Abstract
Experimental evidence has shown a close correlation between the composition and physical state of the membrane bilayer and glucose transport activity via the glucose transporter GLUT1. Cooling alters the membrane lipids from the fluid to gel phase, and also causes a large decrease in the net glucose transport rate. The goal of this study is to investigate how the physical phase of the membrane alters glucose transporter structural dynamics using molecular-dynamics simulations. Simulations from an initial fluid to gel phase reduce the size of the cavities and tunnels traversing the protein and connecting the external regions of the transporter and the central binding site. These effects can be ascribed solely to membrane structural changes since in silico cooling of the membrane alone, while maintaining the higher protein temperature, shows protein structural and dynamic changes very similar to those observed with uniform cooling. These results demonstrate that the protein structure is sensitive to the membrane phase, and have implications for how transmembrane protein structures respond to their physical environment.
Introduction
Glucose transporters (GLUTs), which belong to the sugar transporter branch SLC2A of the major facilitator superfamily (1), are essential membrane proteins in eukaryote cell metabolism and thus are the focus of numerous functional, structural, and drug-discovery studies. The human GLUTs display organ- and membrane-specific distributions with distinct kinetics and substrate specificities (2). With the exception of the myo-inositol/H+ symporter GLUT13, all GLUTs are uniporters that facilitate monosaccharide passive downhill diffusion (3, 4). The GLUTs share an identical structural fold comprising 12 transmembrane helices (TM1–TM12), which have a binding site located in the central region of the transporter delineated by residues contributed from both the N- and C-domains. A substantial endofacial cytosolic linker joins the N- and C-domains, and may play a role in transport function by securing the closure of the inward gate (5).
The glucose transport mechanism of GLUT1 has been studied using biochemical and molecular biology methods such as scanning mutagenesis and fluorescence resonance energy transfer, as well as computational approaches that include molecular-dynamics (MD) simulations (6, 7, 8, 9, 10). Data from most of these studies and recent crystal structures of the GLUT family (11, 12, 13) support a mechanism in which a binding site for glucose is alternatively accessible from either side of the membrane, involving movements of the N- and C-domains over a rotation axis located at the central binding site and perpendicular to the bilayer plane; this is the so-called alternating-access mechanism. However, crystallographic structures of several occluded conformations of the transporter (13), both inward and outward facing, suggest an alternative transport mechanism that relies on the adaptation of the protein to its environment (14). In addition, the endofacial domain is thought to undergo a substantial conformational change during the transport cycle, implying that it serves as a gate at the intracellular side (13).
An alternative model for sugar transport, the so-called multisite model, has been proposed based on cytochalasin-B inhibitor binding studies (8), docking studies (15, 16, 17), and MD simulations (18). According to this model, ligands can diffuse between multiple adjacent sites within a branched network of transiently open tunnels and cavities spanning the transporter. These transient openings within the intramolecular tunnels are triggered by small-scale changes in the carbon skeleton and side groups, or reptations, widening the tunnel bottlenecks without the requirement for any global structural rearrangements.
Changes in the membrane hydrophobic thickness have previously been shown to act as a trigger for the bacterial cold sensor Bacillus subtilis DesK. At cold temperatures, DesK acts as a kinase to autophosphorylate a histidine residue. The phosphoryl residue is transferred to an aspartate in the DNA-binding response regulator. This leads to activation of an acyl lipid desaturatase that desaturates and fluidizes the membrane lipids, thereby returning the membrane to the fluid state (19). This mechanism has been called the sunken-buoy motif (20).
The transport properties of GLUT1 embedded in liposomes have been experimentally evaluated as a function of the composition and structural features of the lipid bilayer (21, 22). It was concluded that although short-chain lipids are able to support glucose transport activity in the gel phase, transport dramatically increases in the membrane fluid state (23). As an example, with 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) membranes, transport activity vanishes during cooling to the gel phase but it increases during premelting and phase transitions, and recovers fully in the fluid phase. However, in the gel phase, inclusion of 20% cholesterol in DPPC membranes was shown to activate transport, although a larger cholesterol enrichment inhibited transport (24). It has been proposed that the cholesterol-dependent increase in transport is largely due to a decrease in membrane microviscosity. The slowing of transport at higher cholesterol concentrations may be due to inhibitor complexes formed with the transporter. The complex role of cholesterol in membrane lipid structure and transporter function is still unresolved.
In this study, MD simulations at the atomistic level were employed to investigate whether the structural changes and interaction patterns of the bilayer phase contribute to structural modifications in the GLUT. Small-scale correlated movements of the Cα backbone atoms and side chains of GLUT1 controlled by the physical state of the membrane were found to direct the passage of glucose, as assessed by in silico docking methods. In particular, the gel membrane phase altered the cavity structures. Additionally, the extramembranous loop regions of the transporter, independently of any membrane constraints, were subject to large structural changes and therefore are likely gating sites for ligand entry into transmembranous domains.
