Abstract
Microtubules tether centrosomes together during interphase 1. How this is accomplished and what benefit it provides to the cell is not known. We have identified a bipolar, minus-end directed kinesin, Kif25, that suppresses centrosome separation. Kif25 is required to prevent premature centrosome separation during interphase. We show that premature centrosome separation leads to microtubule-dependent nuclear translocation, culminating in eccentric nuclear positioning that disrupts the cortical spindle positioning machinery. Kif25’s activity during interphase is required to maintain a centered nucleus to ensure the spindle is stably oriented at the onset of mitosis.
Centrosome separation represents an inaugural step in the entry into mitosis. In many organisms centrosome separation and bipolar spindle formation are driven by the action of molecular motors, namely kinesin and dynein motors 2–4. In interphase, centrosomes are physically held together by a protein linker composed of Rootletin and C-Nap15–7. This linker is resolved at the onset of mitosis by phosphorylation of C-Nap1 by Nek2A facilitating its release from the centrosome and allowing the centrosomes to be driven apart 8,9. However, early studies clearly show a microtubule (MT)-dependent component to centrosome tethering that is not explained by the simple centrosome-linker model 1.
When premature centrosome separation is triggered using epidermal growth factor treatment, cells proceed through mitosis faster yet show fewer chromosome segregation errors 10. Why then, are centrosomes tethered at all during interphase? And what is the contribution of MTs to centrosome tethering? In this study we have answered both of these questions by identifying a centrosome-tethering molecule: the kinesin-14 motor Kif25.
Kinesin-14 motors exhibit minus-end directed MT motility 11 and MT crosslinking activity 12. By combining these two modalities kinesin-14 motors focus poles and coalesce supernumerary centrosomes into a single spindle pole 13–15. Suppression of centrosome separation during interphase by Kif25 represents a unique kinesin-14 function distinct from those previously described.
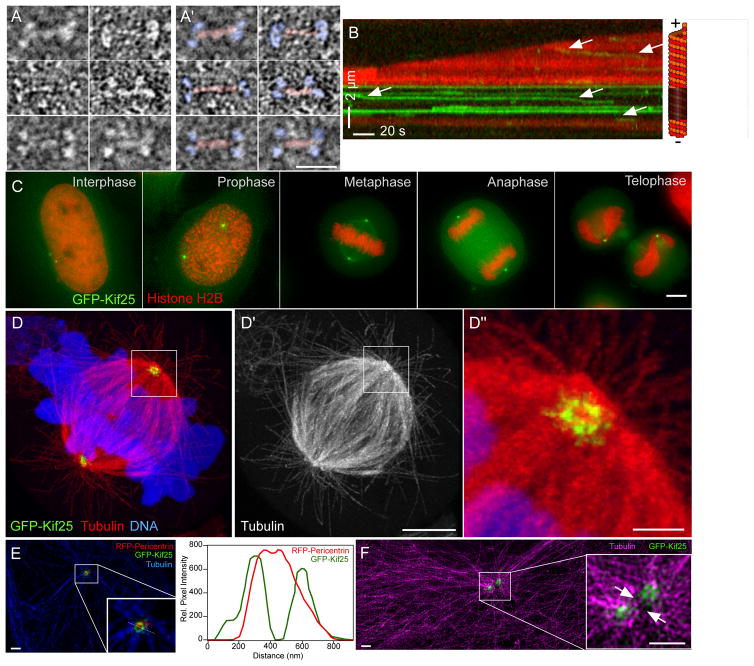
Kif25 is expressed at low levels in a range of human tissues making its presence in HeLa cells ambiguous 16,17. Quantitative-PCR confirmed the presence of Kif25 in HeLa cells (Fig. S1A). We depleted endogenous Kif25 using 2 independent siRNA constructs, again confirming the expression of Kif25 in our human cell line (Fig. S1B). For in vitro characterization of this motor we synthesized the Macaca fascicularis testis, clone QtsA-10923 of Kif25 (GenBank accession number AB168279), possessing almost complete identity with predicted hsKif25 sequences (Fig. S2). This clone comprises the full length Kif25 cDNA sequence as the originally identified Kif25 gene is known to be missing the full N-terminus of the motor 16,18. We determined the native MW of the EGFP-Kif25 motor to be 404 kDa (Fig S1C). The expected MW of Kif25 with attached GFP is 90 kDa therefore the native MW indicates that Kif25 is a tetrameric motor in solution. EM analysis confirmed the structure of the isolated motor as a bipolar tetramer, similar to kinesin-5 motors (Fig 1A) 19. Kif25 retains the ability to crosslink MTs in vitro (Fig. S3) and translocates toward the minus-end of the MT with an average run length of 1.39±0.13 (mean± s.e.m.) μm and velocity of 0.39 ±0.06 μm/s (Fig. 1B). To date, Kif25 is the first identified tetrameric minus-end directed kinesin motor.
Figure 1.
