Abstract
Crystallographic analysis of macromolecules depends on large, well-ordered crystals, which often require significant effort to obtain. Even sizable crystals sometimes suffer from pathologies that render them inappropriate for high-resolution structure determination. Here we show that fragmentation of large, imperfect crystals can provide a simple path for high-resolution structure determination by serial femtosecond crystallography or the cryoEM method MicroED.
Introduction
Large and perfect crystals are desirable for structure determination because they yield a strong signal over background. In reality, crystals of biological material are not perfect. In the mosaic model, a real, imperfect crystal is composed of several small but well-ordered blocks1. These mosaic blocks have a finite size, are misaligned with respect to each other, and may be composed of unit cells with different dimensions2. Depending on the nature and degree of disorder between the mosaic blocks, an imperfect crystal may exhibit a plethora of pathologies, which may hamper subsequent data reduction, limit the resolution of the final model, and can prevent structure determination altogether (Supplementary figures 1–9). Many such defects primarily affect larger crystals, and small crystals may yield superior data quality where diffraction is not limited by the number of diffracting unit cells in the crystal3.
For these reasons, methods such as serial femtosecond crystallography (SFX)4 at an X-ray free-electron laser (XFEL) and the electron cryomicroscopy (cryoEM) method microelectron diffraction (MicroED)5 are actively being developed. These methods can yield structures to resolutions better than 1 Å from crystals that are significantly smaller than those which are required for standard crystallography: ~10,000× smaller in volume for XFEL and ~1,000,000× smaller for MicroED. In fact, in both these techniques large crystals can prove problematic. This is because large crystals can clog up the nozzle of certain SFX sample delivery systems6,7, while in MicroED the large electron scattering cross-section implies that absorption extinguishes diffraction when the sample is too thick8.
Here we show that sonication, vigorous pipetting, or vortexing can be used to break large imperfect crystals into small, single-crystal fragments that are suitable for data collection and atomic structure determination (Figure 2). Delicate samples may benefit from gentler fragmentation by vortexing, while harsher methods such as pipetting and ultimately sonication are required to break more robust crystals. In many cases, the untreated crystals were mosaic, yielded diffraction patterns with multiple lattices, or were otherwise not suitable for standard crystallographic experiments (Figure 2, Supplementary figures 1–9). In the case of the amyloid-forming peptide of tau (VQIVYK), what appeared to be large crystals were in fact crystal bundles that produced low resolution powder-like diffraction. These problems are traditionally overcome by modifying the crystallization conditions to optimize crystal growth and quality a process that can be tedious and labor intensive, particularly when crystal pathologies do not become apparent until data processing. Breaking of large crystals by physical means produced fragments that appeared crystallographically homogeneous and yielded diffraction data at atomic resolution, void of the above artifacts. This was true even without further optimization of the growth conditions for crystals formed by various macromolecules of molecular weights between 0.7 kDa and 34.6 kDa, and solvent contents ranging from 30% to 60% with a range of unit cell dimensions and space groups (Table 1). MicroED has already allowed rapid structure solution from several peptide fragments that resisted solution using microfocus synchrotron sources in spite of many months of crystal optimization (Supplementary figures 2–4), and the approach could be used for samples where large crystals exist but standard crystallographic methods fail, owing to crystal imperfections.
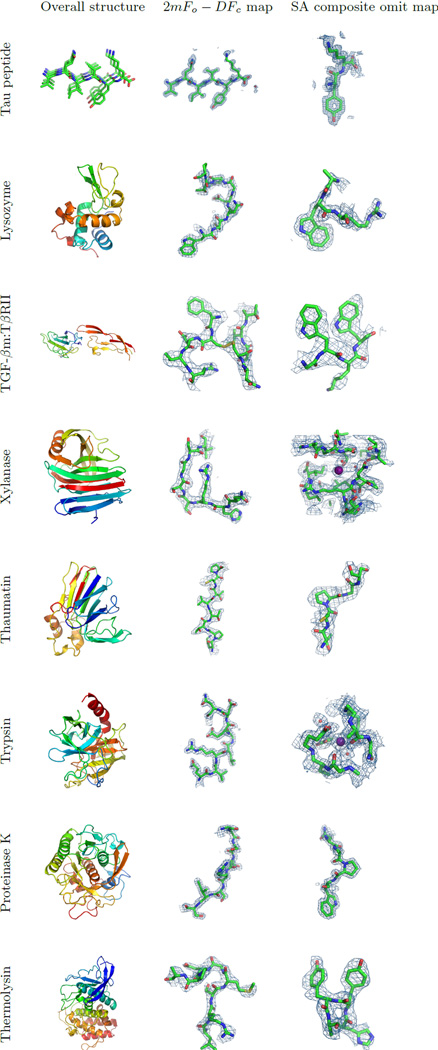
Figure 2. Eight atomic resolution structures determined by MicroED from crystal fragments.
