Abstract
The introduced oriental shrimp Palaemon macrodactylus has recently become widespread in temperate estuaries worldwide. However, this recent worldwide spread outside of its native range arises after a previous introduction to the US Pacific coast, where it was restricted for more than 30 years. Using a phylogeographic approach, the present work investigates the genetic history of the invasion of this decapod worldwide. Japan acted as the main native source area for worldwide introduced populations, but other native areas (likely South Korea and China) may act as source populations as well. The recently introduced European and NW Atlantic populations result from colonization from both Japan and an unknown area of the native range, although colonization from the NE Pacific could not be ruled out. Most introduced populations had higher haplotypic diversity than most native populations. P. macrodactylus has a strong potential to become one of the most widespread introduced species and may become the dominant estuarine shrimp in Europe. The ecological and economic consequences of this invasion remain to be thoroughly evaluated.
Over the last few centuries, but especially since World War II, the geopolitical changes associated with the acceleration and the diversification of transport, have led to an exponential increase in mobility of goods and humans. Today, any mechanism allowing the displacement of water volumes (containing plankton and nektonic larvae) from coastal waters across natural dispersal barriers can potentially facilitate massive invasions of neritic littoral communities1. For this reason, concomitantly to the increase in global trade and especially maritime traffic (the largest carrier of freight in recorded history), hundreds of species are, either intentionally or inadvertently, introduced all over the world.
The role of corridors and vectors of introduction is of fundamental importance in the success of invasive species, as they condition the strength of propagule pressure (i.e. the number and frequency, as well as the quality, of propagules introduced into a new area). For instance, multiple introductions from genetically distinct source populations can result in high genetic diversity in the recipient region through admixture, hybridization and introgression, which in turn may facilitate adaptation2. Conversely, with low propagule pressure, only a subset of the source genetic diversity is expected to be retained, thus potentially limiting either short-term resilience to environmental shifts or longer-term adaptation in the new environment. Well known exceptions have been reported and propagule pressure alone is not a sufficient mechanism to explain successful invasions3,4,5. However, genetic studies may help elucidate source regions, routes of invasion and the estimated number of introduction events, which is critical for the identification of potential future invaders and for the development of effective management strategies to prevent or reduce invasions.
Estuaries are very productive ecosystems providing nursery habitats to many marine and brackish species, often of commercial value. These marine-freshwater ecotones show strong daily fluctuations of physical and chemical parameters and are especially impacted by climate change as are many other aquatic systems (e.g.6). They are also subject to strong anthropogenic activities (e.g. harbours, fish and shellfish farming), which in turn facilitate the introduction of non-indigenous species7,8. For example, in the San Francisco Bay estuary, one new non-indigenous species was recorded (as of 1998) every 14 weeks, and in Europe one-fifth of estuarine species are non-indigenous8,9. In estuaries, shipping (including fouling and ballast water) and aquaculture are undoubtedly the two major introduction vectors but the importance of secondary vectors including natural dispersal might also be crucial in the invasion process10, especially to understand the genetic history of an introduced species.
The decapod family Palaemonidae, also called the true “prawns”11, includes many successful worldwide introduced species such as the marine and estuarine oriental shrimp Palaemon macrodactylus Rathbun, 1902, or the freshwater species Exopalaemon modestus (Heller, 1862) and the giant river prawn Macrobrachium rosenbergii (De Man, 1879). Also known as migrant prawn, or grass shrimp, P. macrodactylus is a caridean shrimp whose native distribution may include most of the Sea of China, a large part of the Sea of Japan, and the Pacific coast of Japan12. It occurs both in estuarine (bay and lagoon) waters as well as in full salinity situations (in the Mediterranean and Argentina13,14,15). In its native range in Japan, life span is two years with a six months breeding season from April to October16, but little is known about its biology, especially in its introduced range. P. macrodactylus is now a well established and often dominant species in many American and European estuaries. It could act as a strong competitor for native species, outperforming them for stress resistance to abiotic environmental factors3,17.
Outside its native range, it was initially collected in San Francisco Bay, California, USA in 1957 and has spread into the Northeast Pacific over the last half-century, as far north as Boundary Bay, British Columbia, Canada, and as far south as the Tijuana Estuary, in southern California. Ashelby, et al.12 provided a detailed history of collections of P. macrodactylus worldwide. Apart from a failed introduction in Australia in the 1960s-1970s, the species had not been recorded outside of the North Pacific Ocean for the next few decades. However, in 1992, it was first detected in Europe (Thames River, UK), where it was progressively recorded in various Atlantic estuaries (e.g. Gironde, Guadalquivir, Scheldt). It is now known from southwestern Spain to the North Sea, and also in the Black Sea where it appeared in 200212,18,19,20,21,22,23,24,25,26,27. It was then collected in 2000 in Argentina25, and in 2001 on the Atlantic coast of the United States28. The step-by-step spread of P. macrodactylus worldwide, with a time lag of several decades between the two main introduction periods (NE Pacific vs. NE Atlantic, SW Atlantic and NW Atlantic) raises the question of the source areas for recently introduced populations. Indeed, the NE Pacific might have also been a source area in combination, or independently, from the native area.
Mitochondrial markers like the cytochrome c oxidase subunit I (COI) marker have been widely used to study the geographic distribution of genetic diversity for both native29,30 and non-indigenous invertebrates such as ascidians or crustaceans, including another Palaemonidae Palaemon elegans31,32,33. They are particularly helpful in describing the fine scale history of species introduction and in revealing cryptic invasions32,34,35.
In this study, our objectives were: 1) to describe the distribution and structure of the genetic diversity of P. macrodactylus in a portion of its native range; 2) to compare the genetic composition of major parts of the introduced range (NE Pacific, NE Atlantic, NW Atlantic, SW Atlantic) between each other and with the native range; 3) to identify sources and routes of introduction; 4) to compare the haplotype diversity between native and introduced populations, and 5) to determine the minimal number of introduction events.
Given the nature of shipping routes likely to disperse the shrimp in ballast water, we hypothesized that recently introduced areas were colonised from both the native range and the long-established NE Pacific population, and/or that other secondary hubs (such as Europe) could act as dispersal centers to other regions (such as the NW Atlantic). We also hypothesized that the genetic diversity in introduced populations would be consistently lower than in the native range.
Results
After alignment and trimming, a fragment of 598 bp of the cytochrome c oxidase subunit I (COI) of Palaemon macrodactylus was used for the analyses. The alignment did not include any indels, and a neutral model of evolution of each sequence dataset could not be rejected (Tajima's D statistics; P > 0.10).
