Abstract
RNA binding proteins can be important modulators of mRNA stability, a critical process that determines the ultimate cellular levels of mRNAs and their encoded proteins. The tristetraprolin or TTP family of RNA binding proteins appeared early in the evolution of eukaryotes, and has persisted in modern eukaryotes. The domain structures and biochemical functions of family members from widely divergent lineages are remarkably similar, but their mRNA “targets” can be quite different, even in closely related species. Recent gene knockout studies in species as distantly related as plants, flies, yeasts and mice have demonstrated crucial roles for these proteins in a wide variety of physiological processes. Inflammatory and hematopoietic phenotypes in mice have suggested potential therapeutic approaches for analogous human disorders.
Post-transcriptional regulation of mRNA stability is a critical aspect of gene expression. While the effects of physiological and pathological signals on cellular gene transcription are widely appreciated, the regulation of mRNA stability is less well understood, despite its crucial role in determining the ultimate levels of mRNA and protein in the cell. Recent work has identified many trans-acting proteins that bind directly to individual mRNAs and regulate their stability, usually after binding to relatively specific and often conserved binding domains in the 3′-untranslated regions (3′UTR) of target mRNAs [1-5].
One such family of mRNA binding proteins, which we will call the tristetraprolin or TTP family, belongs to this class of direct mRNA binding proteins. In general, these proteins bind directly to AU-rich elements in the 3′UTRs of their target mRNAs and promote the removal of the polyA tail, followed by complete mRNA decay. By destabilizing their target mRNAs, these proteins can modulate the effects of gene transcription to help maintain appropriate target transcript and protein levels as part of normal cell and tissue homeostasis [3, 6-10].
Four proteins of this family are expressed in the mouse, and human orthologues exist for three of the four. All four of the mouse genes have been knocked out at the “whole body” level, with remarkably different phenotypes; in several cases, cell- and tissue-specific knockouts have been developed as well [11-16]. Results of these studies have pointed to important physiological roles of these proteins in processes as divergent as innate immunity, establishment of the fetal circulation, hematopoiesis, and placental physiology.
Major questions remain concerning the specificity of action of these proteins, given their widespread expression and similar mechanisms of action. In addition, many aspects of their regulation remain to be deciphered, including the net effects on activity of changes in transcription, mRNA and protein decay, phosphorylation, nucleo-cytoplasmic shuttling, and interacting proteins. Recent overexpression and mutagenesis studies have suggested that these novel pathways might represent targets for therapeutic intervention [17-19]. These considerations suggested that it would be timely to summarize recent research on this interesting protein family.
Phylogenetic distribution of the TTP protein family
The defining characteristic of the TTP protein family is the tandem zinc finger (TZF) domain, the 64 amino acid region of the proteins that mediates direct binding to mRNA. This domain contains two similar zinc fingers of the CCCH class, with internal spacing within each finger of C-x8-C-x5-C-x3-H, with x representing various amino acids. Intra- and inter-finger spacing is constrained in this definition. Leading into each finger is a highly conserved sequence with the general sequence RYTKEL or a variant. An intact TZF domain is sufficient and necessary for high affinity binding to single stranded RNA; remarkably, mutation of a single cysteine or histidine in either finger is enough to completely prevent high-affinity mRNA binding. An NMR structure of the TZF domain from the human TTP family member TIS11D (ZFP36L2) bound to its preferred mRNA target sequence, UUAUUUAUU, showed that the RNA was unstructured in the complex, and the association between the protein and RNA was largely due to a combination of stacking interactions between some of the hydrophobic amino acids and the RNA bases, as well as hydrogen bonding interactions [20, 21].
Proteins containing TZF domains of this type have not been found in prokaryotes, but they have been found in all lineages of eukaryotes [22]. Their presence in eukaryotes ranging from mammals to plants indicates that one or more family members was present in a common ancestor well over a billion years ago. Similarly, their presence in flowering plants through red algae indicates that the TZF domain has remained essentially intact during the ∼ 1.4 billion years of evolution between these extremes (Fig. 1) [23]. Not all members of these lineages express proteins that fit this definition, however; for example, although proteins with intact, characteristic TZF domains are found in most fungi, most species from the most evolved group, the Pezizomycotina subdivision of the Ascomycota, express proteins containing related TZF-like domains that no longer fit the defining consensus [22].
Figure 1. TZF and NOT1 binding domains of TTP family members in eukaryotes.
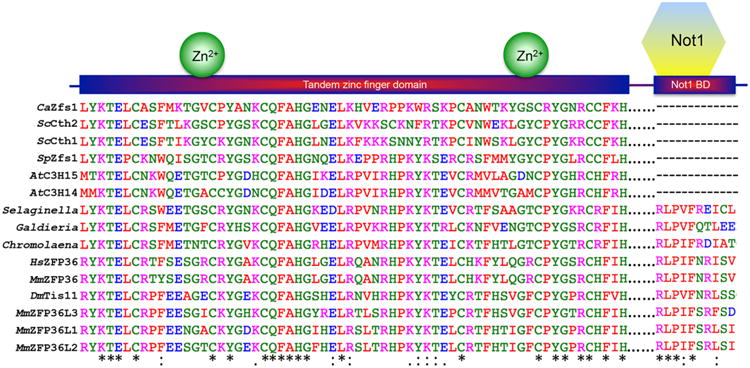
Shown are the amino acid sequences of the TZF domains and their respective predicted C-terminal NOT1 binding domains from TTP family members from the indicated species. NOT1 is CCR4-NOT transcription complex subunit 1, and probable orthologues have been identified in the species described in this figure, even those lacking an obvious C-terminal NOT1 binding domain. The TZF domains are separated by gaps of various sizes from the C-terminal sequences predicted to bind to NOT1. Sequences were aligned and colors were assigned by Clustal Omega based on their physicochemical properties. Asterisks (*) indicate amino acid identity at that site; colons (:) indicate a conserved substitution; and dots (.) indicate a semi-conserved substitution at that site. Abbreviations are as follows: Ca=Candida albicans, Sc=Saccharomyces cerevisiae, Sp=Schizosaccharomyces pombe, At=Arabidopsis thaliana, Sm=Selaginella moellendorffii, Galdieria=Galdieria sulphuraria, Chromolaena= Chromolaena odorata, Hs=Homo sapiens, Mm=Mus musculus, Ds=Drosophila melanogaster.