Left column: ribbon representations of the corresponding macromolecules. The tau peptide model indicates a β-sheet generated by crystal packing. Middle column: 2mFo-DFc charge density maps contoured at 1.5 σ above the mean, showing eight residues in loop regions of the respective structures, where m is the figure of merit and D is derived from coordinate error estimates. For TGF-βm:TβRII one of the disulfide bonds is shown. Right column: SA composite omit maps contoured at 1 σ above the mean, except for the maps from lysozyme and proteinase K, which are contoured at 1.5 σ above the mean. Depressions or holes can be observed in the density of side chains of aromatic residues for tau peptide and lysozyme, and for proline residues in the proteinase K structure. Iodide and calcium ions are visible in the omit maps for xylanase and trypsin, respectively. All figures were generated using PyMol19.
Table 1. Data processing and model refinement statistics for the reported crystal structures.
Data sets were collected and processed as described in the online methods section. The dose rate did not exceed 0.01 e−/Å2/s and the mean per-crystal exposure time is given as 〈Texposure〉 for each sample. Except for tau peptide and TGF-βm:TβRII, reflections were integrated to the corners of the detector; the final resolution cutoff was determined based on CC1/2 and the stability of the refinement procedure.
| Tau peptide (PDB id: 5k7n; EMDB id: EMD- 8216) |
Lysozyme (PDB id: 5k7o; EMBD id: EMD-8217) |
TGF-βm:TβRII (PDB id: 5ty4; EMDB id: EMD- 8472) |
Xylanase (PDB id: 5k7p; EMBD id: EMD-8218) |
Thaumatin (PDB id: 5k7q; EMBD id: EMD-8219) |
Trypsin (PDB id: 5k7r; EMBD id: EMD-8220) |
Proteinase K (PDB id: 5k7s; EMBD id: EMD- 8221) |
Thermolysin (PDB id: 5k7t; EMBD id: EMD-8222) |
|
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Resolution1 (Å) | 14.70–1.10 | 30.58–1.50 | 26.64–2.90 | 25.55–1.90 | 27.73–2.11 | 27.63–1.50 | 20.75–1.30 | 30.14–1.60 |
| Number of crystals | 2 | 7 | 3 | 4 | 3 | 10 | 6 | 4 |
| 〈Texposure〉 (s) | 159.9 | 127.7 | 140.8 | 172.7 | 179.7 | 155.8 | 122.2 | 187.6 |
| Molecular weight (kDa) |
0.7 | 14.4 | 19.1 | 21.0 | 22.2 | 23.4 | 28.9 | 34.6 |
| Data processing | ||||||||
| Resolution1 (Å) | 14.70–1.10 (1.23–1.10) |
30.59–1.80 (2.06–1.80) |
26.64–2.90 (3.07–2.90) |
25.55–2.30 (2.63–2.30) |
27.73–2.50 (2.86–2.50) |
25.86–1.70 (1.79–1.70) |
20.75–1.60 (1.64–1.60) |
30.14–2.50 (2.75–2.50) |
| Space group | C121 | P43212 | P212121 | P212121 | P41212 | P212121 | P43212 | P6122 |
