Abstract
Self-renewing naïve mouse embryonic stem cells (mESCs) contain few mitochondria, which increase in number and volume at the onset of differentiation. KBP (encoded by Kif1bp) is an interactor of the mitochondrial-associated kinesin Kif1Bα. We found that TDH, responsible for mitochondrial production of acetylCoA in mESCs, and the acetyl-transferase GCN5L1 cooperate to acetylate Lys501 in KBP, allowing its recognition by and degradation via Fbxo15, an F-box protein transcriptionally controlled by the pluripotency core factors and repressed upon differentiation. Defects in KBP degradation in mESCs result in unscheduled increase in mitochondrial biogenesis, enhanced respiration and ROS production, and inhibition of cell proliferation. Silencing of Kif1Bα reverts the aberrant increase in mitochondria induced by KBP stabilization. Notably, upon differentiation, Kif1bp−/− mESCs display impaired expansion of the mitochondrial mass and form smaller embryoid bodies. Thus, KBP proteolysis limits the accumulation of mitochondria in mESCs to preserve their optimal fitness, whereas KBP accumulation promotes mitochondrial biogenesis in differentiating cells.
Introduction
Mitochondria are dynamic double-membrane organelles that take part in essential cellular functions, such as aerobic energy production, cell signaling, apoptosis, and calcium homeostasis1, 2. The total mitochondrial content of a cell and the individual activity of each mitochondrion differ from cell type to cell type due to variations in energy requirements. For example, naïve mouse embryonic stem cells (mESCs) – characterized by indefinite self-renewal and wide developmental potency - and differentiated cells vary in their mitochondrial content and activity. Although they perform both glycolysis and oxidative phosphorylation, naïve mESCs display a poor repertoire of mitochondria with immature morphology. The fine tuning of mitochondrial content and function in naïve mESCs is necessary, at least in part, to minimize the production of reactive oxygen species (ROS)3. Upon differentiation, mESCs leave their self-renewal state and expand their mitochondrial network in a process called mitochondrial biogenesis, which is required to adapt to changes in cellular metabolism, volume, and shape3–6. While it is well established that transcription factors, such as PGC-1α and NRF-2, contribute to mitochondrial biogenesis7, 8, little is known about the role of the ubiquitin-proteasome system in controlling the mitochondrial network of mESCs.
Among the targets of transcription factors essential for mESCs such as Oct-3/4 and Sox2, Fbxo15 gained attention as it was originally used to isolate induced pluripotent stem (iPS) cells9, 10. Fbxo15 encodes Fbxo15, one member of more than 70 mammalian F-box proteins, which are the substrate recognition subunits of the SCF (Skp1-Cul1-F-box protein) ubiquitin ligase complexes11, 12. Fbxo15 is unique among the F-box protein family due to its strict mESC expression; however, the function of Fbxo15 in mESCs has remained elusive.
Here, we describe Kif1-Binding Protein (KBP) as a substrate of Fbxo15 in mESCs. In humans, KBP is encoded by the KIAA1279 gene, which is mutated in the Goldberg-Shprintzen syndrome (GSS), an autosomal recessive disorder characterized by neuronal defects, such as microcephaly and mega-colon13. At the cellular level, this disorder is characterized by defects in axonal growth, which can be recapitulated by the expression of mutants of kif1bp (the ortholog of KIAA1279) in zebrafish14. KBP was originally characterized as an interactor of the kinesin Kif1Bα, a motor protein responsible for anterograde mitochondrial transportation15,16. Nonetheless, additional reports17,18 challenged the concept of KBP as a mediator of axonal mitochondrial transportation. Moreover, KBP expression is not limited to neurons, but is ubiquitous and is detected during all developmental stages, albeit at different levels.
Through biochemical, morphological, and functional analyses combined with genetics, we characterized the biological significance of KBP degradation, elucidating a critical control mechanism that limits mitochondrial mass expansion in self-renewing mESCs. The results of these studies are presented below.