Kif25 structure and localization. (A and A′) EM structure of EGFP-Kif25 motor. Panels in A′ are identical to A with the addition of pseudo coloring to show motor domains (blue) and neck regions (red) of the motor (scale bar = 25 nm). (B) Kymograph showing movement of EGFP-Kif25 on dynamic MTs. Arrows indicate motor movement in the direction of the unlabeled seed at the minus end of the MT. Run length and velocity are calculated from n= 97 moving spots pooled from 6 separate movies. (C) EGFP-Kif25 localizes to the centrosome at all stages of HeLa cell cycle (scale bar = 5 μm). (D) Confocal image of expanded HCT-116 cells stably expressing EGFP-Kif25 and stained for α-tubulin. Kif25 localizes to a ring-like structure at the centrosome, zoomed-in view shows Kif25 also on radial spokes around the centrosome (D″) (scale bar 15 μm for D and D′, 1.5 μm for D″). (E) Localization of EGFP-Kif25 relative to RFP-pericentrin in unexpanded LLCPK1 cells imaged using structured illumination microscope (scale bars = 1 μm). Inset, zoomed-in view of area around centrosome in interphase cell. Right, line-scan of fluorescence of EGFP-Kif25 and RFP-pericentrin measured in the inset showing greater diameter of Kif25 relative to pericentrin at the centrosome. (F) SIM image of 2 centrosomes in interphase LLCPK1 cell (scale bars = 1 μm). MTs are labeled in magenta. Inset, zoomed-in view of centrosome region with arrows indicating the presence of MTs between duplicated centrosomes and Kif25 foci at the centrosomes. Four separate transfections of LLCPK cells were fixed, labeled and mounted on separate days. All were imaged individually on the same day and between 3 and 20 images collected from each slide depending on cell density. Representative images are presented from 2 different slides (1E and F). Expansion microscopy was performed on two separate sets of fixed, stained GFP-Kif25 expressing cells on separate days. Twenty-five cells were imaged and a representative cell presented (1D).
Expressed EGFP-Kif25 is centrosomally localized during all stages of the cell cycle (Fig. 1C). Super-resolution expansion microscopy38 localized EGFP-Kif25 to a ring at the centrosome with radial projections (Fig. 1D). The diameter of the EGFP-Kif25 ring measured greater than RFP-pericentrin (Fig. 1E) in co-localized images of unexpanded cells collected with structured illumination microscopy (SIM). Microtubules could be detected passing between duplicated interphase centrosomes that co-localized with EGFP-Kif25 rings, suggesting a direct role for Kif25-mediated centrosomal linkage via crosslinked MTs (Fig. 1F).
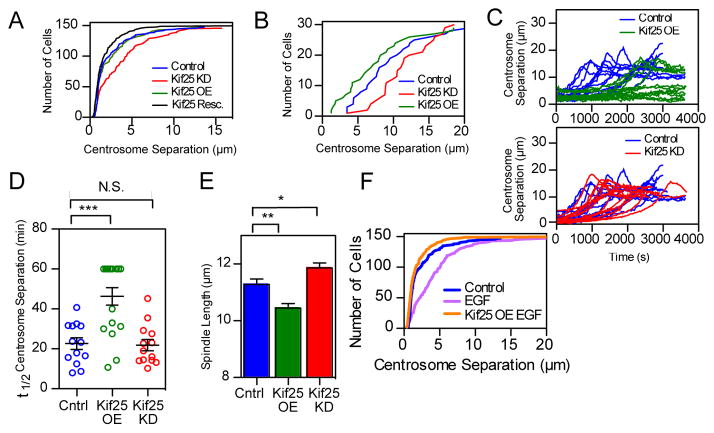
Collectively, the structure, activity and directionality of Kif25 suggest that it may antagonize Eg5. Eg5 drives centrosome separation using plus-end directed motor activity 4,20, therefore we expect Kif25 to suppress centrosome separation. Accordingly, the average distance between duplicated centrosomes in fixed cells increases without Kif25 in interphase and prophase (Figs 2A and 2B, from 2.44 ± 0.21 μm for control to 3.54 ± 0.25 μm for Kif25 KD in interphase, mean± s.e.m., P=0.0009, and from 8.57 ± 0.76 μm in control cells to 11.39 ± 0.69 μm in Kif25 KD cells in prophase, P=0.008). Rescue of Kif25 knockdown by EGFP-Kif25 expression restores control-like interphase centrosome separation distances (1.984 ± 0.21 μm, P=0.036 compared to control). Thus, Kif25 prevents interphase centrosome separation by tethering duplicated centrosomes together. Tethering is dependent on the presence of MTs as nocodazole treatment promotes interphase centrosome separation that cannot be rescued by Kif25 overexpression 1(Fig. S3B). In control and Kif25 knockdown cells, washout of monastrol leads to the rapid establishment of a bipolar spindle 21(Fig 2C, top). Overexpression of Kif25, however, significantly delayed bipolar spindle formation with many cells not separating their centrosomes during the acquisition time (t1/2= 46.3 min) (Fig 2C, top).
Figure 2.
Kif25 suppresses centrosome separation. (A) Asynchronous HeLa cells stained for α- and γ-tubulin to determine centrosome separation. Cumulative frequency plot shows data pooled from 3 separate experiments (n values; control=148 cells, Kif25 KD=146 cells, Kif25 OE=148 cells, Kif25 Rescue=150 cells). Kif25 KD red line, control blue line, Kif25 OE green line, and Kif25 rescued black line, are shown. (B) Same experimental conditions as (A) except imaging prophase cells, 30 cells/condition pooled from 3 independent experiments. (C) Live HeLa cells released from monastrol treatment were used to measure centrosome separation in control (n=13 cells), Kif25 KD (n=13 cells), or Kif25 OE cells (n=15 cells), each trace represents an individual cell. (D) The t1/2 for centrosome separation measured as the time for the spindle to reach ½ the maximum measured length. Each point is a single cell, error bars represent mean±SEM, ***P=0.002 from an unpaired t test. (E) Bipolar spindle length in fixed cells following Kif25 KD or overexpression of EGFP-Kif25. Data are from n=104 control, n=108 Kif25 KD and n=74 Kif25 OE cells/condition pooled from 3 independent repeated experiments. **P=0.001, *P=0.02. (F) Cumulative frequency plot of centrosome separation in EGF-treated HeLa cells for n=148 control, n=148 EGF and n=150 EGF Kif25 cells for each condition pooled from 3 separate experiments. Average centrosome separation for EGF: 4.32±0.3 μm. P < 0.0001 compared to control. EGFP-Kif25 rescue in EGF treated cells average centrosome separation: 1.86±0.2 μm. All measurements are mean ± SEM.