| Unit cell | ||||||||
| a, b, c (Å) | 29.42, 4.99, 37.17 |
76.10, 76.10, 36.80 |
41.53, 71.33, 79.51 |
49.10, 59.02, 70.00 |
57.78, 57.78, 149.70 |
53.12, 56.08, 64.38 |
67.60, 67.60, 101.36 |
90.75, 90.75, 126.13 |
| α, β, γ (°) | 90, 111.55, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| # total reflections1 | 6,185 (463) | 100,693 (282) | 14,911 (2,371) | 38,699 (348) | 51,116 (1,563) | 145,833 (402) | 302,892 (2,044) | 224,846 (314) |
| # unique reflections1 |
3,319 (255) | 14,955 (207) | 3,884 (614) | 10,664 (214) | 12,786 (614) | 23,542 (281) | 46,369 (1,005) | 25,029 (173) |
| CC1/21 | 0.987 (0.639) | 0.901 (0.099) | 0.951 (0.255) | 0.918 (0.201) | 0.848 (0.089) | 0.722 (0.028) | 0.912 (0.056) | 0.847 (0.199) |
| 〈I/σI〉1 | 2.4 (1.1) | 3.7 (1.1) | 3.3 (0.8) | 3.5 (1.2) | 3.5 (2.0) | 2.6 (0.4) | 3.4 (0.9) | 5.6 (3.6) |
| Completeness1 | 83.0 (79.4) | 96.8 (91.8) | 71.9 (71.3) | 82.44 (74.9) | 93.6 (92.2) | 87.5 (56.2) | 96.1 (85.8) | 97.0 (96.8) |
| Multiplicity1 | 1.9 (1.8) | 8.6 (6.1) | 3.8 (3.9) | 4.2 (3.8) | 4.4 (3.9) | 6.9 (3.1) | 8.2 (5.7) | 12.3 (12.2) |
| Refinement | ||||||||
| Rwork1 (%) | 20.97 (21.04) | 23.95 (32.33) | 29.19 (39.51) | 22.95 (35.40) | 25.13 (34.08) | 24.79 (38.72) | 22.35 (36.33) | 28.99 (34.78) |
| Rfree1, 2 (%) | 22.28 (22.43) | 28.42 (37.94) | 32.80 (42.03) | 26.70 (38.95) | 29.45 (38.98) | 28.11 (42.37) | 25.46 (42.25) | 30.96 (36.64) |
| RSCC | 0.84 | 0.89 | 0.72 | 0.85 | 0.86 | 0.89 | 0.91 | 0.86 |
| # residues | 6 | 129 | 166 | 190 | 207 | 223 | 279 | 316 |
| # protein atoms | 53 | 1,001 | 1,327 | 1,481 | 1,551 | 1,629 | 2,029 | 2,432 |
| # water molecules |
2 | 87 | 0 | 23 | 18 | 195 | 221 | 21 |
| # ligand atoms | 0 | 3 | 0 | 2 | 0 | 2 | 2 | 13 |
| 〈ADP〉 (Å2) | ||||||||
| Protein | 12.4 | 13.4 | 47.8 | 25.5 | 20.3 | 13.9 | 8.1 | 4.9 |
| Water | 17.3 | 14.3 | 19.4 | 13.3 | 14.9 | 13.4 | 4.2 | |
| Ligand | 16.7 | 66.5 | 19.6 | 18.9 | 7.5 | |||
| R.m.s.d. bonds (Å) | 0.012 | 0.004 | 0.012 | 0.002 | 0.002 | 0.005 | 0.004 | 0.003 |
| R.ms.d. angles (°) | 0.770 | 0.609 | 1.573 | 0.496 | 0.462 | 0.739 | 0.663 | 0.509 |
| Ramachandran (outliers, favored) (%) |
0.0, 100 | 0.0, 97.6 | 2.5, 89.9 | 0.0, 96.3 | 0.0, 95.1 | 0.0, 96.4 | 0.4, 96.8 | 0.0, 94.9 |
Numbers in parentheses reflect the highest resolution shell for either data collection or refinement.
In all cases the test set comprises approximately 5% of the unique reflections, where possible chosen to match that of the deposited data for the MR search model.