Materials and Methods
System setup
The crystal structure of the human GLUT (GLUT1, PDB: 4PYP) (11) was used as a starting point for the computational work. The structure was resolved in an inward-open conformation, with a glucose derivative bound to the main binding site. For the purpose of the simulations, the crystallographic sugar derivative was not included in the model. The initial GLUT1/DPPC system was generated using the Membrane Builder module (25) with default options in the CHARMM-GUI website (26). Initially, a membrane patch of 100 × 100 Å DPPC lipids was built. The membrane contained 205 molecules of DPPC. This choice was based on a compromise between size and computational resources. Subsequently, the GLUT1 structure was inserted into the membrane patch. To avoid steric clashes, lipids in close contact with the protein were deleted. This resulted in an asymmetric lipid distribution with 100 and 105 lipid molecules in the cytoplasmic and external leaflets, respectively. The combined system was then solvated and neutralized to produce a rectangular simulation box with dimensions of 96 × 96 × 108 Å3 and ∼80,000 atoms. Two independent simulations were run at different temperatures: one above (323.15 K) and one below (308.15 K) the DPPC phase transition temperature of 314.15 K.
MD simulations
The software NAMD2.9 was employed to perform the MD simulations (27). The CHARMM36 force field was used to model the protein and lipids (28). Standard CHARMM parameters were used for ions (29) and the TIP3P model was used for water (30). Pressure was maintained at 1 atm by a Langevin piston (31) with a damping time constant of 50 ps and a period of 200 ps. A semi-isotropic pressure coupling method was used in all of the simulations. For the NAMD calculations, the pressure of the piston acted independently in each dimension but maintained a constant ratio in the x,y axis corresponding to the plane of the membrane. The temperature was maintained constant by coupling the system to a Langevin thermostat with a damping coefficient of 1 ps−1. The particle mesh Ewald (PME) algorithm was used for the evaluation of electrostatic interactions beyond 12 Å, with a PME grid spacing of 1 Å and NAMD defaults for spline and κ values (32). A cutoff at 12 Å was applied to nonbonded forces. Both electrostatics and van der Waals forces were smoothly switched off between the switching distance of 10 Å and the cutoff distance of 12 Å using the default switching function in NAMD. A Verlet neighbor list with a pair-list distance of 13.5 Å was used to evaluate nonbonded neighboring forces within the pair-list distance (33). The lengths of covalent bonds involving hydrogen atoms were constrained by the SETTLE algorithm (34, 35) so that a 2-fs time-step could be used. The multitime-step algorithm Verlet-I/r-RESPA (33, 35) was used to integrate the equations of motion. Both systems were subjected to 10,000 steps of energy minimization, followed by an equilibration consisting of sequential release of various restraints added to the system (26): 1) harmonic restraints to heavy atoms of the protein and ions, 2) repulsive restraints to prevent water from entering into the hydrophobic region of the membrane, and 3) planar restraints to hold the position of the lipid headgroups along the z axis. Subsequently, 400-ns production runs were executed at each temperature.
To ensure that temperature effects could be ascribed exclusively to the phase state of the membrane, two additional simulations in which the protein and bilayer were held at two different temperatures were performed with the GROMACS 5.0 software package (36) (Fig. S1; Table S1). In the first simulation, the temperature of the bilayer was set at 323.15 K to be in the fluid phase, whereas the temperature of the protein was set at 308.15 K, below the bilayer phase transition. In contrast, in the second simulation, the temperature of the protein was set at 323.15 K while it was embedded in a gel DPPC membrane at 308.15 K. Each system was simulated for 400 ns using the same equilibration and production protocols described above.
Analysis of the MD trajectories
The program CAVER 3.0 (37) was used to analyze the size and shape of the available pathways for glucose transit from the GLUT1 inward (IN) and outward (OUT) faces to the glucose-binding site (GBS) at the center of the protein (Fig. 1 B). Aligned coordinate files from trajectories spaced at 1 ns were selected. The algorithm works by constructing a Voronoi diagram to describe the skeleton of the water channels within the framework of the protein structure, followed by a cluster analysis of all the channels identified. For this study, a spherical probe of 0.8 Å radius was selected with a weighting coefficient of 1, clustering threshold of 12, shell radius of 18 Å, and shell depth of 4 Å. The starting point for the calculation was chosen at the center of mass of residues I168, Q282, Q283, N288, and E380 of GLUT1, according to the glucose derivative-protein interactions present in the GLUT1 x-ray structure (PDB: 4PYP) (11).
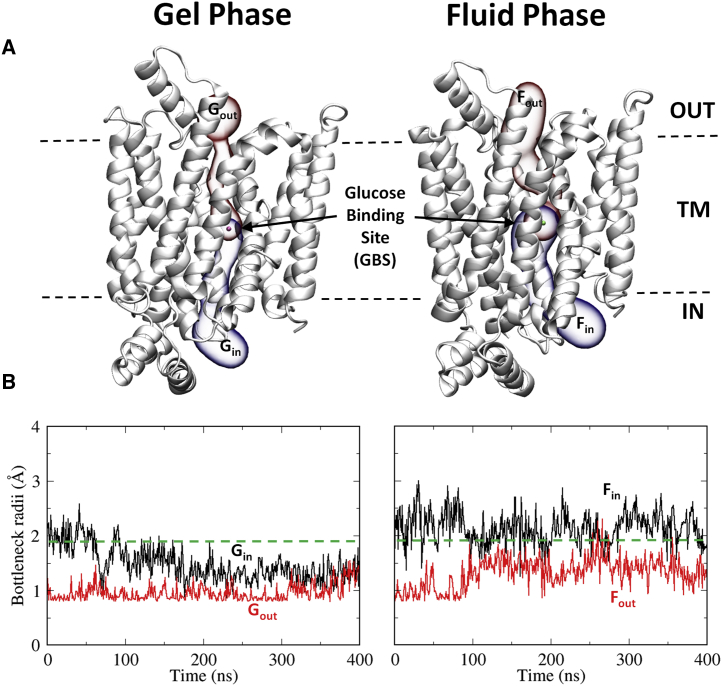
Figure 1.
