569. doi: 10.1126/science.7291995. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30(2):95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Erickson BW. An equilibrium model of the metastable binding sites of a2-macroglobulin and complement proteins C3 and C4. J Biol Chem. 1982;257(20):11864–11867. [PubMed] [Google Scholar]
- Khan SA, Erickson BW. Synthesis of macrocyclic peptide thiolactones as models of the metastable binding-sites of alpha-2-macroglobulin and complement protein C3b. J Am Chem Soc. 1981;103(24):7374–7376. [Google Scholar]
- Khan SA, Sekulski JM, Erickson BW. Peptide models of protein metastable binding sites: competitive kinetics of isomerization and hydrolysis. Biochemistry. 1986;25(18):5165–5171. doi: 10.1021/bi00366a027. [DOI] [PubMed] [Google Scholar]
- Kidmose RT, Laursen NS, Dobo J, Kjaer TR, Sirotkina S, Yatime L, Sottrup-Jensen L, Thiel S, Gal P, Andersen GR. Structural basis for activation of the complement system by component C4 cleavage. Proc Natl Acad Sci USA. 2012;109(38):15425–15430. doi: 10.1073/pnas.1208031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos A, Wende E, Wareham KJ, Monk PN. International Union of Pharmacology. LXXXVII. Complement peptide C5a, C4a, and C3a receptors. Pharmacol Rev. 2013;65(1):500–543. doi: 10.1124/pr.111.005223. [DOI] [PubMed] [Google Scholar]
- Kopacek P, Weise C, Saravanan T, Vitova K, Grubhoffer L. Characterization of an a-macroglobulin-like glycoprotein isolated from the plasma of the soft tick Ornithodoros moubata. Eur J Biochem. 2000;267(2):465–475. doi: 10.1046/j.1432-1327.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Kopacek P, Hajdusek O, Buresova V. Tick as a model for the study of a primitive complement system. Adv Exp Med Biol. 2012;710:83–93. doi: 10.1007/978-1-4419-5638-5_9. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SVL. The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb. Structure. 2009;17(4):611–619. doi: 10.1016/j.str.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci USA. 2000;97(21):11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Gordon N, Hermans S, Lorenz N, Jackson N, Wines B, Spillner E, Christensen JB, Jensen M, Fredslund F, Bjerre M, Sottrup-Jensen L, Fraser JD, Andersen GR. Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc Natl Acad Sci USA. 2010;107(8):3681–3686. doi: 10.1073/pnas.0910565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SK, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6(2):263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SK, Lichtenberg NA, Levine RP. Covalent binding and hemolytic activity of complement proteins. Proc Natl Acad Sci U S A. 1980;77(12):7194–7198. doi: 10.1073/pnas.77.12.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SKA, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 1984;3(8):1819–1823. doi: 10.1002/j.1460-2075.1984.tb02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BV, Williams M, Logarajah S, Baxter RHG. Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles. PLoS Pathog. 2012;8(10):e1002958. doi: 10.1371/journal.ppat.1002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea SM, Johnson S. Putting the structure into complement. Immunobiology. 2012;217(11):1117–1121. doi: 10.1016/j.imbio.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy DJ, Hendrickson WA, Aukhil I, Erickson HP. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258(5084):987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104(5):709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Kolev M, Le Friec G, Leung M, Bertram PG, Fara AF, Subias M, Pickering MC, Drouet C, Meri S, Arstila TP, Pekkarinen PT, Ma M, Cope A, Reinheckel T, Rodriguez de Cordoba S, Afzali B, Atkinson JP, Kemper C. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity. 2013;39(6):1143–1157. doi: 10.1016/j.immuni.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero A, Duquerroy S, Trapani S, Goulas T, Guevara T, Andersen GR, Navaza J, Sottrup-Jensen L, Gomis-Rüth FX. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew Chem Int Ed. 2012;51(13):3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- Mitri C, Jacques J-C, Thiery I, Riehle MM, Xu J, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5(9):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moita LF, Wang-Sattler R, Michel K, Zimmermann T, Blandin S, Levashina EA, Kafatos FC. In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity. 2005;23(1):65–73. doi: 10.1016/j.immuni.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Molina-Cruz A, DeJong RJ, Ortega C, Haile A, Abban E, Rodrigues J, Jaramillo-Gutierrez G, Barillas-Mury C. Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proc Natl Acad Sci USA. 2012;109(28):E1957–1962. doi: 10.1073/pnas.1121183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, Ortega C, van Schaijk BCL, Sauerwein RW, Taylor-Salmon E, Barillas-Mury C. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science. 2013;340(6135):984–987. doi: 10.1126/science.1235264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop Dis. 2010;4(9):e813. doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis D, Holland CH, Lambris JD (2005) Structure of the anaphylatoxins C3a and C5a. In: Morikis D, Lambris JD (eds) Structural Biology of the Complement System. CRC, Boca Raton, pp 161–178
- Mudiganti U, Hernandez R, Brown DT. Insect response to alphavirus infection–establishment of alphavirus persistence in insect cells involves inhibition of viral polyprotein cleavage. Virus Res. 2010;150(1–2):73–84. doi: 10.1016/j.virusres.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Müller-Eberhard HJ. Complement. Annu Rev Biochem. 1975;44:697–724. doi: 10.1146/annurev.bi.44.070175.003405. [DOI] [PubMed] [Google Scholar]
- Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science. 1998;280(5367):1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- Neves D, Estrozi LF, Job V, Gabel F, Schoehn G, Dessen A. Conformational states of a bacterial a2-macroglobulin resemble those of human complement C3. PLoS ONE. 2012;7(4):e35384. doi: 10.1371/journal.pone.0035384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira GA, Lieberman J, Barillas-Mury C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science. 2012;335(6070):856–859. doi: 10.1126/science.1209678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303(5666):2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Müller-Eberhard HJ. Relation of putative thioester bond in C3 to activation of the alternative pathway and the binding of C3b to biological targets of complement. J Exp Med. 1980;152(4):1102–1114. doi: 10.1084/jem.152.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324(5924):258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Upton LM, Sala KA, Christophides GK. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 2011;7(4):e1002023. doi: 10.1371/journal.ppat.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povelones M, Bhagavatula L, Yassine H, Tan LA, Upton LM, Osta MA, Christophides GK. The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9(9):e1003623. doi: 10.1371/journal.ppat.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley JP, Armstrong PB. An endopeptidase inhibitor, similar to mammalian a2-macroglobulin, detected in the hemolymph of an invertebrate, Limulus polyphemus. J Biol Chem. 1983;258(13):7903–7906. [PubMed] [Google Scholar]
- Quigley JP, Armstrong PB. A homologue of a2-macroglobulin purified from the hemolymph of the horseshoe crab Limulus polyphemus. J Biol Chem. 1985;260(23):12715–12719. [PubMed] [Google Scholar]
- Rehman AA, Ahsan H, Khan FH. Alpha-2-macroglobulin: a physiological guardian. J Cell Physiol. 2013;228(8):1665–1675. doi: 10.1002/jcp.24266. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle MM, Kyriacos M, Niaré O, Xu J, Li J, Touré AM, Podiougou B, Oduol F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traoré SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312(5773):577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- Romero A, Romao MJ, Varela PF, Kölln I, Dias JM, Carvalho AL, Sanz L, Töpfer-Petersen E, Calvete JJ. The crystal structures of two spermadhesins reveal the CUB domain fold. Nat Struct Biol. 1997;4(10):783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJC, van Strijp JAG, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10(7):721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp A, Dodds AW, Anderson M, Campbell RD, Willis AC, Law SKA. Covalent binding properties of the human complement protein C4 and hydrolysis rate of the internal thioester upon activation. Protein Sci. 1993;2(5):706–716. doi: 10.1002/pro.5560020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim RB, Sim E. Autolytic fragmentation of complement components C3 and C4 under denaturing conditions, a property shared with a2-macroglobulin. Biochem J. 1981;193(1):129–141. doi: 10.1042/bj1930129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey PM, Barrett AJ. Evolution of a2-macroglobulin. The demonstration in a variety of vertebrate species of a protein resembling human a2-macroglobulin. Biochem J. 1982;205(1):91–95. doi: 10.1042/bj2050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey PM, Barrett AJ. Evolution of a2-macroglobulin. The structure of a protein homologous with human a2-macroglobulin from plaice (Pleuronectes platessa L.) plasma. Biochem J. 1982;205(1):105–115. doi: 10.1042/bj2050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey PM, Fletcher TC, Barrett AJ. Evolution of a2-macroglobulin. The purification and characterization of a protein homologous with human a2-macroglobulin from plaice (Pleuronectes platessa L.) plasma. Biochem J. 1982;205(1):97–104. doi: 10.1042/bj2050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein-Stevenson SL, Foley E, O’Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4(1):e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi G, Guthridge JM, Li D, Young K, Holers VM, Chen XS. Structure of complement receptor 2 in complex with its C3d ligand. Science. 2001;292(5522):1725–1728. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- Tack BF, Harrison RA, Janatova J, Thomas ML, Prahl JW. Evidence for presence of an internal thiolester bond in third component of human complement. Proc Natl Acad Sci USA. 1980;77(10):5764–5768. doi: 10.1073/pnas.77.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen JMH, Isenman DE. A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science. 2011;332(6029):608–611. doi: 10.1126/science.1201954. [DOI] [PubMed] [Google Scholar]
- Varela PF, Romero A, Sanz L, Romão MJ, Töpfer-Petersen E, Calvete JJ. The 2.4 a resolution crystal structure of boar seminal plasma PSP-I/PSP-II: a zona pellucida-binding glycoprotein heterodimer of the spermadhesin family built by a CUB domain architecture. J Mol Biol. 1997;274(4):635–649. doi: 10.1006/jmbi.1997.1424. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Povelones M, Christophides GK. Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics. 2010;11:531. doi: 10.1186/1471-2164-11-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BL, Wang J, Aksoy S. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 2011;9(5):e1000619. doi: 10.1371/journal.pbio.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Lawniczak MKN, Cheng C, Coulibaly MB, Wilson MD, Sagnon NF, Costantini C, Simard F, Christophides GK, Besansky NJ. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci USA. 2011;108(1):244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren CM. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444(7116):217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu Y-Q, Ricklin D, Janssen BJC, Lambris JD, Gros P. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10(7):728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, DeCamp DL, Sprang SR. Structure of a rat a1-macroglobulin receptor-binding domain dimer. Protein Sci. 2000;9(10):1889–1897. doi: 10.1110/ps.9.10.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Dong Z, Duan J, Wang G, Wang L, Li Y, Xiang Z, Xia Q. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS ONE. 2012;7(2):e31168. doi: 10.1371/journal.pone.0031168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Thangamani S, Ho B, Ding JL. The ancient origin of the complement system. EMBO J. 2005;24(2):382–394. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]