Fragmentation was tested on eight proteins: lysozyme, TGF-βm:TβRII, xylanase, thaumatin, trypsin, proteinase K, thermolysin, and a segment of the protein tau (Figure 2, left column). These proteins readily form crystals that are large—too big for MicroED and SFX experiments using liquid injectors—but without crystal growth optimization sometimes exhibit pathologies. These large crystals were broken apart by mechanical means prior to preparation on cryoEM grids. Micro- or nanometer-sized crystal fragments appeared evenly distributed on the grid, and even when grids were densely populated single-crystal data sets could be collected by using the selective area aperture.
MicroED is inherently well suited to study crystal fragments. Electrons interact strongly with matter so that large crystals are not required to detect high-resolution Bragg reflections8. MicroED has been used to determine the structures of the enzymes lysozyme5,9, proteinase K10, catalase11,12, and Ca2+-ATPase12. It was also recently used to solve the 1.4 Å resolution structure of the toxic NAC-core of α-synuclein, where diffraction data were collected from crystals smaller than the wavelength of visible light13. We previously demonstrated that crystals thinner than ~400 nm are suitable for MicroED and routinely yield atomic resolution information. The highest resolution structures reported so far by using MicroED are from four prion protein fragments determined at 1 Å resolution solved by direct phasing methods14.
For each sample, data were collected from 2–10 crystal fragments, and data sets merged for completeness and multiplicity (Table 1). Although we have previously published two structures where a single nanocrystal was sufficient for structure determination9,11 multi-crystal merging is generally preferred. This is partly because the grid on which the crystals are mounted limits the accessible rotation range during data collection, and partly because small, weakly scattering samples require a more intense beam to yield statistically accurate measurements of the Bragg reflections, and may be irreversibly damaged before a complete data set can be obtained. Recording narrow wedges with fewer exposures for each crystal involves a tradeoff between limiting the damage to the sample and maintaining a sufficient dose rate for the signal from high-resolution reflections to be accurately integrated. Because the resolutions obtained by MicroED are comparable or better to what was obtained by X-ray diffraction before sonication, it appears that fragmentation only broke apart the large crystals into small crystal domains but did not otherwise damage the lattice order.
The macromolecular structures in this study were phased using molecular replacement (MR) or by direct methods and refined using electron scattering factors15. In all cases, the refined model fits the calculated density well (Figure 2, middle column) with an overall real-space map correlation coefficient ranging from 0.72 to 0.91 (RSCC; Table 1). The resulting simulated-annealing (SA) composite-omit maps (Figure 2, right column) match their respective models very well, indicating that the data are indeed high quality and unbiased. Further, the SA omit maps for lysozyme reveal depressions or holes in the aromatic rings of amino acid side chains, while for proteinase K toroidal densities were observed even for proline residues at the 1.6 Å resolution cutoff. A well-coordinated calcium ion is clearly visible in the omit map for trypsin, as is one of the iodides in the xylanase structure. Individual atoms are visible in the density for the tau peptide, and several residues show positive 2mFo-DFc peaks for hydrogen atoms; for one residue, modeled hydrogens allowed us to unambiguously place the correct sidechain rotamer.
All structures were determined between 1.1–2.9 Å resolution. We note that the R-factors from model refinement are generally higher in MicroED than typical values obtained in X-ray crystallography at similar resolutions. Others and we have already made such observations in previously published studies5,9,12. We believe that the main reason is lack of adequate electron scattering factors in the crystallographic refinement software. X-rays are scattered by the electron cloud of an atom while electrons are scattered by the atomic Coulomb potential, which arises from the nucleus as well as its surrounding electrons. Current electron scattering factor tables do not adequately account for this difference and this may contribute to the residual between observed and calculated structure factor amplitudes leading to higher than normal crystallographic and free R-factors. We note that the gap between Rwork and Rfree is small indicating no concerns of overfitting (Supplementary figure 10).