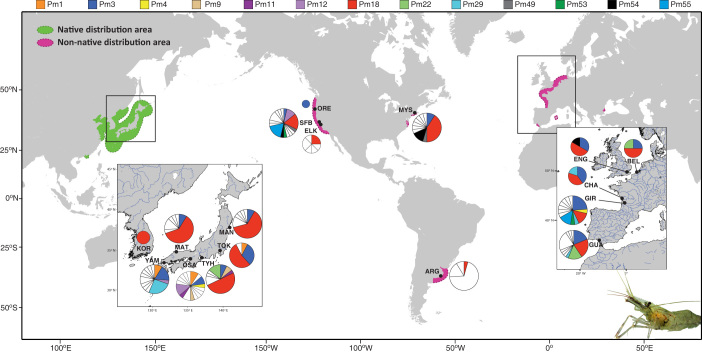
The overall haplotype diversity (h) and nucleotide diversity (π) were estimated to be 0.885 and 0.0077 respectively. A total of 34 polymorphic sites representing 85 haplotypes were found among 286 individuals sampled from 6 native and 10 introduced localities (Figure 1; Table 1). Most of the nucleotide substitutions were synonymous (2 substitutions i.e. 5.9% at position 1 of the codon, and 29 substitutions i.e. 82.3% at position 3) but three non-synonymous changes were identified (3 substitutions i.e. 8.8% at position 1 corresponding to a change from a Leucine to an Isoleucine, a Serine to an Alanine, and an Alanine to a Threonine, respectively). There were 13 haplotypes shared between at least two localities (Figure 1). Among them, three were shared between native Japanese populations (Pm1, Pm9, Pm11), one between Japan and NE Pacific populations (Pm12), three between Japan and Europe (Pm4, Pm22, Pm29), one between NE Pacific and NW Atlantic (Pm49), two between NE Pacific and Europe (Pm53, Pm55), one between NE Pacific, NW Atlantic and Europe (Pm54), and finally two shared among all the different geographical regions (Pm3 and Pm18). Those last two haplotypes were present at high frequencies in introduced populations (Figure 1). The haplotype Pm18 was also found in the native area in South Korea (unknown locality). Indeed, for the 598 bp fragment studied here, Pm18 is strictly identical to the three South Korean barcoding sequences from Genbank (#JX502980-JX502982). Conversely, 34 of the 85 haplotypes were restricted (i.e. private) to the native area in Japan, where the southern locations OSA (see abbreviations in Table 1) and YAM showed the highest haplotypic diversities while northern TOK, MAN and MAT presented the lowest values (Table 1). However, no significant variation of haplotype diversity h or number of private haplotypes with latitude was found (linear regression, p = 0.1097 and p = 0.4235 for h and private haplotypes, respectively).
Figure 1. Locations of sampling sites with COI haplotype frequencies of Palaemon macrodactylus.
Sampling site names are abbreviated as in Table 1. Haplotypes restricted to one sampling location (i.e. private haplotypes) are uniformly represented in white within pie charts while haplotypes shared between locations are color-coded. The size of pie charts is proportional to sampling size. The extension of both native and successful non-native areas are represented according to Ashelby, et al.12. Maps were generated from http://sfb574.geomar.de/gmt-maps.html (Date of access: 09/04/2014).
Table 1. Genetic diversity indices for the oriental shrimp Palaemon macrodactylus. n: number of individuals; Nh: number of haplotypes; Np: number of private haplotypes; h: haplotypic diversity (with standard deviation SD); π: nucleotide diversity (with standard deviation SD); Fs: Fu's Fs value; R2: R2− test. *: 0.05 < p < 0.01; **: 0.01 < p < 0.001; ***: p < 0.001.
| Estuary/river | Location | Area | Code | n | Nh | Np | h ± SD | π ± SD | Fs | R2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Unknown | Unknown, South Korea | Korea | KOR | 3 | 1 | 0 | 1.000 ± 0.000 | 0.000 ± 0.000 | ||
| Toyokawa | Toyohashi, Japan | Japan | TYH | 28 | 8 | 3 | 0.730 ± 0.080 | 0.0032 ± 0.0021 | −1.56 | 0.1179 |
| Yodo and Kishiwada | Osaka, Japan | Japan | OSA | 22 | 17 | 11 | 0.974 ± 0.022 | 0.0099 ± 0.0055 | −7.54** | 0.2016 |
| Tama | Tokyo, Japan | Japan | TOK | 13 | 4 | 3 | 0.654 ± 0.106 | 0.0014 ± 0.0012 | −0.94 | 0.1475 |
| Mangoku-ura inlet | Ishinomaki, Japan | Japan | MAN | 24 | 7 | 5 | 0.605 ± 0.110 | 0.0022 ± 0.0016 | −2.27 | 0.0975 |
| Fushino | Yamaguchi, Japan | Japan | YAM | 23 | 13 | 9 | 0.909 ± 0.042 | 0.0071 ± 0.0041 | −3.78* | 0.1445 |
| Lake Naka-umi | Matsue, Japan | Japan | MAT | 23 | 7 | 5 | 0.625 ± 0.110 | 0.0036 ± 0.0023 | −0.71 | 0.0917 |
| Elkhorn Slough | Monterey Bay, CA | NE Pacific | ELK | 8 | 6 | 5 | 0.929 ± 0.084 | 0.0052 ± 0.0034 | −1.53 | 0.1711 |
| Lake Merritt | San Francisco Bay, CA | NE Pacific | SFB | 28 | 16 | 9 | 0.931 ± 0.028 | 0.0103 ± 0.0060 | −3.62 | 0.22 |
| Lillian Slough | Coos Bay, OR | NE Pacific | ORE | 1 | 1 | 0 | 1.000 ± 0.000 | 0.000 ± 0.000 | ||
| Mystic River | Stonington, CT | NW Atlantic | MYS | 22 | 11 | 7 | 0.823 ± 0.075 | 0.0093 ± 0.0052 | −0.96 | 0.1946 |
| Mar de la Plata harbour | Buenos Aires, Argentina | SW Atlantic | ARG | 23 | 4 | 3 | 0.498 ± 0.111 | 0.0015 ± 0.0012 | −0.08 | 0.1515 |
| Guadalquivir | Trebujena, Spain | Europe | GUA | 32 | 11 | 7 | 0.865 ± 0.030 | 0.0033 ± 0.0021 | −4.10* | 0.0958 |
| Gironde | Vitrezay, France | Europe | GIR | 21 | 13 | 7 | 0.924 ± 0.040 | 0.0119 ± 0.0065 | −1.93 | 0.1799 |
| Charente | Rochefort, France | Europe | CHA | 5 | 3 | 0 | 0.800 ± 0.164 | 0.0023 ± 0.0020 | 0.06 | 0.2848 |
| Zeebrugges harbour | Bruges, Belgium | Europe | BEL | 4 | 3 | 0 | 0.833 ± 0.222 | 0.0017 ± 0.0017 | −0.89 | 0.2500 |
| Thames | Tilbury, UK | Europe | ENG | 6 | 3 | 0 | 0.733 ± 0.155 | 0.0077 ± 0.0051 | 2.73 | 0.3446 |
| Total | 286 | 85 | 0.885 ± 0.015 | 0.0077 ± 0.0042 |
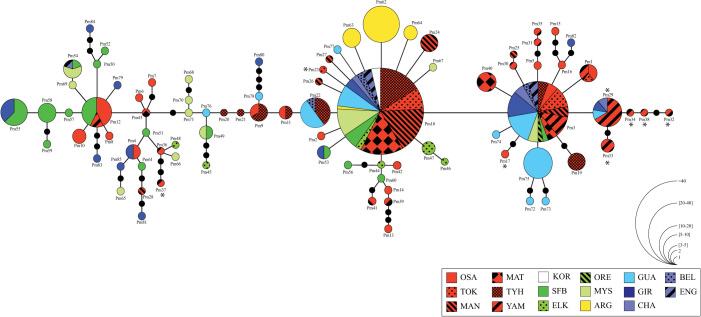
The minimum spanning tree (MST) showed a configuration with three star-like haplogroups centered on the haplotypes Pm3, Pm12, and Pm18 (Figure 2). Each of the haplogroups Pm3 and Pm18 were geographically widely distributed (16 and 14 sampled locations represented, respectively), while the center of diversity of the haplogroup Pm12 was mainly restricted to the OSA and YAM populations in the native area and to SFB, ELK, GIR and ENG in the introduced range. Most non-synonymous substitutions were located at the tips of the network (Figure 2) and restricted to OSA and YAM populations, which presented also the only significant values for the Fu's Fs test in the native range (Table 1).