The predicted structures of the TZF domains from human TTP, the Zfs1 protein from the yeast Schizosaccharomyces pombe, and the family member expressed in the eudicot plant, Chromolaena odorata, exhibit very similar predicted structural organization in complex with the same 9-base RNA sequence, UUAUUUAUU (Fig. 2). Remarkably, replacement of the TZF domain in S. pombe Zfs1 with the TZF domain from either human TTP or the eudicot plant, C. odorata, completely complemented the abnormal flocculation and gene expression phenotypes seen in the Zfs1-deficient yeast (Fig. 3) [24]. These data suggest that there has been strong positive evolutionary pressure to maintain the RNA binding capabilities of the TZF domain over vast periods of evolution.
Figure 2. Comparison of solution structure simulations of the TZF domains from human TTP with the family members from S. pombe and C. odorata.
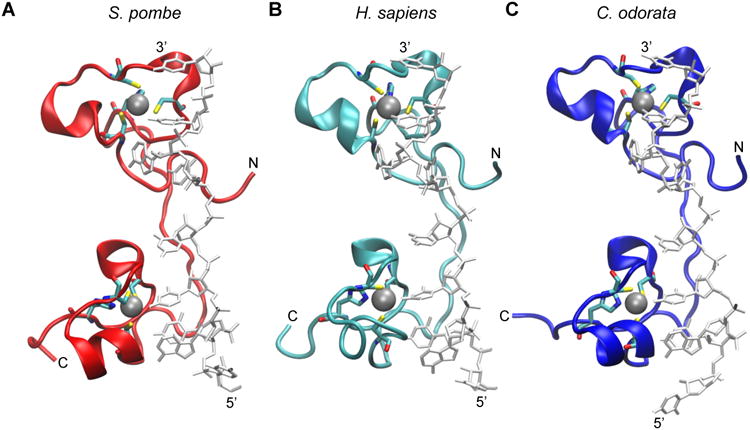
The RNA-bound structure of the TZF domain of ZFP36L2 (TIS11d) (PDB id: 1RGO) [20] was used to construct the models of RNA-bound TZF domains from: (A) S. pombe Zfs1 (GenBank accession number NP_596453); (B) human TTP (NP_003398.1); and (C) the TTP family member from C. odorata (translation of Unigene mRNA sequence GACH01022939.1), as described previously [72]. The peptide backbone of each TZF domain is represented in ribbon form, and the RNA is in stick representation in light grey. Zinc ions (silver spheres) are also shown with their coordinating residues (in stick form). Hydrogens were removed for clarity. The N and C termini of each peptide are indicated, as are the 5′ and 3′ ends of the bound RNA oligonucleotide.
Figure 3. Replacement of the S. pombe Zfs1 TZF domain with the TZF domains from human and plant.
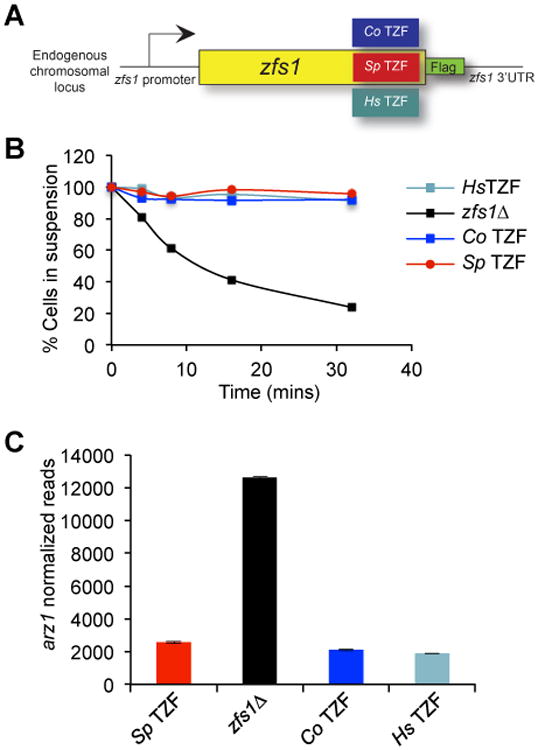
(A) The diagram shows a schematic representation of the construct used to replace the S. pombe Zfs1 TZF domain with the corresponding domains from H. sapiens and C odorata (Sp = S. pombe, Co = C odorata, Hs = H. sapiens) within the endogenous locus. A 3× Flag tag was also integrated into the locus encoding S. pombe Zfs1. (B) Flocculation analysis of the TZF domain replacement strains. Flocculation of the indicated strains was initiated by the addition of CaCl2 and determined using the Helm assay, as described previously [65]. The percentages of cells in suspension were measured based on optical density at 600 nm. Shown for each strain are the means of values from three independent experiments. (C) Shown is a quantitative analysis of the expression of the Zfs1 target transcript from arz1 in the strains expressing the S. pombe TZF domain in which the Zfs1 protein contained the epitope tag (Sp TZF); the Zfs1-deficient strain(zfs1Δ); and the C odorata (Co TZF) and H. sapiens (Hs TZF) TZF domain complementation strains. The y-axis represents normalized RNA counts for the arz1 transcript, shown as mean values from at least four independent isolates, +/- SD. Modified from [24], with permission.