Results
Fbxo15 binds acetylated KBP and targets it for degradation in mESCs
To identify substrates recognized by Fbxo15, we transiently expressed Strep-FLAG-tagged Fbxo15 and Strep-FLAG-tagged Fbxo21 (the latter used as control) in HEK293T cells. Fbxo15 and Fbxo21 complexes were then immunopurified for analysis by Multidimensional Protein Identification Technology (MudPIT), which revealed that Fbxo15 interacted with KBP, Cul1 and Skp1 (Supplementary Fig. 1a and Supplementary Table 1). In contrast, Fbxo21 interacted with Cul1 and Skp1, but not with KBP.
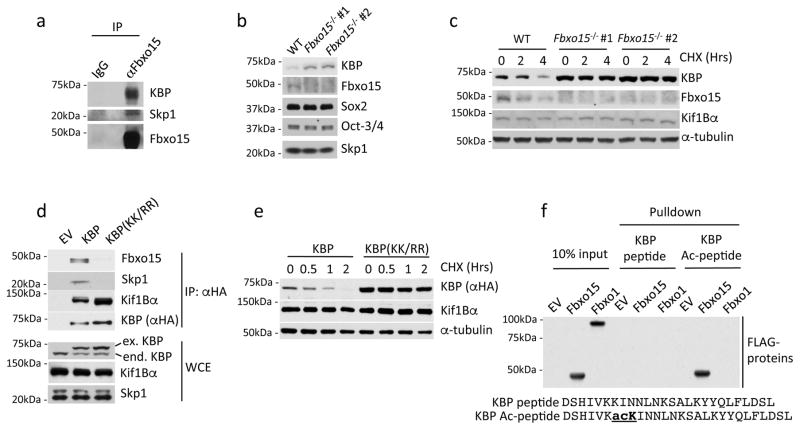
We verified the ability of Fbxo15 to specifically bind endogenous KBP in HEK293T using a collection of other F-box proteins as controls (Supplementary Fig. 1b). Moreover, the interaction between endogenous KBP and endogenous Fbxo15 was confirmed in naïve mESCs (Fig. 1a).
Fig. 1. KBP is an Fbxo15 substrate that is recognized in an acetylation-dependent manner.
a. Self-renewing naïve, mESCs were treated with MG132 for 4 hours prior to lysis. Lysates were immunoprecipitated with either an affinity-purified polyclonal antibody against Fbxo15 or an affinity-purified rabbit IgG, and analyzed by immunoblotting as indicated.
b. Protein extracts from wild type mESCs (WT) and Fbxo15−/− mESCs (two different clones) were immunoblotted for the indicated proteins.
c. Fbxo15−/− mESCs (two different clones) were treated with cycloheximide (CHX) for the indicated times, after which cell extracts were immunoblotted for the indicated proteins. This experiment was performed twice.
d. mESCs were infected with either an empty virus (EV) or lentiviruses expressing HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR). Whole cell extracts (WCE) were immunoprecipitated (IP) with an anti-HA resin, and proteins were immunoblotted as indicated.
e. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR), treated with cycloheximide (CHX) for the indicated times, and total cell lysates were analyzed by immunoblotting as indicated.
f. In vitro-translated FLAG-tagged Fbxo15 and FLAG-tagged Fbxo1 were incubated with beads coupled to the indicated peptides. Beads were extensively washed, and bound proteins were immunoblotted with an anti-FLAG antibody.
Unprocessed original scans of blots are shown Supplementary Fig. 8. Unless otherwise noted, experiments were performed at least three times.
The knockdown of Fbxo15 resulted in increased levels of KBP in mESCs maintained under self-renewing conditions (Supplementary Fig. 1c) to an extent comparable to that observed when mESCs were treated with the proteasome inhibitor MG132 (Supplementary Fig. 1d). The effects of Fbxo15 depletion and proteasome inhibition were not synergistic, indicating that they work in the same pathway. Moreover, silencing of Fbxo15 by two distinct siRNA oligos, used individually, prolonged the half-life of KBP (Supplementary Fig. 1e and, later, Fig. 2b). Similarly, mESCs in which both alleles of Fbxo15 were eliminated using a CRISPR/Cas9-dependent strategy (Supplementary Fig. 1f), also displayed elevated levels and increased stability of KBP (Fig. 1b–c). These results suggest that Fbxo15 targets KBP for proteasomal degradation in self-renewing mESCs.