Figure 2. Minimum spanning network for cytochrome c oxidase subunit I (COI) sequences of the oriental shrimp Palaemon macrodactylus.
Each circle represents a haplotype and its size is proportional to the haplotype frequency. Within the network, each line between haplotypes represents a mutational change. Small black dots indicate unsampled haplotypes inferred from the data. Sampling locations are abbreviated as in Table 1, and localities from the same area share a same background color: red for Japan, white for Korea, green for NE Pacific, light green for NW Atlantic, yellow for SW Atlantic, and blue/violet for Europe. Asterisks indicate an amino acid change.
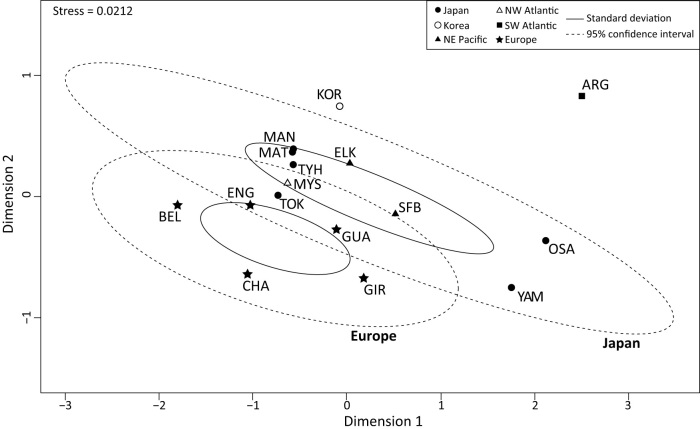
The analysis performed to estimate both pairwise ΦST and Jost's Dest estimates showed similar results, except for MYS where the native locations showed a discrepancy between both estimators (Table 2). For the native area in Asia, YAM and OSA were significantly different from the remaining native locations (Table 2). No significant differences were found among European introduced locations (except for GUA-GIR for ΦST). Conversely, the ARG sampling location (SW Atlantic) showed a clear significant difference with almost all other locations (see Table 2). In the NMDS plot (Figure 3), such a pattern of differentiation of ARG was also found, but the YAM and OSA locations remain within the 95% confidence interval (CI) of the Japanese group. The NMDS representation showed a clear proximity between all invasive and Japanese locations (except for ARG), especially for the European ENG and GUA, the NE Pacific ELK and SFB, and the NW Atlantic MYS, that were all situated within the 95%-CI of the Japanese group (Figure 3).
Table 2. Pairwise ΦST values (below diagonal) and pairwise Jost's Dest values with confidence intervals (above diagonal) for the mtDNA COI of Palaemon macrodactylus. Both significant p-values of ΦST following a FDR correction (set at 0.0091) and confidence intervals of Dest not enclosing 0, are represented in bold. Locality names are abbreviated as in Table 1.
| KOR | OSA | TYH | TOK | MAN | YAM | MAT | ELK | SFB | MYS | ARG | GUA | GIR | CHA | BEL | ENG | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KOR | 1 (1, 1) | 0.212 (0.056, 0.433) | 0.2 (0, 0.534) | 0.104 (0, 0.289) | 1 (1, 1) | 0.115 (0.003, 0.325) | 0.533 (0.127, 0.950) | 0.666 (0.415, 0.924) | 0.303 (0.087, 0.636) | 0.942 (0.808, 1) | 0.615 (0.371, 0.830) | 0.735 (0.476, 0.981) | 0.333 (0, 0.931) | 0.143 (0, 0.853) | 0.211 (0, 0.681) | |
| OSA | 0.24846 | 0.923 (0.780, 0.989) | 0.812 (0.517, 0.975) | 0.964 (0.854, 1) | 0.493 (0.148, 0.747) | 0.961 (0.849, 0.996) | 1 (1, 1) | 0.623 (0.298, 0.832) | 0.917 (0.727, 0.981) | 1 (1, 1) | 0.788 (0.508, 0.938) | 0.534 (0.164, 0.787) | 0.678 (0.264, 0.862) | 0.764 (0.345, 0.903) | 0.793 (0.426, 0.940) | |
| TYH | −0.068 | 0.33763 | 0.055 (0, 0.365) | 0.042 (0, 0.249) | 0.931 (0.782, 0.991) | 0.038 (0, 0.250) | 0.267 (0, 0.783) | 0.458 (0.136, 0.800) | 0.047 (0, 0.367) | 0.944 (0.796, 1) | 0.283 (0.003, 0.592) | 0.489 (0.143, 0.804) | 0.027 (0, 0.689) | 0 (0, 0.334) | 0 (0, 0.507) | |
| TOK | 0.02144 | 0.35821 | 0.08671 | 0.023 (0, 0.344) | 0.725 (0.405, 0.941) | 0.017 (0, 0.338) | 0.355 (0, 0.815) | 0.484 (0.178, 0.798) | 0.044 (0, 0.386) | 0.945 (0.799, 0.998) | 0.271 (0.007, 0.546) | 0.289 (0, 0.653) | 0 (0, 0.348) | 0 (0, 0.410) | 0 (0, 0.275) | |
| MAN | −0.16779 | 0.39492 | 0.05392 | 0.03647 | 0.940 (0.801, 0.997) | 0 (0, 0.156) | 0.33 (0, 0.844) | 0.506 (0.165, 0.843) | 0.073 (0, 0.418) | 0.939 (0.785, 1) | 0.425 (0.144, 0.715) | 0.537 (0.184, 0.847) | 0.048 (0, 0.740) | 0 (0, 0.608) | 0 (0, 0.514) | |
| YAM | 0.22152 | 0.23949 | 0.31052 | 0.26777 | 0.33925 | 0.935 (0.787, 0.994) | 1 (1, 1) | 0.864 (0.652, 0.958) | 0.88 (0.647, 0.979) | 1 (1, 1) | 0.639 (0.350, 0.865) | 0.356 (0, 0.710) | 0.163 (0, 0.721) | 0.662 (0.133, 0.876) | 0.676 (0.271, 0.915) | |
| MAT | −0.1271 | 0.29944 | 0.04852 | 0.01628 | 0.0138 | 0.25975 | 0.319 (0, 0.835) | 0.497 (0.173, 0.826) | 0.063 (0, 0.373) | 0.94 (0.788, 1) | 0.415 (0.127, 0.720) | 0.523 (0.164, 0.837) | 0.033 (0, 0.717) | 0 (0, 0.588) | 0 (0, 0.498) | |
| ELK | 0.08067 | 0.18713 | 0.21876 | 0.31386 | 0.25809 | 0.27023 | 0.13745 | 0.363 (0, 0.678) | 0.164 (0, 0.673) | 0.962 (0.841, 0.995) | 0.47 (0.040, 0.788) | 0.516 (0.100, 0.750) | 0.263 (0, 0.648) | 0 (0, 0.417) | 0.261 (0, 0.693) | |
| SFB | 0.38639 | 0.09225 | 0.48185 | 0.49652 | 0.51632 | 0.41543 | 0.44691 | 0.30331 | 0.316 (0, 0.637) | 0.973 (0.882, 1) | 0.551 (0.269, 0.819) | 0.155 (0, 0.508) | 0.362 (0, 0.765) | 0.166 (0, 0.619) | 0.361 (0, 0.720) | |
| MYS | 0.07518 | 0.04473 | 0.18945 | 0.23799 | 0.24101 | 0.23364 | 0.15807 | 0.04731 | 0.13652 | 0.947 (0.793, 0.998) | 0.309 (0.023, 0.619) | 0.358 (0, 0.722) | 0 (0, 0.577) | 0 (0, 0.403) | 0 (0, 0.356) | |
| ARG | 0.30387 | 0.48765 | 0.31701 | 0.41735 | 0.32369 | 0.46704 | 0.28368 | 0.44839 | 0.57077 | 0.35649 | 0.97 (0.878, 1) | 0.979 (0.912, 0.998) | 0.95 (0.795, 0.994) | 0.935 (0.768, 0.987) | 0.943 (0.792, 0.992) | |
| GUA | 0.11173 | 0.32163 | 0.06484 | 0.12438 | 0.16818 | 0.25039 | 0.12011 | 0.27845 | 0.49807 | 0.22611 | 0.40372 | 0.267 (0, 0.589) | 0 (0, 0.425) | 0 (0, 0.257) | 0.144 (0, 0.520) | |