Discussion
Breaking large crystals into well-formed crystal fragments has a long history in crystallography; indeed X-ray diffraction was discovered from broken up pieces of a copper sulfate crystal16. Sonication17 and vortexing18 have previously been used to prepare crystals of suitable size for XFEL measurements. By breaking up one imperfect large crystal into thousands or even millions of smaller crystallites and recombining a well-diffracting subset during data processing, we have used MicroED to determine the structure of eight different proteins to high resolution. Fragmentation does not increase the effort required for sample screening, and future work on automation is expected to reduce the time for data collection irrespective of how the sample was prepared. Importantly, as demonstrated by the breadth of size and packing of macromolecules whose crystals we investigated, this is a broadly applicable approach where large crystals are available. Fragmentation widens the scope of diffraction methods such as MicroED and SFX to include samples that do not exclusively form tiny crystals but instead form large, imperfect crystals.
Online methods
Large crystals (>500 µm along the longest edge) were grown via hanging-drop vapor diffusion at room temperature using previously established protocols. All enzymes were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Tau peptide (VQIVYK) was dissolved in distilled water and crystals prepared by mixing tau peptide with arachidonic acid and meclocycline sulfosalicylate. Crystals formed with a 1:2 drop peptide to precipitant ratio from 65–70% ethylene glycol in Tris pH 8.5.
Lysozyme (G. gallus) crystals were prepared by equilibrating equal volumes of 50 mg/ml lysozyme in water and 1.7 M sodium chloride, 50 mM sodium acetate pH 4.7.
TGF-βm:TβRII was expressed and purified as described before20,21. Crystals were grown from 0.5 µl of protein (20 mg/ml), 0.25 µl mother liquor, and 0.2 µl seed stock in 100 mM HEPES/NaOH pH 7.5 and 45% MPD.
Xylanase from T. reesei (Hampton Research, Aliso Viejo, CA) was dialyzed against 10 mM bicine pH 9.0, 1 mM magnesium sulfate, 1 mM DTT and combined with a precipitant solution containing 0.3 M sodium iodide, 1.2–1.3 M ammonium sulfate, and 100 mM bicine pH 9.022.
Thaumatin (T. daniellii) crystals were grown from 2 µL of protein (25 mg/ml in water) and Hampton Research Index Reagent 26 (1.1 M ammonium tartate pH 7.0).
Trypsin (B. taurus) was dissolved (60 mg/ml) in 10 mg/ml benzamidine, 3 mM calcium chloride and equilibrated against 4% (w/v) PEG4000, 0.2 M lithium sulfate, 0.1 M MES pH 6.5 and 15% ethylene glycol.
Proteinase K (E. album) crystals were grown by combining 2 µL of protein solution (50 mg/ml) with 2 µL of precipitant solution (1.0–1.3 M ammonium sulfate, 0.1 M Tris pH 8.0)10.
Thermolysin from B. thermoproteolyticus (Hampton Research, Aliso Viejo, CA) crystals were prepared by equilibrating a 160 mg/ml solution of thermolysin (45% dimethyl sulfoxide, 50 mM Tris pH 7.5 and 2.5 M cesium chloride) over 0.5 ml of water23.
Drops containing the crystals were placed in separate microfuge tubes and suspended in crystal mother liquor. A sonicating water bath with electronic control (Elmasonic P30H, Singen, Germany) was set at its lowest power (30% at 37 kHz) for gentle agitation of the crystals in the tube. With the tube sealed, its tip was briefly submerged in the activated water bath for 0.5 s. Alternatively; crystals were fragmented by vigorously pipetting a crystal suspension in mother liquor (trypsin and thaumatin) or vortexed with 0.5 mm disruption glass beads in a 1.5 ml reaction tube for 2 s (TGF-βm:TβRII). A detailed protocol for crystal fragmentation was published online24.