(A) Schematic representation of the glucose pathway in the protein that extends from the intracellular (IN) to the extracellular (OUT) sides of the membrane through the main glucose-binding site (GBS) at the center of the transporter. The permeation pathway connecting the intracellular part up to the main GBS is shown in blue and labeled with the subscript “in.” The pathway connecting the extracellular side with the main GBS is shown in red and labeled with the subscript “out.” G and F refer to the gel and fluid phases of the membrane, respectively. (B) Evolution of the bottleneck radius of the main cavity running along the transporter with time in each membrane phase. The green discontinuous line indicates the minimal radius of a glucose molecule. To see this figure in color, go online.
The membrane thickness was calculated using the MEMBPLUGIN analysis tool in VMD (38).
The numbers of water molecules along the z axis of the central channel in the gel and fluid phases were digitized using ImageJ profiling into 1320 bins representing average numbers per 300 ps per bin. The plots were subdivided into eight 5 Ǻ strips. The water molecules in each bin were cross-correlated between one and eight. Contour maps of the matrices of the regression coefficients Rij were constructed for water molecules in the simulations with the bilayer in the gel and fluid phases.
Docking calculations
Glucose was docked into the GLUT1 transporter via a Lamarckian genetic search algorithm as implemented in the AutoDock 4.2 software package (39). Docking calculations were performed for representative snapshots of the simulations of the transporter embedded in the fluid and gel phases. Ten different snapshots were extracted from the last 100 ns of each simulation, and 100 AutoDock runs were performed for each structure. A grid with dimensions of 80 × 80 × 120 Å3 centered on the GBS was used, with binding modes ranked by a scoring function implemented in the Autodock software. Gasteiger atom charges were assigned to the protein and glucose atoms using AutoDock tools.
Results and Discussion
For each simulation, the carbon backbone root mean-square deviation (RMSD) was calculated relative to the initial crystallographic structure (Fig. S2). In both simulations, the RMSD values were computed for the 400-ns production trajectories taking as a reference the initial x-ray structure. At least 100 ns were needed for the RMSD to plateau. The convergence to a specific value within the resolution of the initial x-ray structure (3.17 Å) suggests that the protein structures remained relatively stable even though the glucose derivative from the crystal structure was removed, which may have influenced the stability of the protein. The RMSD values of the fluid phase system reflect larger structural changes corresponding to movements of the endofacial loops located at the cytoplasmic leaflet. After ∼60 ns, a plateau of ∼2.5 Å was reached in the simulation of the gel phase. A structural comparison of the GBS in the GLUT1 x-ray structure and in the gel/fluid MD trajectories where glucose was absent from the active site revealed small changes in the surrounding residues, in particular Asn288, which flips to interact with another neighbor residue (Fig. S3).
Glucose does not readily permeate phospholipid bilayers, as demonstrated by its osmotic activity, and thus the potential pathways available for glucose to navigate through the transporter were explored while it was embedded in a lipid bilayer, in either the fluid or gel phase. Searches for cavities were computed using the respective MD trajectories. Two main pathways connecting the center of the protein with the endo- and exofacial environments of the membrane were identified (Fig. 1 A). In addition, other cavities connecting the central binding site with the exterior of the membrane were detected. However, these were relatively narrow and observed much less frequently during the simulations than the main inward and outward glucose entry routes. Therefore, it is unlikely that these secondary routes offer viable pathways for glucose entry and release, and hence they were disregarded.
The average and maximum bottleneck radii of the inner (IN) and outer (OUT) permeation branches of the main permeation pathway in the gel and fluid simulations were computed and are reported in Table S2. The values found highlight the effects of the physical phase of the bilayer. The dimensions of the pathway available for glucose to access the central binding site, from either side of the membrane, are reduced when the protein is embedded in a membrane in the gel phase as opposed to the fluid phase. The maximum value of the bottleneck radius found in the inward-facing branch of the pathway, in either the gel (Gin) or fluid (Fin) phase, is wide enough to allow the passage of glucose molecules with a minimal radius of 1.9 Å (40, 41). When the membrane is in the gel phase, the maximum radius of the outer gate was found to be 1.54 Å, smaller than the minimal glucose radius. Therefore, when the membrane is in the gel phase, glucose can only gain access to the central binding site via the endofacial branch of the pathway, whereas in the outward-facing route, glucose translocation is only possible in the fluid phase.