| GIR | 0.11636 | −0.01238 | 0.24753 | 0.25614 | 0.28824 | 0.19313 | 0.2045 | 0.0917 | 0.06902 | −0.01189 | 0.38706 | 0.25023 | 0 (0, 0.366) | 0 (0, 0.487) | 0.12 (0, 0.591) | |
| CHA | 0.17001 | 0.27991 | 0.10819 | 0.00501 | 0.12478 | 0.08362 | 0.03897 | 0.24485 | 0.44415 | 0.18529 | 0.46131 | 0.06387 | 0.17793 | 0 (0, 0.248) | 0 (0, 0.230) | |
| BEL | −0.09128 | 0.22978 | −0.10223 | −0.05863 | −0.09623 | 0.18114 | −0.09405 | 0.1288 | 0.39268 | 0.09319 | 0.35738 | −0.04618 | 0.12043 | −0.00892 | 0 (0, 0.161) | |
| ENG | −0.12809 | 0.10136 | 0.0237 | 0.04535 | 0.06186 | 0.12654 | −0.03945 | 0.01094 | 0.26153 | −0.0204 | 0.33635 | 0.05308 | 0.00727 | −0.02901 | −0.12226 |
Figure 3. Non-metric multidimensional scaling of the different sampling geographic regions based on Jost's Dest values of the oriental shrimp Palaemon macrodactylus.
Solid and dashed lines represent the standard deviations and the 95% confidence interval respectively for those regions that had more than two sampling localities (Europe and Japan).
Several hypotheses about population grouping were tested for the native Japanese locations and for the whole dataset of sampled locations. For the native Japanese populations, without a priori grouping, more variation was attributed to within than to among populations (73 vs. 27%; Table 3). Japanese populations of P. macrodactylus exhibited a moderate overall genetic structure (ΦST = 0.267). According to the above results, we tested whether the southern locations YAM and OSA clustered into one or two genetic entities separated from the group constituted by the remaining Japanese locations (see Table 3). The AMOVA revealed that the genetic component associated with each of these two hypotheses (24 and 37% respectively) was non-significant (despite p-values close to the 0.05-threshold; Table 3). Most of the genetic variation was found within locations (65 and 63% respectively; significant components).
Table 3. Analysis of Molecular Variance (AMOVA) among the Japanese localities of Palaemon macrodactylus.
| Source of variation | df | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| No grouping | ||||
| Among locations | 5 | 63.034 | 0.510 | 26.65 |
| (FST = 0.267, p = 0.000) | ||||
| Within locations | 127 | 178.136 | 1.403 | 73.35 |
| Total | 132 | 241.170 | 1.912 | |
| (YAM-OSA) vs. (TOK-MAN-MAT-TYH) | ||||
| Among groups | 1 | 37.359 | 0.518 | 24.26 |
| (FCT = 0.243, p = 0.065) | ||||
| Among locations within groups | 4 | 25.089 | 0.224 | 10.49 |
| (FSC = 0.139, p = 0.000) | ||||
| Within locations | 127 | 176.936 | 1.393 | 65.24 |
| (FST = 0.348, p = 0.000) | ||||
| Total | 132 | 239.383 | 2.135 | |
| YAM vs. OSA vs. (TOK-MAN-MAT-TYH) | ||||
| Among groups | 2 | 58.066 | 0.815 | 36.55 |
| (FCT = 0.366, p = 0.069) | ||||
| Among locations within groups | 3 | 4.969 | 0.012 | 0.53 |
| (FSC = 0.008, p = 0.010) | ||||
| Within locations | 127 | 178.136 | 1.403 | 62.92 |
| (FST = 0.371, p = 0.000) | ||||
| Total | 132 | 241.170 | 2.229 |
Concerning the whole set of native and introduced populations, we tested the two hypotheses that the recent introductions in Europe, NW and SW Atlantic may preferentially cluster either with Japan and Korea (Asian origin) or with SFB and ELK (NE Pacific origin). The AMOVA (Table 4) showed that the genetic component associated with the Asian origin (24%) was marginally significant (p = 0.052) while the component associated with the NE Pacific origin (0.34%) was clearly non-significant (p = 0.374). In the Asian origin hypothesis, 59% of the genetic variance observed was concentrated within populations, and 17% of the variance was related to between-population differentiation (see Table 4).
Table 4. Analysis of Molecular Variance (AMOVA) among the native and introduced localities of Palaemon macrodactylus.
| Source of variation | df | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| No grouping | ||||
| Among groups | 15 | 200.299 | 0.666 | 28.10 |
| (FST = 0.281, p = 0.000) | ||||
| Within locations | 269 | 458.628 | 1.705 | 71.90 |
| Total | 284 | 658.928 | 2.371 | |
| NE Pacific vs. (Japan-Korea-Europe-NW Atlantic-SW Atlantic) | ||||
| Among groups | 1 | 57.851 | 0.708 | 24.32 |
| (FCT = 0.243, p = 0.052) | ||||
| Among locations within groups | 14 | 142.448 | 0.497 | 17.07 |
| (FSC = 0.226, p = 0.000) | ||||
| Within locations | 269 | 458.628 | 1.705 | 58.61 |
| (FST = 0.414, p = 0.000) | ||||
| Total | 284 | 658.928 | 2.909 | |
| (Japan-Korea) vs. (NE Pacific-Europe-NW Atlantic-SW Atlantic) | ||||
| Among groups | 1 | 17.687 | 0.008 | 0.34 |
| (FCT = 0.003, p = 0.374) | ||||
| Among locations within groups | 14 | 180.199 | 0.654 | 27.78 |
| (FSC = 0.279, p = 0.000) | ||||
| Within locations | 269 | 454.850 | 1.691 | 71.88 |
| (FST = 0.281, p = 0.000) | ||||
| Total | 284 | 652.737 | 2.352 | |
The relationship between genetic and geographical distances was studied using ΦST, [ΦST/(1 - ΦST)], and Dest estimates, before and after log-transformation. No significant isolation-by-distance pattern was observed among the native Japanese locations and among European locations, whatever the estimate and transformation used (Mantel test, p > 0.05).