The solution containing fragmented crystals was then applied to transmission electron microscope (TEM) grids with carbon film support, and plunge-frozen in liquid ethane. Frozen grids were mounted in a Gatan Model 626 cryo-specimen holder and examined using an FEI Tecnai F20 field-emission TEM operated at an accelerating voltage of 200 kV, which corresponds to a de Broglie wavelength of 0.025 Å, and screened for crystals in over-focused diffraction mode. Where a single still shot revealed strong diffraction, data were collected as continuous rotation tilt series9. Individual frames were recorded on a TVIPS TemCam-F416 as 4 s exposures while the stage was rotating at 0.09°/s, except for tau peptide, which was rotated at 0.29°/s during 2 s exposures. For the macromolecular samples, the absolute tilt angle was generally <35° (<65° for tau peptide). Data sets collected from each crystal spanned between 30° and 135°, corresponding to a total dose no greater than 1.2–5.5 e−/Å2 at the given rotation rate. The selective area aperture of the TEM was used to limit the area from which data were collected, making it possible to select a single crystal fragment. The virtual detector distance varied between 730 mm and 3.6 m for the different data sets, corresponding to a maximum resolution between 0.95 Å and 2.0 Å in the detector corners at an acceleration voltage of 200 kV. The movie frames recorded in rolling shutter mode on the TVIPS TemCam-F416 CMOS camera were converted to SMV format, while preserving as much as possible of the metadata necessary for subsequent processing25. Detailed protocols for data collection were published recently26.
Sweeps were corrected to account for negative pixel values10, then indexed and integrated in MOSFLM27 using its graphical interface iMOSFLM28, or in XDS29. To further probe the scattering power of the crystallites, diffraction patterns were initially processed without imposing any resolution cutoffs other than those entailed by the geometry of the experiment. Except for the tau peptide and TGF-βm:TβRII, the high-resolution limits due to factors other than the area of the detector were instead determined case-by-case to give stable processing during refinement of the atomic model. Multi-crystal scaling and merging was performed in AIMLESS30. Structures were phased by molecular replacement using MOLREP31 from the PDB entries 3j6k, 1ktz, 2dfb, 4ek0, 2ptn, 5i9s, and 2tli for lysozyme, TGF-βm:TβRII, xylanase, thaumatin, trypsin, proteinase K, and thermolysin, respectively, or by direct methods using SHELXT32. A free R set comprising approximately 5% of the unique reflections was copied from the deposited data for each search model. The deposited data for the xylanase and trypsin models do not define a free set; instead a new set was chosen using freerflag33.
Maximum likelihood structure refinement was carried out in phenix.refine34 using electron scattering factors15. Explicit water molecules were automatically modeled by phenix.refine and subsequently manually curated in Coot35. SA composite omit maps were computed from the refined models using CNS36 with electron scattering factors37. For these calculations, charged species were manually removed from the phasing models, because they are not included in CNS’s electron scattering library. The SA protocol was defined to exclude ~5% of the structure at a starting temperature of 2500 K. Protocols for data analysis in MicroED were recently published25.
Supplementary Material
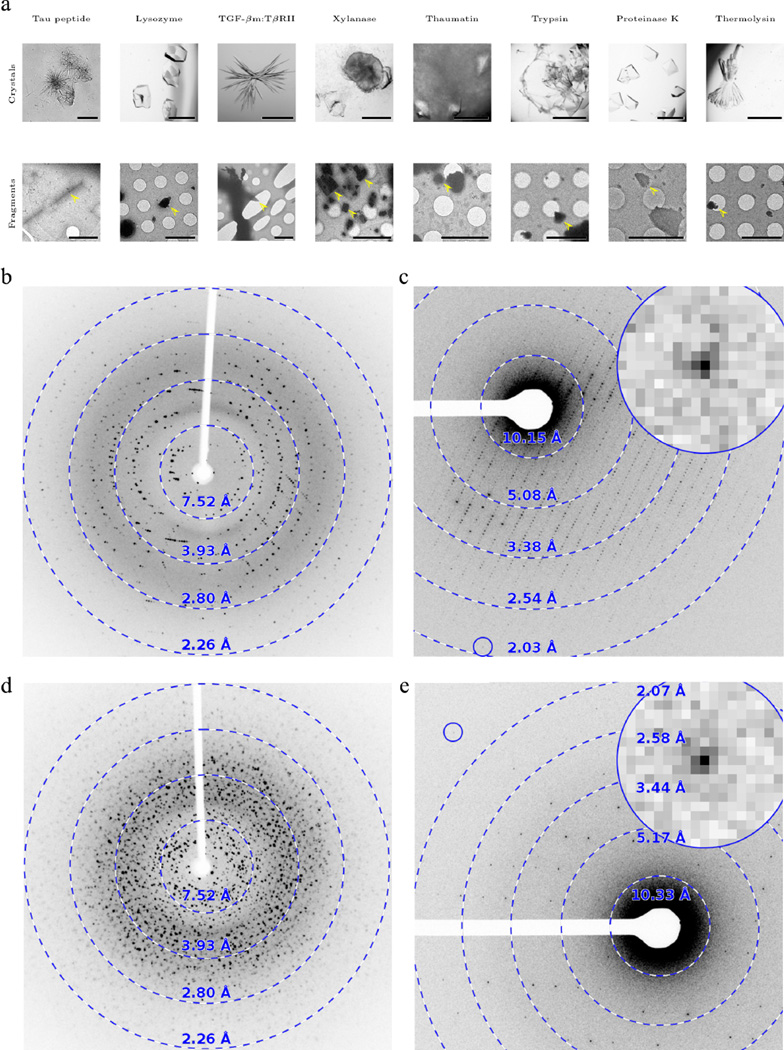
Figure 1. Crystals before and after fragmentation and their X-ray and MicroED diffraction patterns.