Kif25 also impacts pole separation during metaphase (Fig 2E). Kif25 knockdown increases mitotic spindle length while Kif25 overexpression decreases spindle length relative to control cells (Control 11.28±0.19 μm, Kif25 KD 11.86±0.17 μm, P=0.02, Kif25 OE 10.44±0.16, P=0.001, distance mean ± s.e.m). Collectively, it appears that Kif25 plays a major role in antagonizing centrosome separation during interphase to maintain centrosome coupling and, to a lesser extent, during mitosis to impact spindle length.
During canonical centrosome separation Aurora A is activated by cdk1/CyclinB2 leading to downstream activation of Plk1, Mst2 and finally Nek2A which ultimately phosphorylates C-Nap1 triggering centrosome separation (Fig. S3C). To determine if Kif25 knockdown promotes activation of this signaling pathway we measured centrosome localized levels of phospho-Aurora A, phospho-Plk1, Nek2A and C-Nap1 in fixed interphase cells (Fig S3D). Negligible decreases in C-Nap1 and Nek2A levels followed Kif25 knockdown suggested that Kif25 loss is not activating this centrosome-signaling pathway. We further confirmed this by promoting premature centrosome separation with epidermal growth factor (EGF)10, and rescuing this phenotype by overexpression of GFP-Kif25 (Fig 2F). This demonstrates that Kif25 can tether centrosomes in the absence of C-Nap1 and represents a key part of the machinery preventing centrosome separation during interphase.
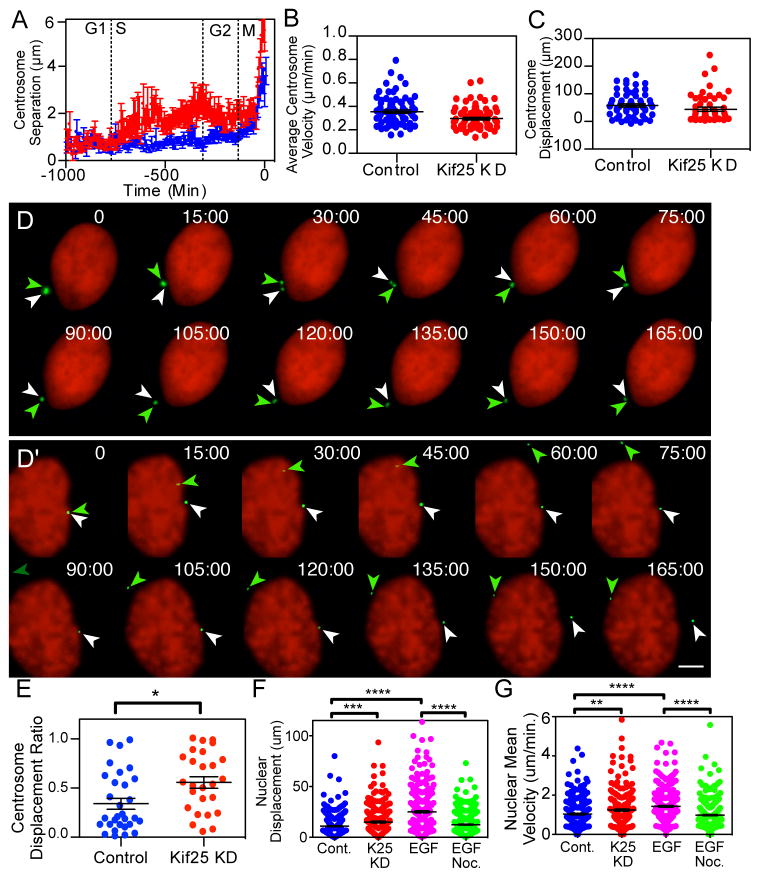
Live cell cycle timing of Kif25-induced premature centrosome separation was determined using PCNA (Proliferating cell nuclear antigen) to monitor cell cycle stage and pericentrin to mark centrosomes 22,23. Control cells exhibit tethered centrosomes until the onset of mitosis whereas Kif25 knockdown causes centrosome separation following the transition to S-phase (Fig 3A). This timing is significant because centrosomes are duplicated during S-phase, suggesting that Kif25 is required to tether centrosomes commensurate with duplication. We measured the speed and displacement of centrosomes following loss of Kif25 (Figs. 3B and 3C). The averaged speed and displacement data for the total population of centrosomes was similar with or without Kif25. However, we did observe differential behavior for individual centrosomes within a single cell. Comparing the displacement lengths of individual centrosomes we found that loss of Kif25 caused one of the centrosomes to move much more than the other (Figs. 3D, 3D′ and 3E, Movies 1 and 2). Because centrosome position is important for nuclear position and movement 24,25 we also measured the nuclear position in cells lacking Kif25. We measured an increase in the average eccentric displacement of the nucleus in cells without Kif25 and also an increase in mean speed of nuclear movement (Figs. 3F and 3G). The increase in nuclear displacement is phenocopied by EGF treatment indicating that premature centrosome separation results in changes in nuclear dynamics. The ability of separated centrosomes to increase nuclear movement is related to their role as microtubule organizing centers as treatment of cells with EGF and nocodazole, to depolymerize MTs, abolishes the nuclear movement phenotypes (Figs. 3F and 3G, Green dots). These results are surprising as nuclear position is reportedly tightly controlled 26.