Within eukaryotes, many representative species express family member proteins that also contain a conserved C-terminal domain (Fig. 1). This domain in human TTP was recently shown to interact with the scaffolding protein NOT1, potentially providing a link between TTP and the larger CCR4-NOT complex, which contains at least two 3′-5′ RNA exonucleases, or deadenylases [25-27]. In addition, the N-terminal region of TTP has been shown to interact directly with the CCR4 subunit of the CCR4-NOT complex [28]. These interactions provide a basis for one proposed mechanism of action of the TTP family proteins, i.e., initial binding to AU-rich elements in mRNA 3′UTRs, followed by recruitment of deadenylases to the neighboring polyA tail, and ultimate mRNA decay [25, 26, 28-30]. The presence of this domain at the C-termini of human TTP and the family member in red algae, as well as in virtually all metazoan family members, suggests that it was present and linked to the TZF domain in an ancient ancestor of most if not all eukaryotes.
Nonetheless, although linked TZF and NOT1 binding domains are found in many primitive plants, the C-terminal NOT1 binding domains are often absent in higher plants. For example, in Arabidopsis thaliana, its two TTP family member proteins, C3H14 and C3H15, terminate abruptly after their TZF domains (Fig. 1) [31]. Similarly, although some primitive fungi express TTP family member proteins that have linked TZF and NOT1 binding domains [22], the proteins from many more advanced fungi, including common laboratory species like S. cerevisiae and S. pombe, also terminate shortly after the TZF domains [32-35]. Nonetheless, the truncated proteins expressed in these yeasts are perfectly capable of promoting the decay of transcripts containing binding sequences. A comparison of the effects of mouse TTP deficiency to those of S. pombe Zfs1 deficiency on target transcript stability is illustrated in Fig. 4. The C-terminal truncated proteins found in yeast and higher plants may interact with some portion of the CCR4-NOT complex through a binding sequence elsewhere in the protein, or promote mRNA deadenylation and decay by interacting with other proteins and/or protein complexes. Outside of the TZF and NOT1 binding domains, there is very little apparent sequence conservation among the family members from different species, or even among different family members from a single species. Other critical sequence regions that are present in the mammalian proteins, and that may need to be accounted for in proteins from other species, include a leucine-rich nuclear export sequence, involved in nuclear-cytoplasmic shuttling [36, 37], and specific phosphorylated serine and threonine residues that are critical for the interaction of the mammalian proteins with 14-3-3- proteins, important for the regulation of protein activity in the cell [29, 38]. In the case of TTP, a conserved domain specific to TTP, the tetraproline (PPPPG) motif, has been implicated in the regulation of translation through its interaction with the translation repression complex, 4EHP-GYF2 [39]. This tetraproline motif has been shown to bind to the GYF2 subunit of 4EHP-GYF2. The loss of this interaction through mutation of the tetraproline motif, or deletion of 4EHP, resulted in increased expression and slower turnover of TTP target mRNAs.
Figure 4. Decay of target transcripts in mouse macrophages and S. pombe cells.
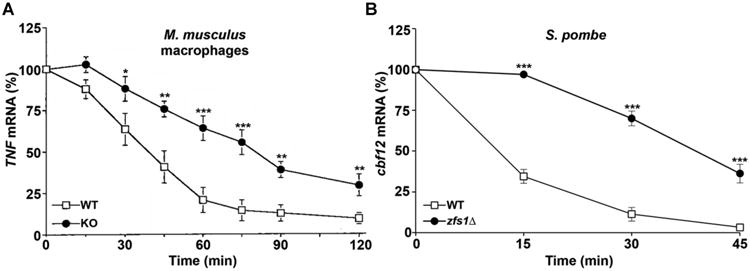
In (A), TNF mRNA decay was measured in macrophages from wild-type and TTP-deficient mice, as described [6], and in (B), Cbf12 mRNA decay was measured in S. pombe wild-type and zfs1Δ cells, as described [65]. mRNA was measured at the indicated times following treatment with the transcriptional inhibitor actinomycin D (A) or thiamine (B). The amount of the target mRNA at time 0 was taken as 100% in each experiment. Results are expressed as the mean relative amount of mRNA ± SEM at each time point (*P < 0.05 when comparing the means by Student's t test; **P < 0.01; ***P < 0.001). Modified from [6, 65], with permission.
Loss of function mutations
Mice
TTP, encoded by the Zfp36 gene in mice and ZFP36 in man, was initially discovered through its rapid induction in fibroblasts responding to insulin, serum or tumor-promoting phorbol esters [40-42]. Its function remained obscure until a knockout (KO) mouse was evaluated [14]. TTP KO mice developed a severe inflammatory syndrome with destructive arthritis, cachexia, myeloid hyperplasia, and autoimmunity [14]. Most aspects of this phenotype could be attributed to excessive activity of tumor necrosis factor alpha (TNF) [14, 43]. Macrophages derived from the KO mice overproduced TNF mRNA and protein in response to lipopolysaccharide [44]. The increases in TNF mRNA and protein were later shown to be consequences of increased TNF mRNA stability, and TTP was found to bind directly to conserved AU-rich elements in the 3′UTR of the TNF mRNA [6]. Studies from bone marrow-derived stromal cells from the same KO mice demonstrated that the transcript encoding granulocyte-macrophage colony-stimulating factor (GM-CSF) was also stabilized in the absence of TTP; in addition, the stabilized GM-CSF mRNA seen in the KO cells contained full-length polyA tails, in contrast to the cells with full complements of TTP, which contained a roughly 50:50 ratio of fully polyadenylated and deadenylated transcripts [7]. These and other studies directly linked TTP to the promotion of deadenylation, the rate-limiting step in mRNA decay in eukaryotes, and the subsequent accelerated decay of the mRNA.