The raggedness index did not yield any significant values whatever the location considered (p > 0.149; not shown), except for KOR due to its low sample size. Fu's Fs-test and the R2-test, performed to examine the dynamics of population growth for P. macrodactylus, especially at native locations, could not reject a model of constant size for locations sampled in this study. There were however a few exceptions for one of the two tests performed (Table 1). However, given the lack of consistency between the tests, we cannot conclude that there was past population expansion.
Discussion
The objective of this study was to describe, using inferences from the widely used COI locus, the genetic structure and recent demography of the introduced oriental shrimp Palaemon macrodactylus in a portion of its native area, but also to provide insights into its invasion history, colonisation and expansion patterns worldwide.
Honshu, the main island of Japan, corresponds to the eastern part of the native distribution of the oriental shrimp, but also represents one of the main potential sources of introduction due to the intense Japanese international trading since World War II, particularly through shipping. Over the sampled area, which included some of the main international harbours of Japan (e.g. Osaka, Tokyo), the haplotypic diversity was very high (range 0.605–0.974; Table 1) and showed a large number of private haplotypes. Such a pattern seems quite frequent in the Palaemonidae family (e.g.33,36,37) but not in decapods in general (see38).
The two southern Honshu locations YAM and OSA showed much higher haplotypic diversities than remaining locations (Table 1). Despite various haplotypes shared with more northern locations, YAM and OSA were quite differentiated (see fixation indexes ΦST and the Jost's differentiation Dest in Table 2 and NMDS in Figure 3). However, the AMOVA failed to isolate them into one or two genetic clusters separated from the remaining locations. Whatever the inference tested, the AMOVA showed that most genetic variance could be explained by variation within locations (>60%; Table 3). The two populations YAM and OSA might be in demographic expansion as shown by the star-like configuration of the MST network with non-synonymous substitutions restricted to its tips and significant Fu's neutrality test (Figure 2; Table 1). However the lack of consistency with the analysis of mismatch distribution and the other neutrality test does not permit to conclude. In the same way, neither a pattern of isolation by distance nor of changing haplotypic diversity with latitude could explain the lower diversity found in northern Japanese locations. The apparent discrepancy between Japanese locations is in fact due to the high frequency of haplotype Pm18 (>50%) in populations other than YAM and OSA. This haplotype was also shared by three South Korean sequences obtained from Genbank (#JX502980-JX502982).
All in all, the results suggest that the Japanese localities act as a single genetic entity, with high rates of gene flow among localities separated by hundreds of kilometres (over 1300 km between MAN and YAM, the two most distant locations). The KOR locality might even be included in this population, but a larger sample size and more localities would be needed to reach a more definitive conclusion.
In a more general way, Palaemonidae shrimps appear to show strong gene flow over large geographical scales. The river shrimp Cryphiops caementarius has high genetic variability with no apparent geographic structuring for distances up to 700 km37. For P. elegans in the European Atlantic and the western Mediterranean, high levels of gene flow were reported even between locations separated by thousands of kilometres33. Although it is tempting to explain such strong gene flow by natural larval dispersal, in regions of high shipping traffic like Eastern Asia, Europe or USA, ballast water dispersal may also act in maintaining or enhancing gene flow, even for native palaemonids. This may be the case of P. macrodactylus in its native range in Japan.
Across the introduced range, the haplotypic diversity compared favourably with that in native Japan. Indeed, for 9 out of 10 introduced populations (except ARG), haplotypic diversity was higher than that in 4 or 5 of the 6 Japanese populations. OSA was the only native population with consistently higher haplotypic diversity than in the introduced range. High haplotypic diversity was found in both recently (1990s) invaded locations in Europe, NW Pacific and SW Atlantic, but also in the NE Pacific with SFB and ELK that were invaded in the 1950s. This suggests that repeated temporal introduction events have occurred between different introduced areas, as previously proposed based on present and historic introduction records12,24. Our results also suggest various introduction events from genetically distinct source populations. This elevated propagule pressure could compensate the loss of genetic diversity via founder effects which may be expected during introduction events2.
The presence of shared haplotypes among very distant locations suggests a recent connection, through human-mediated dispersal. Haplotypes Pm3 and Pm18 were abundant and present in almost all locations (13 and 14 populations respectively over 16 in total). It suggests a common history of all introduced populations with Japan (and also Korea), which is supported by the AMOVA (Table 4), the fixation indexes ΦST (Table 2), and the Jost's differentiation Dest with the NMDS (Figure 3). Interestingly, the absence of Pm18 from the native locations YAM and particularly OSA, a large international harbour, suggests that at least one introduction event originates from northern Japan, although we cannot exclude a Korean origin.
In the NE Pacific, haplotypes Pm3 and Pm18 were both found in SFB, the first introduced location. Haplotype Pm12, only found at low frequency in the native range, is also exclusive to SFB in the introduced range. These haplotypes shared between the native area and SFB suggest that at least one introduction event occurred from Japan to SFB. Elsewhere in the NE Pacific, Pm3 is found only in the northern ORE, and Pm18 only in the southern ELK. Despite the small sample size, this might suggest these two locations were colonised from SFB rather than by direct introduction from Asia.
A time lag of at least 35 years separates the first report of P. macrodactylus outside its native range from the earliest of the reports from Europe, the SW Atlantic, or the NW Atlantic (in 1992 in the Thames River, UK). Given the intensive trading routes among introduced and native areas, this time lag implies that the source of these recently introduced populations might be found in the native area, or the NE Pacific, or both (or, for the NW Atlantic, from Europe). As the NE Pacific might act as a source for more recently introduced populations, it is difficult to identify a genetic signature that allows discrimination among candidate source populations. The presence of the widespread haplotypes Pm3 and Pm18 in these populations does not allow discrimination between an exclusively Japanese or exclusively NE Pacific origin. However, in Europe and the NW Atlantic, we found both a) haplotypes shared exclusively with Japan (Pm22 and Pm29 from TYH and YAM respectively) and b) haplotypes shared exclusively with SFB (Pm49, Pm 53, Pm54 and Pm55). This suggests that European (GUA, GIR, CHA, ENG, BEL) and NW Atlantic (MYS) locations are the result of a combination of introduction events originating from the native area and the NE Pacific, and that the European populations may be the source of the NW Atlantic populations. A common pattern in North Atlantic invasions of Asian species is that Europe was colonised first, followed by ship-mediated transport west secondarily to North America (e.g. the Asian green alga Codium fragile39, the Asian ascidian Styela clava40, or the Pacific bryozoan Tricellaria inopinata41,42).