Figure 3.
Centrosome and nuclear movements following premature centrosome separation. (A) HeLa cells were imaged every 7 min for 72 hrs to determine cell cycle timing of centrosome separation. Traces are the averaged centrosome separation distances for n=39 control cells and n=34 Kif25 KD cells where separation timing was normalized to the initial centrosome separation at the onset of mitosis. (B) Centrosome dynamics in cells were imaged every 30 s for 3 hrs. Centrosomes were analyzed for their (B) average velocity, (C) total displacement of each centrosome measured as the sum of displacements from each frame of the movie and (E) centrosome displacement ratio calculated by dividing the shorter centrosome displacement by the longer centrosome displacement for each cell observed. N=30 individual cells and 60 centrosomes for quantification in B, C and E. *P=0.01 (D and D′) Representative time-lapse images of centrosome movements in control (D) and Kif25 KD (D′) cells. Representative images represent the 30 live cells imaged for each condition quantified in Figures 3B, C, and E. Colored arrows indicate the position of each centrosome at each timepoint. H2B is labeled to visualize the nucleus while Pericentrin marks the centrosome. Scale bar equals 5 μm. (F) Nuclear Displacement and (G) velocity was tracked over time, **P=0.001 ***P=0.0005, ****P<0.0001. n=243 cells per condition. Error bars represent mean±s.e.m., statistics done using unpaired t test.
EGF-dependent premature centrosome separation decreases the time spent in mitosis 10. Unlike EGF treatment, we find that centrosome separation induced by loss of Kif25 does not speed the timing of mitosis (Fig. S4A). This may be related to cellular changes following EGF treatment that are independent of centrosome separation. Conversely, we do see an increase in time spent in mitosis in Kif25 overexpressing cells (Fig. S4A). We determined that this increased time is the result of decreased inter-centromere tension leading to activation of the spindle assembly checkpoint (Figs. S4B and S4C). The checkpoint is never satisfied in many Kif25 overexpressing cells causing cell slippage out of mitosis and significant increases in the number of multinucleate cells (Fig. S4D). We hypothesize that increased Kif25 levels interfere with normal spindle force balance. Testing this will require additional experimentation.
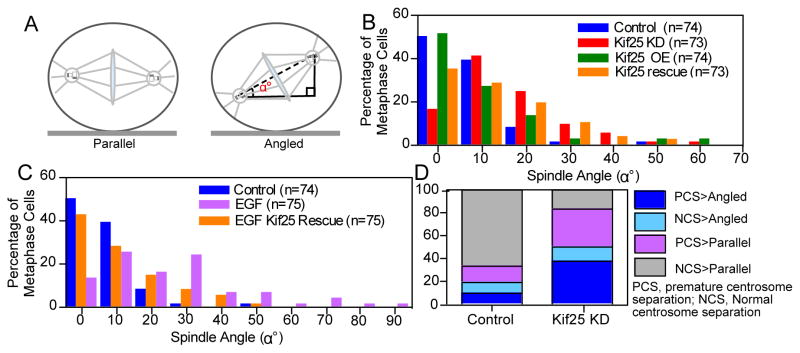
We observed spindle orientation defects in cells lacking Kif25 (Figs. 4A and B). HeLa cells grown in culture align their mitotic spindles parallel to the growth substrate, illustrated by control cells preferentially exhibiting spindle angles within 0–10 degrees (Fig. 4B, blue bars). Loss of Kif25 shifts this distribution toward higher angles (Fig 4B, red bars). We confirmed that the defect in spindle orientation is specific to loss of Kif25 by rescuing cells treated with Kif25 siRNA with co-expressed EGFP-Kif25 (Fig 4B, orange bars).
Figure 4.
Spindle orientation is altered following premature centrosome separation. (A) Schematic showing the difference between parallel and angled spindles and the measurement of spindle orientation angle, α°, relative to the coverslip. (B) Histogram of measured spindle orientation profiles of cells fixed and labeled with α- and γ-tubulin following Kif25 KD, Kif25 OE, and EGFP-Kif25 rescue following siRNA KD. (C) Histogram of measured spindle orientation profiles of cells fixed and labeled with α- and γ-tubulin following EGF treatment and EGF-treated cells overexpressing EGFP-Kif25. (D) Parsed data showing the relationship between centrosome separation state and spindle angle during mitosis. Data is initially separated based on whether cells show normal centrosome separation or premature centrosome separation then each cell is classified based on whether the mitotic spindle is parallel to the coverslip or angled. Data is from live asynchronously cycling cells transiently expressing mRFP-Pericentrin and GFP-H2B (n=26 cells, Kif25 KD; n=25 cells Control).
Spindle orientation is controlled through interactions of astral MTs and force generators located at the cell cortex 27,28. Changes in cell or spindle size could alter this interaction. We used utrophin 29 to label the cell periphery and ruled out changes in cell length, width or height as causes of the spindle orientation defect (Figs. S5A and B). Increasing spindle length by depleting Kif18A 30, also had no effect on orientation (Fig. S5C).