These initial studies led to the basic model that has been used in subsequent loss of function studies in many organisms: In the absence of a TTP family member, its direct mRNA targets accumulate in cells, usually accompanied by increases in the encoded proteins. In the case of the original mouse KO for TTP, much of the phenotype could be explained by overexpression of TNF mRNA and its encoded protein; however, many other likely target mRNAs have been identified since then, some of which undoubtedly contribute to the original phenotype [3].
Orthologues of TTP have been found in virtually all vertebrates, including man, with the single exception of birds [22, 45]. It is apparently absent in all bird lineages, despite its presence in current reptiles. At some point early in bird evolution, the chromosomal locus containing the ZFP36 gene was probably disrupted. An interesting question that may never be answered is whether TTP was expressed in dinosaurs; however, its presence in modern reptiles, including alligators, suggests that it was present in a common ancestor of birds, dinosaurs, and modern reptiles.
In the mouse, three other family members are expressed in addition to TTP, and all but one are widely expressed. In cell co-transfection studies, all four family members seem to behave similarly, in terms of RNA binding, and promotion of mRNA deadenylation and decay promotion [46, 52]. However, the phenotypes of mice deficient in each of the family members are remarkably different.
Zinc finger protein 36-like 1 (ZFP36L1; also known as TIS11b, cMG1, BRF1, and ERF1) is the most closely related by amino acid sequence to the single protein expressed in close invertebrate relatives of vertebrates, such as Ciona instestinalis, and it is likely that most of the other family members in modern vertebrates are derived from this original ancestor by genome duplication. Disruption of Zfp36l1 results in lethality by embryonic day 11 (E11), apparently due to failure of fusion of the allantois with the chorion, leading in turn to absence of the umbilical circulation [13]. Zfp36l1 mRNA was absent in the chorionic plate, but was highly expressed in the developing allantois. A candidate target transcript is that encoding vascular endothelial growth factor (Vegf-a) [47]. This transcript contains multiple potential TTP family member binding sites, has been shown to bind directly to ZFP36L1 [48], and its translation was enhanced in Zfp36l1 knockout mice [47].
The third family member in mice and humans is zinc finger protein 36-like 2 (ZFP36L2; also known as TIS11D, BRF2, or ERF2). An initial loss of function mutant resulted in decreased expression of a protein lacking the N-terminal 29 amino acids, but still containing the TZF domain [49]. These mice appeared normal but had complete female infertility, resulting in embryonic failure at approximately the two-cell stage. Further studies with these mutant mice suggested that ZFP36L2 could bind directly to the mRNA encoding luteinizing hormone receptor (LHR), which is necessary for hormonal functioning during reproduction [50].
In contrast, mice with a complete KO for Zfp36l2 died within several weeks of birth [12]. These mice exhibited marked decreases in all hematopoietic cell lineages, with death likely due to the observed thrombocytopenia and hemorrhage. There were decreases in the numbers of hematopoietic progenitor cells derived from both fetal liver and yolk sac, suggesting that ZFP36L2 plays an important role in hematopoietic stem cell function during mouse development and later in life.
ZFP36L3 is only expressed in certain species of rodents. It is thought to have arisen in an ancestor of these species about 25-30 million years ago through a process of retrotransposition involving Zfp36l2 [51]. It is an intronless, imprinted X-chromosome gene whose expression is limited to placenta and yolk sac in the mouse [52]. It is the only mouse family member that appears to reside permanently in the cytoplasm, in contrast to the nucleo-cytoplasmic shuttling behavior of the other proteins. A complete KO of Zfp36l3 in mice resulted in decreased neonatal survival rates [11], as well as the accumulation of many transcripts that were stabilized in studies of trophoblastic stem cells derived from the KO mice, many of which are important for placental function.
Although ZFP36L3 is not found outside of these rodent species, it will be important to determine whether any of the other family members have “taken over” its placental functions in man and other mammals.
Recent studies have begun knocking out individual genes and combinations of genes in whole mice and in selected cell types. For example, TTP has been knocked out specifically in myeloid cells [15, 53]. Although these animals appeared healthy, they were hypersensitive to low doses of LPS, which caused the development of lethal endotoxin shock and huge elevations in serum TNF concentrations. These results suggested that cells other than myeloid cells must contribute to the complete syndrome, an idea that is supported by subsequent examination of cytokines produced by fibroblasts [54] and dendritic cells [55].
An interesting question concerns the specificity of the KO phenotypes, since all the mammalian members of the family behave similarly in biochemical assays of RNA binding, deadenylation, and mRNA decay. Single and double conditional knockouts in lymphocytes have recently been created [56]. Thymic development was normal in the single knockouts; however double knockout mice had perturbed thymic development and T-lymphoblastic leukemia. Furthermore, double knockout mice exhibited elevations of the mRNA encoding the oncogenic transcription factor Notch1. Both ZFP36L1 and ZFP36L2 were shown to physically interact with the optimal binding sequence present in the Notch1 mRNA 3′UTR and suppress its expression. Later studies of Zfp36l1 and Zfp36l2 knockouts in pro-B cells by the same group revealed a critical role in B cell development [57]. ZFP36L1 and ZFP36L2 together were crucial for maintaining cell quiescence in order for proper B cell progression through the cell cycle [57].