Analysis of the time course of the bottleneck radii during the 400-ns MD simulations highlights the larger effect of the gel bilayer on the transporter structure in the context of glucose translocation (Fig. 1 B). The outward-facing branch of the pathway has a bottleneck radius close to the minimal radius of 0.8 Å used for the search of cavities, with a maximal 0.5 Å deviation from this value. In contrast, the inward pathway evolves from an open state with a bottleneck radius greater than the 1.9 Å minimal glucose radius. However, the tunnel structure becomes progressively narrower until complete closure occurs at ∼180 ns of the MD simulation. When the fluid membrane is considered, the bottleneck radius of the inward branch of the pathway is generally wider and more stable, having values exceeding the minimum glucose radii for almost the entire simulation. In contrast, the outward branch of the pathway is accessible only during short periods of time, for example, from 270 to 285 ns (Fig. 1 B).
Therefore, in the gel state, glucose cannot access the central binding site because both the inward and outward branches are too narrow. Crucially, however, in the fluid phase, glucose can occasionally gain access to the inner parts of the transporter from the external solution. Since access through the inward branch of the pathway is almost continuously open, it is evident that when the membrane is in the fluid state, opening of the outward branch is the main rate factor limiting glucose transit.
To further investigate the frequency with which these bottlenecks attain sufficient width to permit glucose exchange, only time intervals with a bottleneck radius smaller than the minimal glucose radius of 1.9 Å were analyzed (Fig. S4). The inward branch of the pathway, when embedded in a fluid bilayer, spontaneously opens and closes to glucose access, with a maximum open interval of almost 50 ns. In contrast, the outward-facing branch of the pathway for the majority of time remains closed, although it widens for short intervals. These observations validate the multisite model for sugar transport (8, 40, 42), in which glucose molecules can transit along a network of moderately high-affinity binding sites in the absence of large-scale global rearrangements of the protein structure, i.e., without any alternating access contribution. A recent computational study of GLUT1 glucose transport also supports a combination of the multisite model and the classical alternating-access mechanistic model as the key determinant for sugar translocation (18).
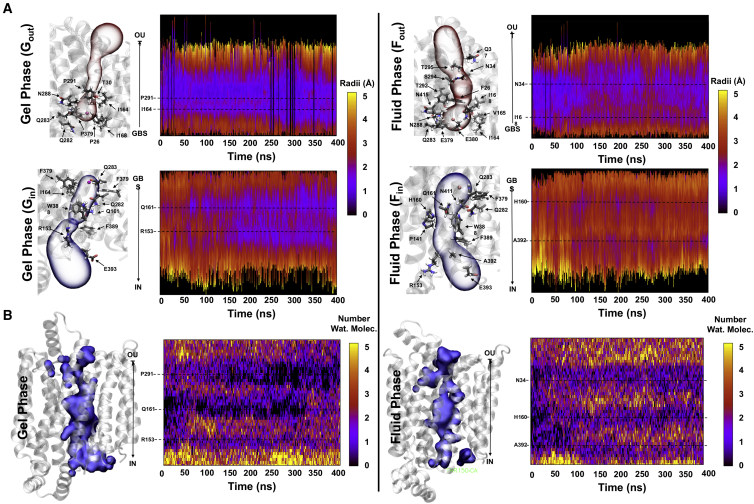
Representative heat maps of the time-dependent evolution of the radius of the pathway along the protein are illustrated in Fig. 2. Two bottleneck regions for the GLUT1 inward branch of the pathway and an extensive constricted zone in the outward branch of the pathway are observed. The key bottleneck residues defining each branch are shown in Fig. 2. For simplicity, only bottleneck residues involved in more than 80% of the snapshots analyzed are shown. From all the residues identified, a set of four create the main channel bottleneck observed in the inward-facing branch of the pathway in the gel state: R153, Q161, W388, and F389. Residues T30, I164, N288, and F291 define the bottleneck region of the outward branch. In the fluid phase, residues F26, N34, I168, Y292, S294, and T295 define the bottleneck region of the outward branch of the pathway, and residues P141, R153, H160, Q161, W388, and F389 the inward branch of the pathway. R153 is a key bottleneck residue for the inward branches of the pathways regardless of the bilayer phase. However, its side-chain position and interactions differ depending on the physical state of the bilayer. When GLUT1 is embedded in a fluid membrane, R153 forms a salt-bridge interaction with E243, which keeps both residues locked in a relatively stable conformation. In the gel phase, small conformational changes affecting the endofacial TM helix prevent formation of the R153-E245 salt bridge, resulting in a decrease of the tunnel width (Fig. S4).
Figure 2.
(A) Time evolution of the radius of the pathways for the Fin, Fout, Gin, and Gout branches. The color code reflects the width at each point in the pathway. Key bottleneck residues are indicated for each branch. F and G correspond to the fluid and gel phases, respectively. “In” denotes the branch facing the intracellular side of the bilayer, and “out” denotes the extracellular side. (B) Time evolution of the number of water molecules within the GLUT1 channel. A continuous surface representation of the channel hydration for each GLUT1 MD trajectory is shown, and the tertiary structure of the transporter is shown in transparent representation. To see this figure in color, go online.