We reasoned that the spindle positioning defects are the direct result of premature centrosome separation and tested this by treating cells with EGF and measuring spindle angle profiles in mitotic cells (Fig. 4C). Cells treated with EGF show severe defects in spindle orientation similar to loss of Kif25 (Fig. 4C, pink bars) that was rescued by expressed EGFP-Kif25 (Fig. 4C, orange bars). This shows for the first time the direct influence of premature centrosome separation on spindle orientation during the ensuing mitosis.
We parsed our live cell data to directly correlate centrosome separation and spindle orientation (Fig. 4D). In control cells the majority of the population display normal centrosome separation and form a properly oriented parallel spindle (grey bars, 66% of cells). The remaining control cells display a roughly equal distribution between three classes; normal centrosome separation and angled spindles (light blue bars, 10% of cells), premature centrosome separation and a parallel spindle (pink bars, 14% of cells), or premature centrosome separation and angled spindles (dark blue bars, 10% of cells). Depletion of Kif25 causes a significant decrease in normal centrosome separation>Parallel cells (grey bars, 17% of cells). Cell exhibiting normal centrosome separation and angled spindles does not change relative to control cells (light blue bars, 13% of cells), suggesting a baseline level of spindle misorientation via alternate pathways. Importantly, cells displaying premature centrosome separation have an equal probability of forming a parallel or angled spindle (pink and dark blue bars in Kif25 KD, 33 and 37%, respectively). This suggests that premature centrosome separation randomizes spindle orientation.
We also quantified positional data as cells rounded at the onset of mitosis as this time point would have the greatest effect on the position of chromosomes and assembling bipolar spindles. We see no change in the position of the centrosomes with respect to the center-of-mass of the cell and the center-of-mass of the nucleus (Fig S5D). However nuclear position shows a small non-significant change in cells lacking Kif25 (Fig. S5D). Parsing the data shows that only cells that exhibited a significant displacement of the nucleus from the cell centroid prior to mitosis went on to produce angled spindles (Figs. S5E and S5F). Thus, the position of the interphase nucleus influences the orientation of the forthcoming mitotic spindle31.
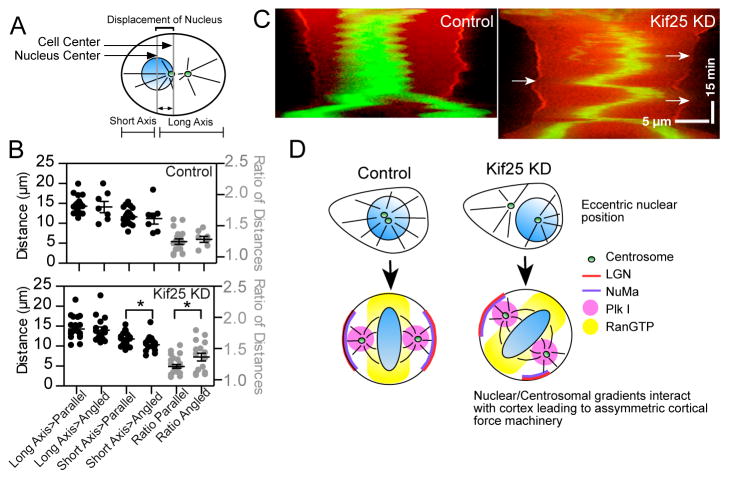
We measured the distance from the nuclear center to the cortex, using the future cell division axis as a reference 32. Nuclear position at cell rounding was parsed as before (Fig 5A) between parallel or angled mitotic spindles. Control cells do not show any significant differences in cells with parallel vs. angled mitotic spindles (Fig. 5B, top). Following loss of Kif25 the short axis distance between the nucleus and the cortex is significantly shorter in cells that subsequently form angled mitotic spindles (Fig. 5B, bottom). This is further reflected in the distance ratios where Kif25 KD cells with angled mitotic spindles show a higher ratio of long axis to short axis distance. This confirms that loss of Kif25 and premature centrosome separation promotes eccentric interphase nuclear positioning and that this, more than any other parameter, correlates with misoriented spindles.
Figure 5.
Eccentric nuclear position at mitosis onset randomizes spindle orientation. (A) Schematic showing measurements made to quantify the position of the nucleus in pre-mitotic cells. (B) Nuclear position was determined using the center-of-mass of the nucleus and measuring the distance from the cell cortex along the future cell division axis for each individual cell. A ratio of long axis-to-short axis was taken as a second measurement of nuclear centering (control cells; n=18 cells parallel group, n=7 angled group. Kif25 KD; n=22 parallel group, n=15 angled group). *P=0.025 using unpaired t test. Data are Mean ± s.e.m. (C) Representative cell kymographs showing spindle oscillations following Kif25 KD. Cells expressing tdTom-LGN and EGFP-H2B were selected while in prophase followed by imaging every 30 s. Arrows show the loss of LGN at the cortex is concurrent with close chromosome approach. Scale bars, 5 μm in X-axis and 15 min in Y-axis. (D) Model of how loss of Kif25 leads to spindle misorientation. During the normal cell cycle Kif25 suppresses centrosome separation prior to the onset of mitosis using its MT cross-linking activity (upper left). Centrally localized centrosomes and nucleus at the onset of mitosis allow Plk1 gradients from the centrosome (Pink circle) and RanGTP from the metaphase plate (Yellow) to symmetrically localize cortical force generation machinery. Without Kif25, centrosome tethering is lost and premature centrosome separation (upper right) leads to changes in centrosome movements and nuclear repositioning causing the inappropriate establishment of the spindle orientation machinery (bottom right).