Drosophila melanogaster
Because of the complexity introduced by having multiple, possibly redundant proteins in vertebrates, loss of function studies have also been conducted in organisms that have fewer expressed family member proteins. For example, in the fruit fly Drosophila melanogaster, as well as in other insects examined, only a single-family member is expressed, termed Tis11 [58]. Studies in cultured Drosophila S2 cells in which Tis11 was knocked down revealed a large number of stabilized transcripts, many of which contained potential binding sites [59]. Transfection of cultured mammalian cells with cDNAs expressing normal and mutant Drosophila Tis11 showed that the fly protein could bind to the optimal mammalian binding site on RNA, and could promote mRNA deadenylation and decay in a manner almost identical to human TTP [60]. As shown before in transfection experiments with human TTP, the activity of the fly protein was partially dependent on the presence of the conserved C-terminal NOT1 binding domain. Knock-down and KO studies in adult flies demonstrated the accumulation of many potential targets, but there was almost no overlap between the transcripts accumulating in the fly and in the S2 cells [60]. It will be interesting to determine whether the putative targets identified in S2 cells will correspond more closely to flies at a different developmental stage.
Yeasts
In yeast, TTP family members have been studied in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and the human pathogens Candida albicans and Candida glabrata. In all four organisms, the TTP family members lack obvious C-terminal NOT1 binding domains. S. cerevisiae expresses two TTP family members, Cth1 and Cth2 [32, 33]. S. cerevisiae uses these proteins to regulate a set of transcripts involved in iron homeostasis, as determined by studies in which one or both family members were deleted [61]. Similar to S. cerevisiae, the sole TTP family member expressed in C. glabrata, Cth2, plays an essential role in the downregulation of iron homeostasis associated transcripts [62]. S. pombe, like most other yeasts, expresses a single protein of this type, named Zfs1 [34]. Many Zfs1 target mRNAs identified in this species encoded cell surface glycoproteins known to be involved in cell-cell adhesion; indeed, the Zfs1-deficient cells exhibited abnormal cell-cell interactions leading to flocculation, a reversible condition in which cells aggregate spontaneously and form “flocs” [63]. The increased flocculation observed in zfs1Δ mutants is due, at least in part, to a direct effect on the turnover of cbf12 mRNA, which encodes a transcription factor that regulates expression of several transcripts encoding adhesion proteins [64, 65]. These data suggested that a major physiological function of Zfs1 in S. pombe is to regulate the levels of cell surface proteins involved in cell-cell interactions.
The human pathogenic fungus C. albicans expresses a single, considerably shorter TTP family member, also termed Zfs1, and deficiency mutants exhibited accumulation of many potential target transcripts [35]. However, unlike Cth1 and Cth2 in S. cerevisiae, Zfs1 does not seem to play a role in iron metabolism in either S. pombe or C. albicans, and Zfs1 does not seem to be important for cell-cell adhesion in C. albicans.
Plants
Plants from red algae through eudicots express at least one TTP family member. However, in Arabidopsis thaliana, as in most eudicots examined, an obvious C-terminal NOT1 binding domain is not present in the two family members expressed in this species, C3H14 and C3H15 (Fig. 1). Deletion of both C3H14 and C3H15 resulted in plants with thinner stem secondary walls, along with the upregulation of several key genes involved in secondary wall biosynthesis [66].
TTP family member target specificity
One of the striking findings from the mouse KO models of the four individual genes was the “non-overlap” of the phenotypes. Although direct TTP targets are becoming better understood, the full spectra of targets for the other proteins in different cells and tissues under different experimental conditions remain to be determined. We have estimated that the prevalence in transcriptomes of one or more instances of the minimal optimal 7-mer binding site, UAUUUAU, is approximately 22% in S. pombe, 17% in C. albicans, 27% in Drosophila, 23% in mice, and 26% in humans [35, 60, 65]. However, in the organisms in which gene knockout experiments have been performed, there are many fewer accumulating transcripts that contain such AU-rich binding elements than the total number of 7-mer-containing mRNAs in that organism. This is true for organisms in which only a single protein is expressed; even more complicated is the question of overlapping target sequences when several proteins are expressed, as in mammals, where three or four expressed family member proteins can inhabit the same cell type. What accounts for target selection and specificity?
Several factors may be at play. As demonstrated in the original structure of a TTP family member TZF domain bound to its optimum UUAUUUAUU target sequence, and illustrated in Fig. 2, no secondary structure was present in the RNA in the complex [20]. One possible contributor to target selectivity could therefore be the presence or absence of mRNA secondary structure at the specific binding site. Another possibility could involve inhibition of TTP binding to the ideal UUAUUUAUU target sequence by other ARE binding proteins, such as HuR [67-69]. Other factors could include subcellular localization of the mRNA; stoichiometry of the protein:mRNA ratios; phosphorylation or other modification of TTP family member proteins, or protein-protein interactions that might affect RNA binding; subcellular localization of TTP family members in restricted sites; and others.