A large proportion of private haplotypes was found in all populations, whether native or introduced (Figures 1 and 2). The native area demes YAM and OSA showed a particularly high number of private haplotypes resulting in high haplotype diversity (Table 1). While predictable in the native area, the presence of so many private haplotypes at each introduced location was unexpected. This is especially the case for ELK and ARG, where they dominate in numbers and frequency. However, the private haplotypes are not very divergent from shared ones, and especially from Pm3 and Pm18 (Figure 3). So many private haplotypes in introduced populations suggests unsampled native locations (from Korea, China or other parts of Japan) contributed as source populations, suggesting multiple introductions into each area. The number of private haplotypes found suggests the contribution of unsampled native populations to P. macrodactylus invasion is significant (Fig. 1 and 2), although Japan is likely to be the main source of invasion. The geographically restricted character of private haplotypes implies various independent introduction events from different places around the world. A reduction in haplotypic diversity is common in very recently introduced populations (e.g.31,32). However, the present study illustrates how propagule pressure, through repeated introduction events and/or large inoculum size, can compensate for deleterious effects of bottlenecks and re-establish equivalent or even higher levels of diversity2. More detailed insights into colonization pathways may be found using faster evolving markers (e.g. microsatellites or SNPs). These could be used to evaluate how multiple introductions can convert among-population genetic diversity in the native range to within-population genetic diversity in the introduced range, inducing potentially novel allelic combinations that can drive phenotypic diversification or alter the adaptive potential of invasive populations35.
The hypothesis of repeated introduction events suggests that a viable corridor has existed for many years from the Northwest Pacific to other sites worldwide via ballast water. The risk of marine invasions by global shipping has recently been evaluated by ecoregions and showed strong connections between native and introduced areas where P. macrodactylus is present43,44. The probable native distribution of P. macrodactylus includes three countries that have experienced exponential economic development over the last 60 years: the main islands of Japan, South Korea and northeaster China. The huge international trade between Japan and South Korea since the 1950s is mainly through shipping traffic. Over the last two decades, China has also exponentially increased its export shipping, with Shanghai (the world's busiest harbour) being within P. macrodactylus' native range. In addition, most recently invaded localities (as well as SFB) are in the vicinity of large, international harbours (e.g.12,13,45). The present study strongly supports the hypothesis that ballast water in international shipping is the original and main introduction vector of P. macrodactylus worldwide.
However, other dispersal mechanisms, both natural and anthropogenic, are likely to contribute to the invasion. Coastal vessel traffic, both commercial and private, is likely to be important. Commercial cargo vessels do not, for example, take up and release ballast water solely in ports and harbours. For many different reasons, such as adjusting trim and stability, ballast water may be released at sea as a vessel travels along a coastline (legally permissible everywhere for safety reasons, and permissible in most countries because coastal movement, as opposed to transoceanic carriage, of ballast water is not regulated), and thus plankton, including larvae (meroplankton) could drift into small lagoons and embayments that do not have international or other large cargo vessel traffic. Smaller vessels, including fishing boats and recreational craft, may have ballast tanks as well. Older fishing vessels may also have “bait wells” with flow-through water systems, in which shrimp can survive (J. T. Carlton, personal observations). Old tires used as bumpers (fenders) on small coastal vessels may also be capable of transporting larger errant species, such as crabs, shrimp, and fish, in the tire well. These shrimp, which may reach 7 cm or more in length, can be an attractive bait species for fishermen, and thus could also be transported some distances in bait buckets and then released. In short, a plethora of secondary dispersal vectors are available to non-indigenous species once they have arrived from overseas in ballast water. Larval transport by coastal currents must also occur between international harbours and adjacent estuaries. For example, the Guadalquivir River where the species was first recorded in 1997 is located in the vicinity of Cádiz international harbour. From the Guadalquivir, P. macrodactylus may have spread naturally to the Guadiana River, 100 km westward20,21.
Overall, this study emphasizes the strong invasive potential of Palaemon macrodactylus. P. macrodactylus has the potential to become the dominant estuarine shrimp in Europe and one of the most widespread aquatic introduced species. The ecological and economic consequences of this invasion remain to be thoroughly evaluated. In the coming decades, this late 20th century and early 21st century surge of new populations of P. macrodactylus around the world will likely reach an asymptote or cease entirely (although this shrimp will continue to spread along coastlines where it is now established, and perhaps expand even more due to global climate change and warming coasts). Following an early event that transported this shrimp from Asia to SFB, likely during an increase in military shipping activity during the Korean War, P. macrodactylus then spread many decades later within a short period of time in the North Atlantic (North America and Europe) and South Atlantic (Argentina), almost certainly due to increased global trade. As a result of many ballast water-mediated invasions, the United Nations International Maritime Organization, and numerous individual nations, have introduced increasingly rigorous controls over ballast water discharge. If these controls are effective, future invasions of P. macrodactylus to novel locations may decline or cease.
Methods
Sampling, DNA extraction and sequencing
Sampling of the oriental shrimp Palaemon macrodactylus was undertaken using hand-nets or keep-nets and collected samples were preserved in ethanol. The species was sampled from 6 localities in its native area (Japan) and 10 localities from the introduced range spanning almost all the known introduced localities (Table 1, Fig. 1). We made every attempt to obtain specimens from China. This included contacting over 30 Chinese specialists in the native range. The species was reported as rare due to intense exploitation, and difficult to sample (Changfang Zhou, pers. Comm.).
Total genomic DNA was extracted from pleopods using a Chelex 10% or a salt-extraction protocol46. A fragment of the mitochondrial DNA (mtDNA) coding for the cytochrome c oxidase subunit I (COI) gene was amplified using the crustacean decapod specific primer pair CrustCOIF and DecapCOIR47. Amplifications were carried out in a 15 μl reaction volume, with 1–2 μl of genomic DNA, 2 μl of 10× buffer, 0.16 μl dNTP (200 mM), 0.8 μl of MgCl2 (50 mM) and 0.6 units of polymerase (Bioline). PCR was performed using a hotstart and consisted in an initial denaturation step at 94°C for 5 min, followed by 30 amplification cycles (94°C for 30 s, 52°C for 30 s and 72°C for 1 min) and a final elongation step at 72°C for 5 min. Sequencing was performed by Macrogen Europe Company (the Netherlands). Sequences were then manually edited using BioEdit v. 7.1.948. All the haplotypes were deposited into the EMBL database (Accession numbers: HG792276-HG792360). We also included in our dataset the three COI sequences of P. macrodactylus from South Korea available in public databases (Genbank Accession numbers: JX502980-JX502982). Nucleotide sequences were translated into amino acid sequences using the Drosophila mitochondrial genetic code49.
Genetic analyses
To identify sequences that do not fit the neutral theory model we used Tajima's D statistic50. Genetic diversity within locations was characterized by standard diversity indices of haplotype diversity (h) and nucleotide diversity (π)51. A linear regression analysis was performed in R 3.0.1 (http://CRAN.R-project.org/) to test whether variation in genetic diversity of Japanese locations was associated with latitude. A minimum spanning network was also generated51,52. Due to its extremely reduced sampling size (n = 1), the ORE location was not included in all downstream analyses.
The best-fit nucleotide substitution model under the corrected Akaike Information Criterion was determined53. Accordingly, population pairwise fixation indices (ΦST; performed using 10,000 permutations) were calculated51 using the Jukes-Cantor (JC) substitution model. The p-values were corrected for multiple comparisons using the false discovery rate (FDR) control according to Benjamini and Yekutieli54 method (B-Y), as described in Narum55 and with an experiment-wise error rate set at 0.05. We also calculated the Jost's Dest56 using 1,000 bootstrap replicates. Although this measure does not incorporate genetic distances between haplotypes, it provides an unbiased measure of genetic differentiation between samples56,57. A non-metric multidimensional scaling (NMDS)58 was used to graphically represent the differentiation among locations based on Dest estimates.
In order to determine the hierarchical structure of COI variation, an analysis of molecular variance (AMOVA) was conducted based on 10,000 random permutations51. Various a priori hypotheses about population grouping were tested for native Japanese localities, but also for both native and introduced localities.