It is noteworthy that R153 is an absolutely conserved amino acid in the GLUT family, whereas F26, N34, P141, Q161, I164, I168, N288, Y292, and W388 are conserved in six or more GLUT members. Mutations of R153 and outward-facing T295 residues are related to glucose-deficiency diseases (43, 44), and W388 has been shown to be critical for binding of ligands such as cytochalasin B and forskolin (45).
Analysis of the evolution of the size of the translocation pathway for glucose in GLUT1 (as described above) illustrates that in the gel phase the passage connecting the extracellular and intracellular sides of the GLUT1 transporter is closed. The inner regions of both the inward and outward branches of the permeation pathway are inaccessible. Under these circumstances, glucose transit via the inner branch of the GLUT1 pathway toward the central GBS would therefore be improbable. In contrast, when the transporter is embedded in a lipid bilayer in the fluid phase, in silico docking analysis shows that glucose can bind at any position along the permeation pathway, further confirming that passage across the transport via a staged diffusion process is a possible transit mechanism (Fig. S5). These flexible structures corroborate the multisite model of sugar transport in the XylE transporter based on multiple static crystal conformers (46). Experiments have demonstrated that ATP binding within this endofacial linker region retards the net glucose influx, possibly by causing partial occlusion of the aperture shielding the internal transporter vestibule from the cytosolic solution (47).
To confirm that temperature effects on the transporter are not the cause for the change in size of the bottleneck radius, additional simulations were performed in which the transporter and the lipid bilayer were coupled to independent thermostats set at different temperatures. In the simulation where the membrane was held at the gel-phase temperature and the temperature of the GLUT1 transporter was raised above that temperature, both the inward and outward branches of the tunnel still displayed the narrow bottleneck radius observed for GLUT1 embedded in a gel-phase bilayer (Table S2).
Additionally, simulation of a GLUT1 transporter embedded in a fluid membrane at a temperature below the gel-phase temperature displayed open tunnels for the inward and outward branches. Together, these results eliminate protein temperature as the principal modulator of the bottleneck radius, and indicate that the force induced by the interaction with the membrane surface tension is critical to the size of the intramolecular voids that are crucial for the performance of the protein as a TM transporter.
The results reveal a considerable similarity between the structural oscillations of the GLUT1 transporter in a fluid or gel membrane, although the latter restricts the magnitude of these movements. A comparison of the protein B-factors in the gel and fluid bilayers reveals an overall similarity of the thermal fluctuations of the residues in both gel and fluid phases, although these fluctuations are more constrained in the gel phase (Fig. S6). In a fluid membrane, the largest protein fluctuations occur in the linker regions between TM helices, especially in the endofacial domain. The gel phase reduces the global thermal fluctuations of the embedded protein in comparison to the fluid state, mainly affecting the C-terminal region of the transporter.
Although previously, membrane lipids were thought to play mainly a supportive role in biological transport processes, it has now become evident that they critically modulate protein function (48, 49). In this study, several membrane properties were analyzed and compared in the fluid and gel phases, including the bilayer thickness (defined as the distance between the average positions of the headgroups in the upper and lower leaflets). The average thickness of the membrane in the gel phase evolves over time to a higher value compared with that of a fluid membrane (42.5 Å vs. 40 Å) (Fig. S7 A). This reflects a higher degree of order of the hydrophobic lipid tails in the gel phase. The computed values agree closely with those obtained from relative electron densities calculated through the bilayer normal of multilamellar dispersions of the phospholipids, examined by synchrotron x-ray diffraction methods (gel, 42.4 Å; fluid, 40.2 Å; Fig. S8). Although a fluid membrane can accommodate the protein without alteration of the lipid structure, gel membranes are perturbed by the presence of the protein (Fig. S7 B). This is probably a consequence of the greater membrane thickness in the gel phase, due to denser lipid packing, and the interactions of the hydrophobic lipid tails and the TM segments of the proteins. To further characterize the lipid structure in both phases, order parameters for the lipids surrounding the protein and the bulk lipids were computed using the last 100 ns of the trajectories (Fig. S7 D). Lipid tails showed higher-order parameters in the gel phase than in the fluid phase, as expected, with lipids surrounding the protein displaying higher mobility. In the fluid phase, lipids showed lower-order parameters and were not perturbed by the presence of the protein. The bilayer in which GLUT1 was interpolated at 35°C exhibited differences observed in the bilayers of the pure lipid in the absence of the protein. Thus, although the order parameters indicate that the hydrocarbon chains are in a gel configuration, there is disorder of the chains at the protein-lipid interface. The characteristic tilt of the hydrocarbon chains with respect to the bilayer normal tends to a more vertical orientation, and there is a variation in membrane thickness in the vicinity of the protein that is not consistent with a periodic ripple structure of the lipid.
To check whether the properties of the lipids in the external and cytoplasmic leaflets are affected differently by the presence of the protein, the average order parameters of the acyl chain and area/lipid were derived from the entire 400 ns and last 100 ns trajectories. The order parameters along the length of the chains were significantly greater in the external leaflet compared with the cytoplasmic leaflet in the respective gel and fluid phases throughout the simulations. This is consistent with the calculated areas per lipid, which were significantly smaller in the external leaflet than in the cytoplasmic leaflet (see Table S3).