One way to approach the question of mRNA target specificity is to use a species that expresses only a single TTP family member and compare the apparent mRNA targets identified by gene disruption studies with the orthologous mRNA from closely related species. In S. pombe, we compared seven well-validated Zfs1 target transcripts to the orthologous transcripts from the three other sequenced species in the Schizosaccharomyces genus: S. japonicus, S. octosporus, and S. cryophilus [70]. All four species express Zfs1, with high level conservation of the TZF domains. Somewhat surprisingly, we found relatively little target conservation among these related species [65].
In a similar experiment, validated Zfs1 targets from the human pathogen C. albicans were compared to their orthologous sequences from other members of the “CTG clade”, a group of related species that translate CUG as serine instead of leucine [35, 71]. All of these species expressed Zfs1 proteins that were closely related to the C. albicans protein [35]. When we examined three of the validated Zfs1 target transcripts from C. albicans in two closely related species, C. dubliniensis and C. tropicalis, we found that the number of potential 7-mer binding sites was markedly lower in the orthologues from C. dubliniensis than in C. albicans (Fig. 5). Two of the three orthologous transcripts from C. tropicalis contained no binding sites; this is remarkable because both C. dubliniensis and C. albicans are thought to be descended from either C. tropicalis or a closely related species as a common ancestor.
Figure 5. Evolutionary relationships of Zfs1 targets within members of the CTG clade.
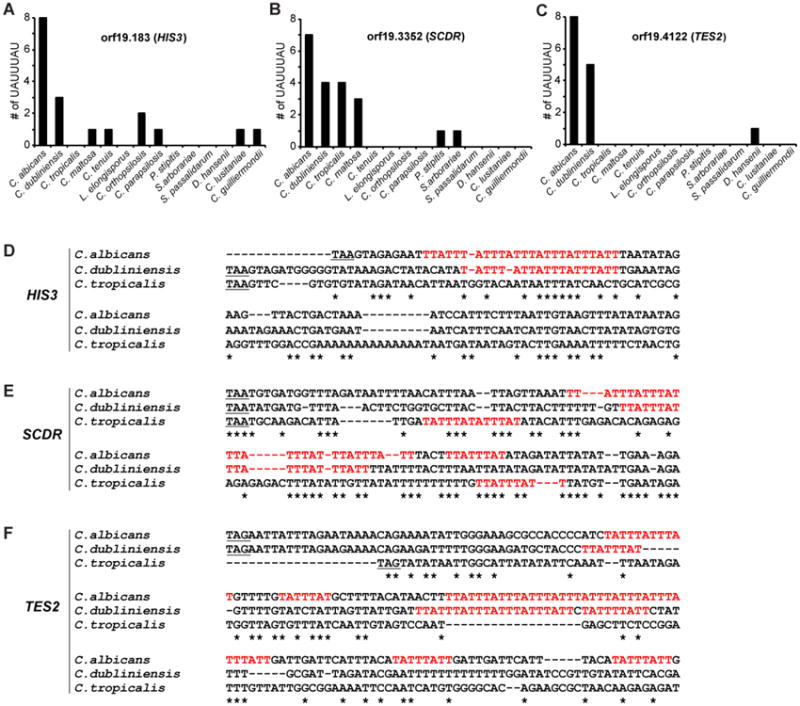
Shown are the numbers of potential 7-mer binding sites within 500 base pairs of the stop codons in transcripts from the orthologues of C. albicans HIS3 (A; orf19.183), SCDR (B; orf19.3352), and TES2 (C; orf19.4122). The order of species along the X-axis roughly reflects increasing evolutionary distance of the species from C. albicans, based on previous phylogenetic comparisons. (D-F) Alignments performed with Clustal Omega of the sequences of the 3′UTRs of (D) HIS3, (E) SCDR, and (F) TES2 from the indicated CTG clade species are shown, including the potential binding 7-mers in red. The alignment begins at the stop codon (underlined) and continues at least 60 bases after the 3′-most 7-mer identified in any of the species. Asterisks (*) indicate sequence identity. Reproduced from [35], with permission.
Furthermore, when we examined the orthologous sequences of the validated transcripts from C. albicans in more distantly related members of the CTG clade, we found that the number of 7-mer binding sites rapidly decreased with increasing evolutionary distance from C. albicans (Fig. 5).
Similar results were obtained when 3′UTR sequences of putative Tis11 target transcripts were examined among many species from the Drosophila genus [60]. The conclusion that emerges from these studies is that, while TTP family member protein sequences are expressed and conserved in closely related species, at least within the TZF domains, the mRNAs that they bind to and act upon are often very different. It appears that evolutionary pressures have “directed” each species to come up with its own panel of TTP family member targets, possibly as part of the whole process of speciation.
Concluding Remarks
The phylogenetic data summarized here suggest that one or more TTP family member proteins was first expressed in an early eukaryote more than 1.5 billion years ago. Comparative studies of TZF domains from eukaryotes as divergent as plants, yeasts and mammals have shown that the high affinity binding of the proteins to their ideal target sequence, UUAUUUAUU, has been maintained in many descendants of that ancestral eukaryote. In addition, the ability of these family members to promote decay of transcripts containing binding sites has been preserved in species ranging from plants to mammals, despite the absence of a conserved C-terminal NOT1 binding domain in most fungi and eudicot plants. Despite the requirement of a version of the AU-rich sequence shown above for TTP family member binding to RNA, the presence of such a sequence in the 3′UTR of transcripts is not sufficient to guarantee their behavior as targets. How each organism, and, in some cases, cell type within an organism, has developed its own personal sets of target transcripts, and how these targets are distinguished among the much larger number of transcripts containing potential binding sequences in the cell, is an interesting question for future study.