To test for correlation between genetic and geographic distance (km), we performed Mantel tests with 10,000 permutations59. Analyses were performed using the ΦST, the Slatkin's [ΦST/(1 − ΦST)] and the Dest values previously calculated. For genetic and geographic distance matrices, both non-transformed and respective log-transformed values were used. Geographical distances were calculated as the minimum coastline distances between adjacent sampling locations.
To explore the historical demography of populations, especially the native ones, we used the ‘raggedness index’, based on mismatch distributions, looking at the fit of the observed mismatch distribution to a model of sudden population expansion60,61. To test if COI evolved in a neutral manner we performed50 the Fu's Fs-test62 and the R2-test63.
Acknowledgments
We are sincerely grateful to all people who helped with sampling in different parts of the world. For Europe, we would like to thank: R. Lopez Luque, P. Boets, J. Cuesta, C. Ruel, C. Diaz, A.-M. Vincent, M. Girardin, A. Shaw, C. Ashelby, N. Petkov, B. Georgiev, P. Zehtindjiev. For sampling in North and South America: G. Velazquez, L. Haram, K. Durkin, J. Schaefer, J. Chapman, M. Fountain and the Elkhorn Slough National Estuarine Research Reserve, CA. For sampling in Japan: H. Ariyama, Y. Koda, T. Asaka, M. I. Rico, M. Sekine, K. Shimada, W. Doi, Dr Usami, R. Harada and the Kirara-hama Natural Park, Yamaguchi, Japan. And special thanks to C. Zhou for the attempt to obtain samples from China. This work was funded by a project from the Spanish Ministry of Economy and Competitiveness (CGL2010-16028) and a CSIC JAE postdoctoral fellowship to C.L.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.L., C.R. and A.J.G. conceived the ideas. C.L., A.S. and N.P. ran the genetic labwork. C.L. prepared all the figures. C.L., A.S., N.P., M.O., M.B., J.T.C., C.R. and A.J.G. contributed to sampling collection worldwide, manuscript text, and reviewed the manuscript.
References
- Carlton J. T. & Geller J. B. Ecological roulette - The global transport of nonindigenous marine organisms. Science 261, 78–82 (1993). [DOI] [PubMed] [Google Scholar]
- Roman J. & Darling J. A. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464 (2007). [DOI] [PubMed] [Google Scholar]
- Lejeusne C., Latchere O., Petit N., Rico C. & Green A. J. Do invaders always perform better? Comparing the response of native and invasive shrimps to temperature and salinity gradients in south-west Spain. Estuar. Coast Shelf Sc. 136, 102–111 (2014). [Google Scholar]
- Kolbe J. J. et al. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181 (2004). [DOI] [PubMed] [Google Scholar]
- Kolbe J. J., Leal M., Schoener T. W., Spiller D. A. & Losos J. B. Founder Effects Persist Despite Adaptive Differentiation: A Field Experiment with Lizards. Science (2012). [DOI] [PubMed] [Google Scholar]
- Lejeusne C., Chevaldonné P., Pergent-Martini C., Boudouresque C. F. & Pérez T. Climate change effects on a miniature ocean: the highly diverse, highly impacted Mediterranean Sea. Trends Ecol. Evol. 25, 250–260 (2010). [DOI] [PubMed] [Google Scholar]
- Ruiz G. M., Carlton J. T., Grosholz E. D. & Hines A. H. Global invasions of marine and estuarine habitats by non-indigenous species: Mechanisms, extent, and consequences. Am. Zool. 37, 621–632 (1997). [Google Scholar]
- Cohen A. N. & Carlton J. T. Accelerating invasion rate in a highly invaded estuary. Science 279, 555–558 (1998). [DOI] [PubMed] [Google Scholar]
- Reise K., Olenin S. & Thieltges D. W. Are aliens threatening European aquatic coastal ecosystems? Helgol. Mar. Res. 60, 77–83 (2006). [Google Scholar]
- Tepolt C. K. et al. European green crabs (Carcinus maenas) in the northeastern Pacific: genetic evidence for high population connectivity and current-mediated expansion from a single introduced source population. Divers. Distrib. 15, 997–1009 (2009). [Google Scholar]
- Holthuis L. B. FAO species catalogue, Vol. 1. Shrimps and prawns of the world. An annotated catalogue of species of interest to fisheries. Vol. 1 271 (FAO Fisheries Synopsis No. 125 Volume 1, 1980). [Google Scholar]
- Ashelby C. W., Johnson M. L. & De Grave S. The global invader Palaemon macrodactylus (Decapoda, Palaemonidae): an interrogation of records and a synthesis of data. Crustaceana 86, 594–624 (2013). [Google Scholar]
- Spivak E. D., Boschi E. E. & Martorelli S. R. Presence of Palaemon macrodactylus Rathbun 1902 (Crustacea: Decapoda: Caridea: Palaemonidae) in Mar del Plata harbor, Argentina: first record from southwestern Atlantic waters. Biol. Invasions 8, 673–676 (2006). [Google Scholar]
- Torres A. P. et al. First record of Palaemon macrodactylus Rathbun, 1902 (Decapoda, Palaemonidae) in the western Mediterranean. Mediterr. Mar. Sci. 13, 278–282 (2013). [Google Scholar]
- Vázquez M. G., Ba C. C. & Spivak E. D. Life history traits of the invasive estuarine shrimp Palaemon macrodactylus (Caridea: Palaemonidae) in a marine environment (Mar del Plata, Argentina). Sci. Mar. 76, 507–516 (2012). [Google Scholar]
- Omori M. & Chida Y. Life history of a caridean shrimp Palaemon macrodactylus with special reference to the difference in reproductive features among ages. B. Jpn. Soc. Sci. Fish. 54, 365–375 (1988). [Google Scholar]
- González-Ortegón E., Cuesta J., Pascual E. & Drake P. Assessment of the interaction between the white shrimp, Palaemon longirostris, and the exotic oriental shrimp, Palaemon macrodactylus, in a European estuary (SW Spain). Biol. Invasions 12, 1731–1745 (2010). [Google Scholar]
- Ashelby C. W., Worsfold T. M. & Fransen C. First records of the oriental prawn Palaemon macrodactylus (Decapoda: Caridea), an alien species in European waters, with a revised key to British Palaemonidae. J. Mar. Biol. Assoc. U.K. 84, 1041–1050 (2004). [Google Scholar]
- Béguer M., Girardin M. & Boet P. First record of the invasive oriental shrimp Palaemon macrodactylus Rathbun, 1902 in France (Gironde Estuary). Aquat. Invas. 2, 132–136 (2007). [Google Scholar]
- Chicharo M. A. et al. Alien species in the Guadiana estuary (SE-Portugal/SW-Spain): Blackfordia virginica (Cnidaria, Hydrozoa) and Palaemon macrodactylus (Crustacea, Decapoda): potential impacts and mitigation measures. Aquat. Invas. 4, 501–506 (2009). [Google Scholar]
- Cuesta J. A., González-Ortegón E., Drake P. & Rodríguez A. First record of Palaemon macrodactylus Rathbun, 1902 (Decapoda, Caridea, Palaemonidae) from European waters. Crustaceana 77, 377–380 (2004). [Google Scholar]
- d'Udekem d'Acoz C., Faasse M., Dumoulin E. & De Blauwe H. Occurrence of the Asian shrimp Palaemon macrodactylus in the southern bight of the North Sea, with a key to the Palaemonidae of north-western Europe (Crustacea: Decapoda: Caridea). Nederlandse Faunistische Mededelingen 22, 95–111 (2005). [Google Scholar]