How could nuclear position alter spindle orientation? Previous work has shown that chromosomes and centrosomes have considerable influence on mitotic spindle positioning 31,33. In these studies, chromosomes approaching too closely to the cell cortex promote release of cortical LGN. LGN is a crucial component of the cortical force generation machinery that localizes the force generator dynein 34. We used LGN to visualize the establishment of the force generation machinery during mitosis as well as H2B to visualize chromosome position (Fig. 5C, Movie 3). In cells depleted of Kif25 chromosomes are offset from the cell center while the cell is still rounding at the onset of mitosis (Fig. 5C, top left, Movie 4). Establishment of the cortical force machinery can be visualized as an increased intensity of LGN at the cortex of the rounding cell. Importantly, in the vicinity of the offset chromosomes it is clear that LGN is being excluded from this region of the cortex. The cell is then unable to establish a metastable spindle position. The movement of the metaphase plate towards each cortex coincides with the release of LGN from the proximal cortex and the strengthening of the LGN staining on the opposite cortex. Similar spindle oscillations were observed in 16/21 recorded Kif25 KD cells and only 4/25 control cells. This result is consistent with other studies that have experimentally introduced chromosomes near the cell cortex 31,33.
Collectively, our data shows that premature centrosome separation promotes eccentric positioning of the nucleus within the cell. If this is not spontaneously corrected by nuclear envelope breakdown the spindle will not achieve a metastable position in the center of the cell (Fig. 5D) and spindle orientation will be stochastic. Centered positioning of the centrosomes and chromosomes in a normal cell promotes a centralized mitotic spindle, which is important for the equal division of chromosomes and other cellular organelles into each of the two daughter cells. Because spindle position during mitosis is metastable, eccentric positioning of the spindle at mitotic onset is, in most cases, unrecoverable. This model accounts for our observation that following premature centrosome separation there is an equal probability that cells will form a parallel or an angled spindle. The difference in these two populations comes down to the position of the nucleus as the cell enters mitosis, which in the case of Kif25 KD is randomized. This highlights the importance of nuclear positioning at the cell center for the correct symmetrical establishment of the cortical force generation machinery.
Materials and Methods
Cell culture, transfection, and immunofluorescence
No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. Cells were purchased from ATCC and are tested monthly for viral and mycoplasma contamination. HeLa cells were grown in MEM Alpha medium (Invitrogen) supplemented with 10% FBS (Hydroclone;Thermo Fisher Scientific) in the presence or absence of Pen-Strep. LLCPK-1 cells were cultured in RPMI-1640 (Gibco|Thermofisher, Waltham, MA) +10% FBS (Hyclone, Logan, UT). Cells were transfected with 1.5 μg EGFP-Kif25 plasmid DNA and 2 μg all other constructs, unless specified otherwise, by electroporation using Nucleofector II (Lonza, Basel, Switzerland) according to the manufacturer’s instructions. Cells were transfected with 24 nM Control (negative control siRNA1, Ambion) or Kif25 siRNA using Lipofectamine RNAiMAX (Invitrogen) and incubated for 36–48 hours prior to experiments. For Kif25 depletion, two siRNAs targeted to distinct regions of the Kif25 gene were used independently to assess Kif25 KD phenotypes (targeting sequences (1) 5′-AGUGGAAGUUUACAAUAAU-3′ and (2) 5′-CAGAGUGACUUAGGAAUUA-3′ (Ambion)). Both siRNA constructs produced identical spindle orientation profiles and the number 2 construct was used in all further experiments. For fixed cell assays cells were plated on 12 mm coverslips and fixed in 1% PFA in −20°C methanol for 10 minutes. Coverslips were then blocked with 20% donkey serum for 1 hour. Cells were labeled with the following primary antibodies: mouse-anti-α-tubulin (1:100; Sigma DM1α), mouse-anti-γ-tubulin (1:1000; sigma GTU-88), mouse-anti-Plk1 (1: 200;Santa Cruz Biotechnology Sc-17783), rabbit-anti-NuMA (1:500; Novus NB500-174), rabbit-anti-phospho-Aurora A (1:100; Cell Signaling Tech 3079), mouse-anti-C-Nap1 (1:100; Santa Cruz Biotechnology 3090540), rabbit-anti-nek2A (1:100; kind gift of Elmar Schiebel), mouse-anti-phospho T210-Plk1 (1:300; Abcam 2A3) for 1 hr at room temperature. Anti-mouse and anti-rabbit antibodies conjugated to fluorescein or rhodamine (Jackson Laboratories) were used at 1:100 for 1 hr at room temperature. Stained cells were mounted in Vectashield with DAPI (Vector).
Samples used for super-resolution imaging were transfected with GFP-Kif25 and RFP-Pericentrin using a Nucleofector II (Lonza, Basel, Switzerland). Cells were fixed after 24 hours of protein expression in 37°C, PBS containing 3.2% paraformaldehyde (EM Sciences, Hatfield, PA) and 0.1% gluteraldehyde (EM Sciences, Hatfield, PA) for 10 minutes and then reduced in 0.1% Sodium Borohydride (Sigma, St. Louis, MO). After blocking in full strength FBS (Hyclone, Logan, UT), cells were labeled with YL1/2 (Abcam, Cambridge, MA) antibody against detyrosinated tubulin and Cy5-donkey anti-rat (Jackson Immunochemical, West Grove, PA) secondary antibodies. Labeled cells were post-fixed in 0.2% glutaraldehyde (EM Sciences, Hatfield, PA) in PBS.
DNA plasmid construction
The gene coding for the Macaca fascicularis testis cDNA clone QtsA-10923 was designed to be siRNA resistant and synthesized (Bio Basic Inc., Ontario, Canada). The Kif25 clone was conjugated to both EGFP and td-Tomato fluorophores with both constructs showing identical localization at the centrosome.