Although the evolutionary history of this protein family is useful in unraveling their molecular mechanisms of action, the accumulated data from mouse knockouts point to their major importance in the physiology of mammals, including humans. This information will be useful in understanding the effects of potential loss of function mutations or other disease-modifying variants in man. Perhaps of more widespread importance, the discovery of the physiological roles played by these proteins in mammalian physiology suggests the possibility of harnessing these novel pathways in the development of new therapies for conditions as diverse as disorders of human immunity and inflammation, hematopoiesis, fetal development, and placental function.
Outstanding questions.
The transcriptomes surveyed contain approximately 20% of transcripts that have at least one UAUUUAU minimal TTP family member binding site in their 3′-untranslated regions, yet the number of apparent “targets” for the TTP family proteins expressed in those species is much lower. What determines whether a given AU-rich element containing mRNA will be a true target?
In organisms such as mammals that express more than one TTP family member in the same cells, what factors determine which protein will exert the dominant effect on mRNA stability? In cells that express more than one such protein, will other family members compensate for the deficiency of one or more of the others?
In the absence of apparent C-terminal NOT1 binding domains, how do yeasts and higher plant TTP family members stimulate mRNA deadenylation and decay? Similarly, in proteins that normally contain linked tandem zinc finger domains and C-terminal Not1 binding domains, is there residual activity in proteins lacking the C-terminal domain?
What are the similarities and differences in the mechanisms of action of TTP family member proteins from lineages as diverse as mammals, protists and plants?
What are the accessory proteins that TTP family members interact with to mediate target specificity and/or mRNA target decay?
How do birds regulate the decay of AU-rich element-containing cytokine mRNAs in the apparent absence of TTP?
Since ZFP36L3 is expressed only in the placenta and yolk sac of a subset of rodents, do other members of the protein family take over its functions in non-rodent mammals?
What are the post-translational modifications that are crucial for family member activity?
Given the severity of the four knockout phenotypes in seen in mice, can we use the pathways identified in these studies to design novel therapeutic approaches for the relevant diseases, such as chronic immune and inflammatory disorders and defects in hematopoiesis?
Trends.
Regulation of gene expression occurs at several stages, from transcriptional initiation to mRNA translation. Post-transcriptional control of gene expression by RNA binding proteins plays an essential role in this process.
Tristetraprolin (TTP) family members are defined by tandem CCCH zinc finger domains that bind to AU-rich elements found in the 3′untranslated regions of target mRNAs, and stimulate their turnover.
TTP family members from evolutionarily distant species also contain a conserved C-terminal sequence that, in human TTP, can bind to the CCR4-NOT deadenylase complex through an interaction with the NOT1 subunit, providing a potential mechanism for the action of these proteins to destabilize mRNA.
Gene knockout studies from organisms as distant as plants and mice demonstrate that these proteins regulate important physiological pathways that can differ among species.
Acknowledgments
We thank the members of our laboratories for helpful discussions, and Drs. Jessica Williams and Dori Germolec for comments on the manuscript. Supported in part by the Intramural Research Program of the NIEHS, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman R, et al. RNA binding proteins as regulators of immune cell biology. Clin Exp Immunol. 2016;183:37–49. doi: 10.1111/cei.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Ortin JE, et al. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White EJ, et al. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reznik B, Lykke-Andersen J. Regulated and quality-control mRNA turnover pathways in eukaryotes. Biochem Soc Trans. 2010;38:1506–1510. doi: 10.1042/BST0381506. [DOI] [PubMed] [Google Scholar]
- 6.Carballo E, et al. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 7.Carballo E, et al. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 8.Stoecklin G, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaba A, et al. Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J Immunol. 2012;189:2089–2093. doi: 10.4049/jimmunol.1201126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao W, et al. Tristetraprolin regulates interleukin-6 expression through p38 MAPK-dependent affinity changes with mRNA 3′ untranslated region. J Interferon Cytokine Res. 2011;31:629–637. doi: 10.1089/jir.2010.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stumpo DJ, et al. Deficiency of the placenta- and yolk sac-specific tristetraprolin family member ZFP36L3 identifies likely mRNA targets and an unexpected link to placental iron metabolism. Development. 2016;143:1424–1433. doi: 10.1242/dev.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stumpo DJ, et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stumpo DJ, et al. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol Cell Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 15.Qiu LQ, et al. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. J Immunol. 2012;188:5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan IM, et al. Deletion of tristetraprolin caused spontaneous reactive granulopoiesis by a non-cell-autonomous mechanism without disturbing long-term hematopoietic stem cell quiescence. J Immunol. 2011;186:2826–2834. doi: 10.4049/jimmunol.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patial S, et al. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc Natl Acad Sci U S A. 2016;113:1865–1870. doi: 10.1073/pnas.1519906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smallie T, et al. Dual-Specificity Phosphatase 1 and Tristetraprolin Cooperate To Regulate Macrophage Responses to Lipopolysaccharide. J Immunol. 2015;195:277–288. doi: 10.4049/jimmunol.1402830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross EA, et al. Dominant Suppression of Inflammation via Targeted Mutation of the mRNA Destabilizing Protein Tristetraprolin. J Immunol. 2015;195:265–276. doi: 10.4049/jimmunol.1402826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson BP, et al. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 21.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Blackshear PJ, Perera L. Phylogenetic distribution and evolution of the linked RNA-binding and NOT1-binding domains in the tristetraprolin family of tandem CCCH zinc finger proteins. J Interferon Cytokine Res. 2014;34:297–306. doi: 10.1089/jir.2013.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pires ND, Dolan L. Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond B Biol Sci. 2012;367:508–518. doi: 10.1098/rstb.2011.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells ML, et al. Functional equivalence of an evolutionarily conserved RNA binding module. J Biol Chem. 2015;290:24413–24423. doi: 10.1074/jbc.M115.673012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabian MR, et al. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandler H, et al. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basquin J, et al. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell. 2012;48:207–218. doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clement SL, et al. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchese FP, et al. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, et al. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics. 2008;9:44. doi: 10.1186/1471-2164-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Q, Herschman HR. The yeast homologue YTIS11, of the mammalian TIS11 gene family is a non-essential, glucose repressible gene. Oncogene. 1995;10:487–494. [PubMed] [Google Scholar]
- 33.Thompson MJ, et al. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene. 1996;174:225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- 34.Kanoh J, et al. Schizosaccharomyces pombe zfs1+ encoding a zinc-finger protein functions in the mating pheromone recognition pathway. Mol Biol Cell. 1995;6:1185–1195. doi: 10.1091/mbc.6.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells ML, et al. Post-transcriptional regulation of transcript abundance by a conserved member of the tristetraprolin family in Candida albicans. Mol Microbiol. 2015;95:1036–1053. doi: 10.1111/mmi.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips RS, et al. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J Biol Chem. 2002;277:11606–11613. doi: 10.1074/jbc.M111457200. [DOI] [PubMed] [Google Scholar]
- 37.Twyffels L, et al. A masked PY-NLS in Drosophila TIS11 and its mammalian homolog tristetraprolin. PLoS One. 2013;8:e71686. doi: 10.1371/journal.pone.0071686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoecklin G, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu R, et al. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016;22:373–382. doi: 10.1261/rna.054833.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai WS, et al. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 41.DuBois RN, et al. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- 42.Ma Q, Herschman HR. A corrected sequence for the predicted protein from the mitogen-inducible TIS11 primary response gene. Oncogene. 1991;6:1277–1278. [PubMed] [Google Scholar]
- 43.Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98:2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- 44.Carballo E, et al. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (-/-) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J Clin Invest. 1997;100:986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai WS, et al. Life without TTP: Apparent absence of an important antiinflammatory protein in birds. Am J physiol Regul Integr comp Physiol. 2013;305:R689–R700. doi: 10.1152/ajpregu.00310.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai WS, et al. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell SE, et al. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 48.Ciais D, et al. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 49.Ramos SB, et al. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 50.Ball CB, et al. The RNA-binding protein, ZFP36L2, influences ovulation and oocyte maturation. PLoS One. 2014;9:e97324. doi: 10.1371/journal.pone.0097324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gingerich TJ, et al. Emergence and evolution of Zfp36l3. Mol Phylogenet Evol. 2016;94:518–530. doi: 10.1016/j.ympev.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackshear PJ, et al. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 53.Kratochvill F, et al. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol. 2011;7:560. doi: 10.1038/msb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu LQ, et al. Tristetraprolin (TTP) coordinately regulates primary and secondary cellular responses to proinflammatory stimuli. J Leukoc Biol. 2015;97:723–736. doi: 10.1189/jlb.3A0214-106R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molle C, et al. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. J Exp Med. 2013;210:1675–1684. doi: 10.1084/jem.20120707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodson DJ, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galloway A, et al. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science. 2016;352:453–459. doi: 10.1126/science.aad5978. [DOI] [PubMed] [Google Scholar]
- 58.Ma Q, et al. The Drosophila TIS11 homologue encodes a developmentally controlled gene. Oncogene. 1994;9:3329–3334. [PubMed] [Google Scholar]
- 59.Spasic M, et al. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS genetics. 2012;8:e1002433. doi: 10.1371/journal.pgen.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi YJ, et al. The Drosophila Tis11 protein and its effects on mRNA expression in flies. J Biol Chem. 2014;289:35042–35060. doi: 10.1074/jbc.M114.593491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puig S, et al. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 62.Gerwien F, et al. A Novel Hybrid Iron Regulation Network Combines Features from Pathogenic and Nonpathogenic Yeasts. MBio. 2016;7 doi: 10.1128/mBio.01782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soares EV. Flocculation in Saccharomyces cerevisiae: a review. J Appl Microbiol. 110:1–18. doi: 10.1111/j.1365-2672.2010.04897.x. [DOI] [PubMed] [Google Scholar]
- 64.Prevorovsky M, et al. Cbf11 and Cbf12, the fission yeast CSL proteins, play opposing roles in cell adhesion and coordination of cell and nuclear division. Exp Cell Res. 2009;315:1533–1547. doi: 10.1016/j.yexcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Wells ML, et al. Posttranscriptional regulation of cell-cell interaction protein-encoding transcripts by Zfs1p in Schizosaccharomyces pombe. Mol Cell Biol. 2012;32:4206–4214. doi: 10.1128/MCB.00325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chai G, et al. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J Exp Bot. 2015;66:2595–2609. doi: 10.1093/jxb/erv060. [DOI] [PubMed] [Google Scholar]
- 67.Tiedje C, et al. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukherjee N, et al. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014;15:R12. doi: 10.1186/gb-2014-15-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sedlyarov V, et al. Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol Syst Biol. 2016;12:868. doi: 10.15252/msb.20156628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santos MA, et al. The genetic code of the fungal CTG clade. C R Biol. 2011;334:607–611. doi: 10.1016/j.crvi.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 72.Lai WS, et al. Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin. J Biol Chem. 2014;289:565–580. doi: 10.1074/jbc.M113.466326. [DOI] [PMC free article] [PubMed] [Google Scholar]