- González-Ortegón E., Cuesta J. A. & Schubart C. D. First report of the oriental shrimp Palaemon macrodactylus Rathbun, 1902 (Decapoda, Caridea, Palaemonidae) from German waters. Helgol. Mar. Res. 61, 67–69 (2007). [Google Scholar]
- Lavesque N. et al. Recent expansion of the oriental shrimp Palaemon macrodactylus (Crustacea: Decapoda) on the western coasts of France. Aquat. Invas. 5, S103–S108 (2010). [Google Scholar]
- Micu D. & Niţă V. First record of the Asian prawn Palaemon macrodactylus Rathbun, 1902 (Caridea: Palaemonoidea: Palaemonidae) from the Black Sea. Aquat. Invas. 4, 597–604 (2009). [Google Scholar]
- St.Raykov V., Lepage M. & Pérez-Domínguez R. First record of oriental shrimp, Palaemon macrodactylus Rathbun, 1902 in Varna Lake, Bulgaria. Aquat. Invas. 5, S91–S95 (2010). [Google Scholar]
- Worsford T. M. & Ashelby C. W. Additional UK records of the non-native prawn Palaemon macrodactylus (Crustacea: Decapoda). Mar. Biodiv. Rec. 1, e48 (2006). [Google Scholar]
- Warkentine B. E. & Rachlin J. W. Palaemon macrodactylus Rathbun 1902 (Oriental Shrimp) in New York: Status Revisited. Northeast. Nat. 19, 173–180 (2012). [Google Scholar]
- Lejeusne C. & Chevaldonné P. Brooding crustaceans in a highly fragmented habitat: the genetic structure of Mediterranean marine cave-dwelling mysid populations. Mol. Ecol. 15, 4123–4140 (2006). [DOI] [PubMed] [Google Scholar]
- Chevaldonné P., Rastorgueff P. A., Arslan D. & Lejeusne C. Molecular and distribution data on the poorly-known, elusive, cave mysid Harmelinella mariannae (Crustacea: Mysida). Mar. Ecol. (in press) [Google Scholar]
- Bock D. G., Zhan A., Lejeusne C., MacIsaac H. J. & Cristescu M. E. Looking at both sides of the invasion: patterns of colonization in the violet tunicate Botrylloides violaceus. Mol. Ecol. 20, 503–516 (2011). [DOI] [PubMed] [Google Scholar]
- Lejeusne C., Bock D. G., Therriault T. W., MacIsaac H. J. & Cristescu M. E. Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol. Invasions 13, 635–650 (2011). [Google Scholar]
- Reuschel S., Cuesta J. A. & Schubart C. D. Marine biogeographic boundaries and human introduction along the European coast revealed by phylogeography of the prawn Palaemon elegans. Mol. Phylogenet. Evol. 55, 765–775 (2010). [DOI] [PubMed] [Google Scholar]
- Mackie J. A., Darling J. A. & Geller J. B. Ecology of cryptic invasions: latitudinal segregation among Watersipora (Bryozoa) species. Sci. Rep. 2, 871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller J. B., Darling J. A. & Carlton J. T. Genetic Perspectives on Marine Biological Invasions. Annu. Rev. Mar. Sci. 2 (2010). [DOI] [PubMed] [Google Scholar]
- Botello A. & Alvarez F. Genetic variation in the stygobitic shrimp Creaseria morleyi (Decapoda: Palaemonidae), evidence of bottlenecks and re-invasions in the Yucatan Peninsula. Biol. J. Linn. Sco. 99, 315–325 (2010). [Google Scholar]
- Dennenmoser S., Thiel M. & Schubart C. D. High Genetic Variability With No Apparent Geographic Structuring in the mtDNA of the Amphidromous River Shrimp Cryphiops caementarius (Decapoda: Palaemonidae) in Northern-Central Chile. J. Crustacean Biol. 30, 762–766 (2010). [Google Scholar]
- Garcia-Merchan V. H. et al. Phylogeographic patterns of decapod crustaceans at the Atlantic-Mediterranean transition. Mol. Phylogenet. Evol. 62, 664–672 (2012). [DOI] [PubMed] [Google Scholar]
- Provan J., Murphy S. & Maggs C. A. Tracking the invasive history of the green alga Codium fragile ssp. tomentosoides. Mol. Ecol. 14, 189–194 (2005). [DOI] [PubMed] [Google Scholar]
- Dupont L., Viard F., Davis M., Nishikawa T. & Bishop J. Pathways of spread of the introduced ascidian Styela clava (Tunicata) in Northern Europe, as revealed by microsatellite markers. Biol. Invasions 12, 2707–2721 (2010). [Google Scholar]
- Cook E. J. et al. Distribution of the invasive bryozoan Tricellaria inopinata in Scotland and a review of its European expansion. Aquat. Invas. 8, 281–288 (2013). [Google Scholar]
- Johnson C. H., Winston J. E. & Woollacott R. M. Western Atlantic introduction and persistence of the marine bryozoan Tricellaria inopinata. Aquat. Invas. 7, 295–303 (2012). [Google Scholar]
- Kaluza P., Kolzsch A., Gastner M. T. & Blasius B. The complex network of global cargo ship movements. J. R. Soc. Interface 7, 1093–1103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebens H., Gastner M. T. & Blasius B. The risk of marine bioinvasion caused by global shipping. Ecol. Lett. 16, 782–790 (2013). [DOI] [PubMed] [Google Scholar]
- Warkentine B. E. & Rachlin J. W. The First Record of Palaemon macrodactylus (Oriental Shrimp) from the Eastern Coast of North America. Northeast. Nat. 17, 91–102 (2010). [Google Scholar]
- Aljanabi S. M. & Marinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25, 4692–4693 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske P. R., McQuaid C. D., Froneman P. W. & Barker N. P. Impacts of marine biogeographic boundaries on phylogeographic patterns of three South African estuarine crustaceans. Mar. Ecol.-Prog. Ser. 314, 283–293 (2006). [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sympos. Ser. 41, 95–98 (1999). [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X. & Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497 (2003). [DOI] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Excoffier L. & Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- Teacher A. G. F. & Griffiths D. J. HapStar: automated haplotype network layout and visualization. Mol. Ecol. Res. 11, 151–153 (2011). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Meth 9, 772–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
- Narum S. R. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv. Genet. 7, 783–787 (2006). [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026 (2008). [DOI] [PubMed] [Google Scholar]
- Gerlach G., Jueterbock A., Kraemer P., Deppermann J. & Harmand P. Calculations of population differentiation based on GST and D: forget GST but not all of statistics!. Mol. Ecol. 19, 3845–3852 (2010). [DOI] [PubMed] [Google Scholar]
- Oksanen J. et al. vegan: Community Ecology Package v. R package version 2.0-7. (2013)).
- Jensen J. L., Bohonak A. J. & Kelley S. T. Isolation by distance, web service. BMC Genet. 6, 6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A. D. & Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569 (1992). [DOI] [PubMed] [Google Scholar]
- Schneider S. & Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial. Genetics 152, 1079–1089 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S. E. & Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100 (2002). [DOI] [PubMed] [Google Scholar]