RT-PCR
Total cell RNA was isolated using an RNeasy mini kit (Qiagen). Reverse transcription was carried out using a Quantitect Reverse transcription kit (Qiagen) and primers specific for Kif25 (cat. No QT00024934, Qiagen) and Kif18A (cat. No QT00042455, Qiagen) as a positive control. RT-PCR reactions used the SYBR Green PCR kit (Qiagen) according to the manufacturers instructions. Samples lacking cDNA or reverse transcriptase were used as negative controls, no signal was observed in either case.
Western Blot
HeLa cells were lysed in 1X Laemmli sample buffer 48 hr after addition of siRNA. Lysates was passed through a 21-gauge needle, boiled for 10 min, and separated on 4%–12% acrylamide gradient gels by SDS-PAGE. Proteins were transferred to nitrocellulose membrane and analyzed by western blot with polyclonal anti-Kif25 antibodies (1:500;Santa Cruz Biotechnology, SC-133709) and monoclonal anti-α-tubulin antibodies (1:1000; Sigma, T6199). Proteins were visualized by chemiluminescence.
Hydrodynamic analysis of EGFP-Kif25
For gel filtration, EGFP-Kif25 (280 micrograms) plus standard proteins thyroglobulin, apoferritin and BSA (Sigma) were run on a 40 ml bed volume, 1.4 cm diameter Sephacryl S-400-HR column in buffer A (300 mM KCl, 200 mM imidazole-HCl pH 7.0, 25 mM KPO4, 2 mM MgCl2, 0.1 mM EGTA, 10% glycerol, 0.01 mM ATP, 5 mM beta-mercaptoethanol, 0.1% CHAPS) at 0.25 ml/min, 4°C. One mL samples of eluted fractions were precipitated with TCA, neutralized with NH4OH, dissolved in 0.05 ml SDS sample buffer, and analyzed by PAGE. Thyroglobulin was quantified by western blot, using anti-thyroglobulin antibody, following transfer to nitrocellulose membrane of the same gel used to quantify other standards. Velocity sedimentation was carried out using EGFP-Kif25 (35 micrograms) plus standard proteins apoferritin, beta-amylase and BSA. Samples were centrifuged on a 5–20% sucrose gradient in buffer A in an SW 55Ti rotor 5 hours at 210 K g, 4°C. Recovered fractions were analyzed by SDS-PAGE. EGFP-Kif25 molecular weight was calculated as previously described 35.
Negative stain electron microscopy
To prepare negative stain samples, purified GFP-Kif25 was diluted 100-fold in buffer A, applied to carbon-coated grids, and stained with 0.7% uranyl formate. Electron microscopy was performed on a Morgagni microscope (FEI co.) operating at 100 kV, and images were acquired at 22,000X magnification on an Orius SC1000 CCD camera (Gatan, Inc.).
Drug Treatments
For monastrol release experiments cells were incubated in media containing 100 μM monastrol for 2 hr, then quickly washed with fresh 37°C CO2-independent media (Invitrogen) 3X prior to imaging, as previously described (21). To measure mitotic timing cells were transfected with either EGFP-Kif25 or Kif25 siRNA prior to thymidine block for synchronization. Resting inter-centromere length in live cells was measured following treatment with 20 μM Nocodazole for 2 hr to depolymerize MTs. EGF-induced centrosome separation was carried out by treating asynchronous HeLa cells with 50 ng/ml recombinant human EGF (Invitrogen) for 4 hrs, as previously described 10.
In vitro MT-crosslinking and motility assays
EGFP-Kif25 protein was expressed and purified from sf9 cells. MT-crosslinking assays were carried out in flow cells using taxol stabilized MTs, grown from purified bovine brain tubulin labeled with Alexa 568 in BRB80 + 1mg/ml casein, 2 mM GTP, 100 μM ATP, 10 μM taxol and 10% DMSO. Purified EGFP-Kif25 was used at 500 nM and spun at 80,000 x g for 5 minutes to clear aggregates then mixed with labeled taxol-stabilized MTs in the flow cell and incubated 5 minutes prior to imaging. Kif25 motility was carried out by binding 0.06 mg/ml GMPCPP Cy5 labeled MT seeds to coverslips using rigor kinesin. Dynamic MTs were grown by flowing in 1.05 mg/ml Alexa-568 labeled tubulin. EGFP-Kif25 was used at a concentration of 150 nM in motility experiments. 97 motile spots were analyzed to determine the average velocity and run length of the motor. Imaging was carried out using a Personal Deltavision microscope (Applied Precision) outfitted with 4-laser TIRF capabilities, Olympus 60 x 1.49 NA TIRF objective and Ultimate focus (Applied Precision) at 37°C.
Fixed and live cell imaging
Fixed cells were imaged on a Nikon upright microscope equipped with a CCD camera and a 60x 1.4 NA lens (Nikon) or a Deltavision system equipped with a CCD camera and a 60x 1.42 NA lens (Olympus). Selected images were deconvolved using a Deltavision image-processing workstation (Applied Precision). Live cells were plated on a 35-mm glass coverslip coated with poly-d-lysine (MatTek Corporation) for 24–48 hr prior to imaging. Before imaging, cells were switched to 37°C CO2-independent media (Invitrogen) with 10% FBS. Live-cells were imaged either with a Deltavision RT system (Applied Precision) equipped with a CCD camera and a 60x 1.42 NA lens (Olympus) and a 37°C environmental chamber (Applied Precision) or on a Nikon Biostation IM-Q for long-term imaging.