(a) Top row: light micrographs of imperfect crystals before fragmentation. Scale bars are 500 µm, except for tau peptide and TGF-βm:TβRII where the scale bar is 50 µm. Bottom row: electron micrographs of fragmented crystals (scale bars are 5 µm). The X-ray diffraction patterns exhibit multiple lattices (b; thaumatin) or even powder-like diffraction (d; trypsin). These pathologies are not present in crystallite fragments investigated by MicroED (c, e). The inset shows a close-up of the spot indicated by the blue circle. X-ray diffraction patterns were collected on a Cu-Kα home over a 0.37° rotation range; MicroED patterns were recorded as detailed in the methods section. Further examples are shown in Supplementary figures 1–9.
Acknowledgments
The Gonen laboratory is supported by funds from the Howard Hughes Medical Institute. The Eisenberg laboratory is supported by HHMI, NIH (1R01-AG029430, DE), and DOE (DE-FC02-02ER63421, DE). The Calero laboratory is supported by NIH, GC, RO1GM120292, GC, RO1DK102495, GC, and BioXFEL-STC1231306, GC. The Hinck laboratory is supported by NIH R01-GM58670, APH and R01-CA172886, APH.
Footnotes
Author contributions
DS and TG designed experiment; MJdlC and FER prepared lysozyme, xylanase, thaumatin, trypsin, proteinase K, and thermolysin samples; JR and DE prepared tau peptide samples; SCW, SKK, CSH, APH, and CG prepared TGF-βm:TβRII samples; MJdlC, DS, JR, and SCW collected data; MJdlC, JH, PS, MRS, DC, SCW, and GC, analyzed data, refined and determined models; JH, JR, and MRS prepared figures; MJdlC, JH, and TG wrote the manuscript with contributions from all authors.
Competing financial interests statement
The authors declare no competing financial interests.
Accession codes
Data availability statement
Atomic coordinates and structure factors were deposited to the Protein Data Bank (PDB; 5k7n, 5k7o, 5ty4, 5k7p, 5k7q, 5k7r, 5k7s, and 5k7t) and the Electron Microscopy Data Bank (EMD; EMD-8216, EMD-8217, EMD-8472, EMD-8218, EMD-8219, EMD-8220, EMD-8221, and EMD-8222), and the raw data were uploaded to the Structural Biology Data Grid38 (10.15785/SBGRID/284, 10.15785/SBGRID/285, 10.15785/SBGRID/368, 10.15785/SBGRID/286, 10.15785/SBGRID/287, 10.15785/SBGRID/288, 10.15785/SBGRID/289, 10.15785/SBGRID/290).