Distinct from the thermodynamic pressure of the system, where any differences in pressure tend to zero, local differences in surface tension within the lipid bilayer arise as a result of the particular composition and phase state of the lipid, where the bilayer thickness, curvature, and pressure profile generate differences in lateral pressure that constrain the area occupied by proteins interpolated into the bilayer (46). As a means of evaluating a possible correlation between the membrane phase and the bottleneck width, the membrane area and the bottleneck radius of the pathway were correlated (Fig. S9 A). The correlation coefficient between the membrane area of the gel phase and the GLUT1 inward channel (Gin) was found to be 0.66. This indicates that for the Gin branch of the pathway, the membrane areas exert sufficient pressure to compress the transporter, thereby narrowing the GLUT1 inner pathway and impeding sugar transport (Fig. S9 B). Similarly, by correlating the bottleneck radius with the membrane thickness, a correlation coefficient of −0.54 for the Gin channel was obtained. This supports the view that only the gel phase of the membrane affects the dynamics of the transporter sufficiently to lead to occlusion of the channel, particularly in those regions located near the GLUT1 inward face. In this case, the correlation indicates that an increase in the membrane thickness narrows the GLUT1 Gin tunnel, probably by exerting a higher pressure on the channel gate through the lipid headgroups, possibly aided by the observed distortions created within the bilayer plane. Clearly, these results lend further support to the view that the gel phase of the membrane influences the protein, leading to a narrower glucose translocation pathway that restricts glucose transport.
The degree of hydration of the protein was studied. As a channel with a radius of 1–2 Å, it contains some water under ambient conditions, and a change in the mean number of water molecules inside the protein or their distribution is likely to affect the accessibility of glucose. In this respect, the numbers and distribution of water molecules within the channel (Fig. S10), as well as the correlation between the channel size and the number of water molecules (i.e., the degree of hydration), were computed over time. Water molecules diffused freely along long sections of the main tunnel in the simulations where the lipid membrane was in the fluid phase as opposed to the gel phase. The main tunnel was interconnected by neighboring segments, whereas in the gel phase the connections did not extend across the entire channel due to compression of the protein at certain times.
The average numbers of water molecules within cavities along the z axis of the central channel were subdivided into eight 5-Ǻ-wide zones in both gel and fluid phases. They were then digitized and averaged during successive 300-ps intervals encompassing the entire 400-ns simulation time course shown in Fig. S12. The variation in water density in each of the eight zones was correlated with the remaining seven zones, and a correlation map of all the regression coefficients was obtained for the gel- and fluid-phase waters. The findings indicate that water diffuses more freely along longer tunnel segments in the fluid phase than in the gel phase. In the fluid phase, the entire channel length is connected by neighboring highly correlated segments. In contrast, in the gel phase, the regions of high connectivity are interrupted due to longer closure times in the bottleneck regions. This segmentation can be ascribed to external compression exerted by the gel membrane. In the gel phase, it is apparent that water from the central zones is displaced toward expanded external vestibules, so there is relatively little net change in the total number of water molecules within the central channel regions as a result of compression of the transporter.
It is evident from the dynamics of the number of water molecules occupying the central channel that there are three bottleneck positions with two intermediate cavities. The upper two bottlenecks appear to be more temperature sensitive than the lowest (internal) bottleneck. The fluid-gel phase transformation will result in a much higher percentage closure time of the bottlenecks to water, and presumably to glucose. The reduced temperature effect transforms the interpretation with regard to the mechanism of glucose transport.
The alternating-access model proposes one choke point in the transport process; we have a series of three. The effect of cooling is normally interpreted as being due to a slowing of the rate processes of the transporter inversions. Here, it is evident that cooling results mainly in narrowing of the channel, particularly at the bottlenecks. Therefore, slowing of transport is consistent with reduced rates of staged diffusion, and thus with the model proposed recently in (40).
It is important to recognize that this work only covers a symmetrical DPPC membrane, whereas lipid organization in the human plasma membrane is asymmetric and highly heterogeneous. Therefore, much more complex interactions can be expected. In this regard, a recent article by Hresko et al. (50) highlighted the activation effect of anionic phospholipids on the turnover rate of GLUT3 and GLUT4 transporters, whereas the substrate affinity remained unaltered, providing evidence of a direct interaction of the studied phospholipids with the transporter. Anionic phospholipids are found exclusively on the endofacial leaflet of mammalian lipid bilayers, thus supporting a crucial role for bilayer composition in transporter activity.
Determining the extent to which the membrane phase alters glucose diffusion through the central channel awaits further simulations. The role of the more salient single-point mutations within the channel also awaits investigation.