CenpB was imaged as previously described to calculate inter-centromere distance 30. Mad1 persistence and spindle assembly starting in prophase were carried out using the continuous Z-sweep function in the Deltavision software imaging 4 μm sections every 30 s. Quantification of MAD1 persistence time was measured as the time elapsed between the spindle reaching 8 μm in length and the disappearance of the last Mad1 foci. We chose 8 μm as the cutoff because once the spindle reached this length it always formed a bipolar structure. Mitotic timing was measured using a 20x 0.75 NA lens (Olympus) taking images every 5 min.
For long-term observation of centrosome and nuclei movements images were taken on the Biostation IM-Q every 7 minutes for 72 hours. Centrosome and Nuclear movements were tracked using CellProfiler 2.0 36(Cellprofiler.org) to select and track the relevant structures. Centrosome velocity and distance measurements were carried out on >60 individual centrosomes from >30 cells. Nuclei displacement and velocity was measured in asynchronous interphase cells tracking 500 nuclei per condition.
For super-resolution SIM imaging labeled cells were imaged on a Deltavision OMX Blaze V4 instrument (GEHealthcare, Issaquah). Reconstructed images were further evaluated using Fiji.
Spindle orientation analysis
Asynchronous HeLa cells fixed and were labeled with α- and γ-tubulin following Kif25 KD, Kif25 OE, and EGFP-Kif25 rescue following siRNA KD. Cells were imaged by taking 0.25 μm slices through the cell. The angle and length of the mitotic spindle was determined using an automated MATLAB protocol to identify the spindle poles 37 and automatically calculate length and distance based on their x-, y-, and z-axis separation.
Nucleus and centrosome localization analysis
Interphase HeLa cells were analyzed using Fiji to identify the centroid of the cell and the centroid of the nucleus for each cell. Centrosome locations were manually selected. The linear distance between each position was then calculated. Data plotted as mean ± SEM.
Statistics and Reproducibility
All statistical analysis was carried out using a Student’s t test in Prism (version 5.0) to compare the significance of two data sets. Quantitative experiments were repeated in at least 3 independent experiments with further experiments done if necessary to reach desired population number. Other experiments were performed at least twice, as indicated in the figure legends.
Data availability
All data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant GM69429 to L.W.; National Institutes of Health Fellowship F32GM105099 and American Cancer Society Fellowship PF1409901CCG to J.D.; NIH grant GM118396 to J.K. and Burroughs Wellcome Career Award to J. V.
Footnotes
Author contributions: JD and LW are responsible for planning project and experiments as well as experimental analysis and writing the manuscript. MW performed and analyzed experiments. EL and JK performed and analyzed experiments using EM for Kif25 structure. AH and JV performed experimental and analyzed experiments using super-resolution microscopy.
The authors declare no competing financial interests.
References
- 1.Jean C, Tollon Y, Raynaud-Messina B, Wright M. The mammalian interphase centrosome: two independent units maintained together by the dynamics of the microtubule cytoskeleton. Eur J Cell Biol. 1999;78:549–560. doi: 10.1016/S0171-9335(99)80020-X. [DOI] [PubMed] [Google Scholar]
- 2.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 3.Raaijmakers JA, et al. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. The EMBO Journal. 2012 doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry AM, et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mardin BR, et al. EGF-induced centrosome separation promotes mitotic progression and cell survival. Dev Cell. 2013;25:229–240. doi: 10.1016/j.devcel.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–8. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mountain V, et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–366. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goshima G, Nédélec F, Vale R. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endow SA, Komma DJ. Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd-GFP fusion proteins. J Cell Sci. 1996;109(Pt 1):2429–2442. doi: 10.1242/jcs.109.10.2429. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto S, Matsushima M, Nakamura Y. Identification, genomic organization, and alternative splicing of KNSL3, a novel human gene encoding a kinesin-like protein. Cytogenet Cell Genet. 1998;83:25–29. doi: 10.1159/000015159. [DOI] [PubMed] [Google Scholar]
- 17.Maliga Z, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- 18.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Kashina AS, et al. A bipolar kinesin. Nature. 1996;379:270–2. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith E, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011;30:2233–2245. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J Cell Biol. 2012;197:231–237. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–80. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kisielewska J, Lu P, Whitaker M. GFP-PCNA as an S-phase marker in embryos during the first and subsequent cell cycles. Biol Cell. 2005;97:221–229. doi: 10.1042/BC20040093. [DOI] [PubMed] [Google Scholar]
- 24.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;144:414–426. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Laan L, et al. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burkel BM, Von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tame MA, Raaijmakers JA, Afanasyev P, Medema RH. Chromosome misalignments induce spindle-positioning defects. EMBO Rep. 2016;17:317–25. doi: 10.15252/embr.201541143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyre E, et al. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol. 2011;193:141–154. doi: 10.1083/jcb.201101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erickson H. Size and shape of protein molecules at the nanometer level determined by sedimentation gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpenter AE, et al. CellProfiler: image analysis software for identifying quantifying and cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decarreau J, Driver J, Asbury C, Wordeman L. Rapid measurement of mitotic spindle orientation in cultured Mammalian cells. Methods Mol Biol. 2014;1136:31–40. doi: 10.1007/978-1-4939-0329-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chozinski TJ, Halpern AR, Okawa H, Kim HJ, Tremel GJ, Wong RO, Vaughan JC. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods. 2016;13(6):485–8. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author on reasonable request.