References
- 1.Darwin CG. XCII. The reflexion of X-rays from imperfect crystals. Philos. Mag. Ser. 6. 1922;43:800–829. [Google Scholar]
- 2.Nave C. A Description of Imperfections in Protein Crystals. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:848–853. doi: 10.1107/s0907444998001875. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S, et al. Small is beautiful: protein micro-crystallography. Nat. Struct. Biol. 1998;5 Suppl:634–637. doi: 10.1038/1325. [DOI] [PubMed] [Google Scholar]
- 4.Schlichting I. Serial femtosecond crystallography: the first five years. IUCrJ. 2015;2:246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi D, Nannenga BL, Iadanza MG, Gonen T. Three-dimensional electron crystallography of protein microcrystals. Elife. 2013;2:e01345. doi: 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weierstall U. Liquid sample delivery techniques for serial femtosecond crystallography. Philos Trans R Soc L. B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0337. 20130337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sierra RG, et al. Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 9.Nannenga BL, Shi D, Leslie AGW, Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods. 2014;11:927–931. doi: 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hattne J, Shi D, de la Cruz MJ, Reyes FE, Gonen T. Modeling truncated pixel values of faint reflections in MicroED images. J. Appl. Crystallogr. 2016;49:1029–1034. doi: 10.1107/S1600576716007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T. Structure of catalase determined by MicroED. Elife. 2014;3:1–11. doi: 10.7554/eLife.03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yonekura K, Kato K, Ogasawara M, Tomita M, Toyoshima C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proc. Natl. Acad. Sci. 2015;112:3368–3373. doi: 10.1073/pnas.1500724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya MR, et al. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc. Natl. Acad. Sci. 2016;113:11232–11236. doi: 10.1073/pnas.1606287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colliex C, et al. International Tables for Crystallography. C, (International Union of Crystallography) 2006 [Google Scholar]
- 16.Laue M Von. Concerning the detection of X-ray interferences. Nobel Lect. 1915:347–355. at < http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:Concerning+the+detection+of+X-ray+interferences#0>. [Google Scholar]
- 17.Stevenson HP, et al. Transmission electron microscopy for the evaluation and optimization of crystal growth. Acta Crystallogr. Sect. D Struct. Biol. 2016;72:603–615. doi: 10.1107/S2059798316001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bublitz M, et al. Structural studies of P-type ATPase–ligand complexes using an X-ray free-electron laser. IUCrJ. 2015;2:409–420. doi: 10.1107/S2052252515008969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrödinger LLC. The PyMOL Molecular Graphics System. 2014 at < https://www.pymol.org>. [Google Scholar]
Methods-only references
- 20.Zúñiga JE, et al. Assembly of TβRI:TβRII:TGFβ Ternary Complex in vitro with Receptor Extracellular Domains is Cooperative and Isoform-dependent. J. Mol. Biol. 2005;354:1052–1068. doi: 10.1016/j.jmb.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Kim SK, et al. An engineered TGF-β monomer that functions as a dominant negative to block TGF-β signaling. J. Biol Chem. doi: 10.1074/jbc.M116.768754. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe N, Akiba T, Kanai R, Harata K. Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:784–792. doi: 10.1107/S0907444906017379. [DOI] [PubMed] [Google Scholar]
- 23.English AC, Done SH, Caves LSD, Groom CR, Hubbard RE. Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol. Proteins. 1999;37:628–640. [PubMed] [Google Scholar]
- 24.de la Cruz MJ, et al. Micro- and nanocrystal preparation for MicroED and XFEL serial crystallography by fragmentation of imperfect crystals. Protoc. Exch [Google Scholar]
- 25.Hattne J, et al. MicroED data collection and processing. Acta Crystallogr. Sect. A Found. Adv. 2015;71:353–360. doi: 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi D, et al. The collection of MicroED data for macromolecular crystallography. Nat. Protoc. 2016;11:895–904. doi: 10.1038/nprot.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie AGW, Powell HR. Evolving Methods for Macromolecular Crystallography. Evolving Methods for Macromolecular Crystallography. Vol. 245. Netherlands: Springer; 2007. [Google Scholar]
- 28.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D. Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 32.Sheldrick GM. SHELXT – Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 37.Gonen T, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer PA, et al. Data publication with the structural biology data grid supports live analysis. Nat. Commun. 2016;7:10882. doi: 10.1038/ncomms10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.