Conclusions
MD simulations of the GLUT1 transporter embedded in a gel or fluid DPPC membrane bilayer were performed to study the effects of the bilayer physical state on the dimensions and dynamics of the glucose translocation pathways within GLUT1. Hydrated DPPC bilayers form gel phases at temperatures between those of the lamellar crystal phase (<7°C) and lamellar liquid-crystal phase (>41°C). Here, we have demonstrated that the presence of GLUT1 causes a disturbance of the bilayer structure at temperatures where a gel phase of the pure phospholipid is observed. The gel phase of DPPC has been well characterized by a range of biophysical techniques, including x-ray diffraction (e.g., the data used to prepare Fig. S8) and MD simulations (51). Here, inhomogeneity of the lipids is observed in the vicinity of GLUT1 due to the protein. When embedded in a fluid membrane, as opposed to being in a gel phase, GLUT1 exhibited a larger bottleneck radius for both the main endofacial and exofacial branches of the primary pathway connecting the extracellular and intracellular sides of the bilayer. The external branch of the pathway in the fluid phase was found to be principally closed, with bottleneck values smaller than the minimal glucose radius. However, transient open conformations that could allow glucose passage were also detected. Overall, these results confirm the viability of a multisite model for glucose translocation in which sugar molecules diffuse through a network of binding sites while the overall global conformation of the protein is conserved. Although the differences in diameter observed in the simulations are small, our contention that the diameter is a factor in limiting passage through the channel is not unreasonable. This is consistent with the report by Fu et al. (18) showing that unhydrated glucose can negotiate the narrowest dimensions of the channel by spontaneous diffusion.
Author Contributions
J.I.-F. and C.D. performed the simulations and analyzed the data. J.I.-F., P.J.Q., R.J.N., and C.D. planned the simulations, interpreted the data, and wrote the manuscript.
Acknowledgments
ARCHER, the UK National Supercomputing Service; the Hartree Center; and the National Service for Computational Chemistry Software are acknowledged for providing computational resources.
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/L01825X/1).
Editor: Scott Feller.
Footnotes
Eleven figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30156-X.
Supporting Material
References
- 1.Pao S.S., Paulsen I.T., Saier M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolescu A.R., Witkowska K., Cheeseman C. Facilitated hexose transporters: new perspectives on form and function. Physiology (Bethesda) 2007;22:234–240. doi: 10.1152/physiol.00011.2007. [DOI] [PubMed] [Google Scholar]
- 3.Mueckler M., Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med. 2013;34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F.Q., Keating A.F. Functional properties and genomics of glucose transporters. Curr. Genomics. 2007;8:113–128. doi: 10.2174/138920207780368187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng D., Yan N. GLUT, SGLT, and SWEET: structural and mechanistic investigations of the glucose transporters. Protein Sci. 2016;25:546–558. doi: 10.1002/pro.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robichaud T., Appleyard A.N., Carruthers A. Determinants of ligand binding affinity and cooperativity at the GLUT1 endofacial site. Biochemistry. 2011;50:3137–3148. doi: 10.1021/bi1020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Zutter J.K., Levine K.B., Carruthers A. Sequence determinants of GLUT1 oligomerization: analysis by homology-scanning mutagenesis. J. Biol. Chem. 2013;288:20734–20744. doi: 10.1074/jbc.M113.469023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carruthers A., DeZutter J., Devaskar S.U. Will the original glucose transporter isoform please stand up! Am. J. Physiol. Endocrinol. Metab. 2009;297:E836–E848. doi: 10.1152/ajpendo.00496.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., Terry D., Javitch J.A. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Ke M., Gong H. Protonation of Glu(135) facilitates the outward-to-inward structural transition of fucose transporter. Biophys. J. 2015;109:542–551. doi: 10.1016/j.bpj.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng D., Xu C., Yan N. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510:121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 12.Nomura S., Sakamaki S., Tsuda-Tsukimoto M. Discovery of canagliflozin, a novel C-glucoside with thiophene ring, as sodium-dependent glucose cotransporter 2 inhibitor for the treatment of type 2 diabetes mellitus. J. Med. Chem. 2010;53:6355–6360. doi: 10.1021/jm100332n. [DOI] [PubMed] [Google Scholar]
- 13.Deng D., Sun P., Yan N. Molecular basis of ligand recognition and transport by glucose transporters. Nature. 2015;526:391–396. doi: 10.1038/nature14655. [DOI] [PubMed] [Google Scholar]
- 14.Quistgaard E.M., Löw C., Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol. 2016;17:123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham P., Afzal-Ahmed I., Naftalin R.J. Docking studies show that D-glucose and quercetin slide through the transporter GLUT1. J. Biol. Chem. 2006;281:5797–5803. doi: 10.1074/jbc.M509422200. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham P., Naftalin R.J. Implications of aberrant temperature-sensitive glucose transport via the glucose transporter deficiency mutant (GLUT1DS) T295M for the alternate-access and fixed-site transport models. J. Membr. Biol. 2013;246:495–511. doi: 10.1007/s00232-013-9564-5. [DOI] [PubMed] [Google Scholar]
- 17.Naftalin R.J. Osmotic water transport with glucose in GLUT2 and SGLT. Biophys. J. 2008;94:3912–3923. doi: 10.1529/biophysj.107.122531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X., Zhang G., Gao Q. Mechanistic study of human glucose transport mediated by GLUT1. J. Chem. Inf. Model. 2016;56:517–526. doi: 10.1021/acs.jcim.5b00597. [DOI] [PubMed] [Google Scholar]
- 19.Saita E., Albanesi D., de Mendoza D. Sensing membrane thickness: lessons learned from cold stress. Biochim. Biophys. Acta. 2016;1861:837–846. doi: 10.1016/j.bbalip.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Cybulski L.E., Martín M., de Mendoza D. Membrane thickness cue for cold sensing in a bacterium. Curr. Biol. 2010;20:1539–1544. doi: 10.1016/j.cub.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 21.Connolly T.J., Carruthers A., Melchior D.L. Effects of bilayer cholesterol on human erythrocyte hexose transport protein activity in synthetic lecithin bilayers. Biochemistry. 1985;24:2865–2873. doi: 10.1021/bi00333a008. [DOI] [PubMed] [Google Scholar]
- 22.Tefft R.E., Jr., Carruthers A., Melchior D.L. Reconstituted human erythrocyte sugar transporter activity is determined by bilayer lipid head groups. Biochemistry. 1986;25:3709–3718. doi: 10.1021/bi00360a035. [DOI] [PubMed] [Google Scholar]
- 23.Carruthers A., Melchior D.L. Human erythrocyte hexose transporter activity is governed by bilayer lipid composition in reconstituted vesicles. Biochemistry. 1984;23:6901–6911. doi: 10.1021/bi00321a096. [DOI] [PubMed] [Google Scholar]
- 24.Yuli I., Wilbrandt W., Shinitzky M. Glucose transport through cell membranes of modified lipid fluidity. Biochemistry. 1981;20:4250–4256. doi: 10.1021/bi00518a003. [DOI] [PubMed] [Google Scholar]
- 25.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 27.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 31.Feller S.E., Zhang Y.H., Brooks B.R. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 32.Darden T., York D., Pedersen L. Particle mesh Ewald—an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 33.Verlet L. Computer experiments on classical fluids. I. Thermodynamical properties of Lennard-Jones molecules. Phys. Rev. 1967;159:98. [Google Scholar]
- 34.Miyamoto S., Kollman P.A. SETTLE—an analytical version of the Shake and Rattle algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 35.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular-dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 36.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 37.Chovancova E., Pavelka A., Damborsky J. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guixà-González R., Rodriguez-Espigares I., Selent J. MEMBPLUGIN: studying membrane complexity in VMD. Bioinformatics. 2014;30:1478–1480. doi: 10.1093/bioinformatics/btu037. [DOI] [PubMed] [Google Scholar]
- 39.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham P., Naftalin R.J. Reptation-induced coalescence of tunnels and cavities in Escherichia coli XylE transporter conformers accounts for facilitated diffusion. J. Membr. Biol. 2014;247:1161–1179. doi: 10.1007/s00232-014-9711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnett C.B., Naidoo K.J. Ring puckering: a metric for evaluating the accuracy of AM1, PM3, PM3CARB-1, and SCC-DFTB carbohydrate QM/MM simulations. J. Phys. Chem. B. 2010;114:17142–17154. doi: 10.1021/jp107620h. [DOI] [PubMed] [Google Scholar]
- 42.Naftalin R.J. Alternating carrier models of asymmetric glucose transport violate the energy conservation laws. Biophys. J. 2008;95:4300–4314. doi: 10.1529/biophysj.108.136366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klepper J., Voit T. Facilitated glucose transporter protein type 1 (GLUT1) deficiency syndrome: impaired glucose transport into brain—a review. Eur. J. Pediatr. 2002;161:295–304. doi: 10.1007/s00431-002-0939-3. [DOI] [PubMed] [Google Scholar]
- 44.Klepper J., Leiendecker B. GLUT1 deficiency syndrome—2007 update. Dev. Med. Child Neurol. 2007;49:707–716. doi: 10.1111/j.1469-8749.2007.00707.x. [DOI] [PubMed] [Google Scholar]
- 45.Garcia J.C., Strube M., Mueckler M.M. Amino acid substitutions at tryptophan 388 and tryptophan 412 of the HepG2 (Glut1) glucose transporter inhibit transport activity and targeting to the plasma membrane in Xenopus oocytes. J. Biol. Chem. 1992;267:7770–7776. [PubMed] [Google Scholar]
- 46.Battle A.R., Ridone P., Martinac B. Lipid–protein interactions: lessons learned from stress. Biochim. Biophys. Acta. 2015;1848:1744–1756. doi: 10.1016/j.bbamem.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Leitch J.M., Carruthers A. ATP-dependent sugar transport complexity in human erythrocytes. Am. J. Physiol. Cell Physiol. 2007;292:C974–C986. doi: 10.1152/ajpcell.00335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.daCosta C.J.B., Dey L., Baenziger J.E. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat. Chem. Biol. 2013;9:701–707. doi: 10.1038/nchembio.1338. [DOI] [PubMed] [Google Scholar]
- 49.Sadiq S.K., Guixa-Gonzalez R., Selent J. Molecular modeling and simulation of membrane lipid-mediated effects on GPCRs. Curr. Med. Chem. 2013;20:22–38. [PubMed] [Google Scholar]
- 50.Hresko R.C., Kraft T.E., Hruz P.W. Mammalian glucose transporter activity is dependent upon anionic and conical phospholipids. J. Biol. Chem. 2016;291:17271–17282. doi: 10.1074/jbc.M116.730168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redmill P.S., McCabe C. Molecular dynamics study of the behavior of selected nanoscale building blocks in a gel-phase lipid bilayer. J. Phys. Chem. B. 2010;114:9165–9172. doi: 10.1021/jp1039942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.