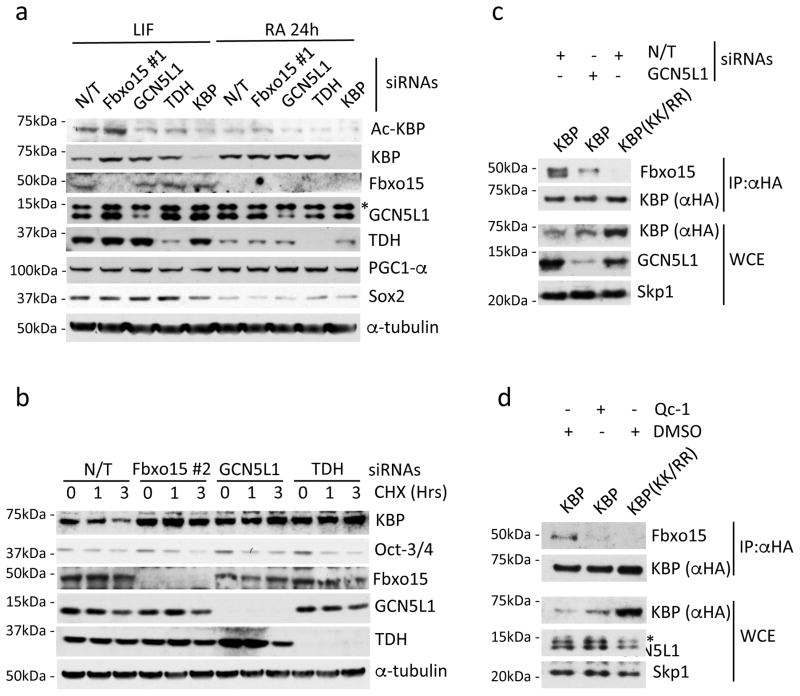
Fig. 2. GCN5L1 and TDH are necessary for the Fbxo15-mediated degradation of KBP in mESCs.
a. mESCs were transfected with either a nontargeting (N/T) siRNA or siRNAs to Fbxo15 (oligo #1), GCN5L1, TDH, or KBP. Cells were either maintained in LIF-containing medium or induced to differentiate for 24 hours (24h) after LIF withdrawal and exposure to retinoic acid (RA). Cells were then collected and lysed for immunoblotting as indicated. The asterisk denotes an unspecific band.
b. mESCs were transfected with a nontargeting (N/T) siRNA or siRNAs to Fbxo15 (oligo #2), GCN5L1, or TDH and treated with cycloheximide (CHX) for the indicated times. Cells were then collected and lysed for immunoblotting as indicated.
c. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and then transfected with the indicated siRNAs. Whole cell extracts (WCE) were immunoprecipitated (IP) with an anti-HA resin, and immunocomplexes were probed with antibodies to the indicated proteins.
d. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and then treated for 12h with either DMSO or Qc-1. Whole cell extracts (WCE) were immunoprecipitated (IP) with anti-HA resin, and immunocomplexes were probed with antibodies to the indicated proteins. The asterisk denotes an unspecific band.
Unprocessed original scans of blots are shown Supplementary Fig. 8. Unless otherwise noted, experiments were performed at least three times.
In most instances, F-box proteins recognize substrates only when one or two residues in their “degron” (i.e. the degradation motif) are phosphorylated11. To identify the degron in KBP that is required for its interaction with Fbxo15, we generated a number of human KBP deletion mutants and identified a highly conserved stretch of 24 amino acids that is necessary for Fbxo15 binding (Supplementary Fig. 2a–f). We mutated all potential phospho-sites in this region (Ser500, Ser512, Tyr516, Tyr517, and Ser523) to alanine, either individually or in different combinations, but no mutation impaired the binding of KBP to Fbxo15 (Supplementary Fig. 2a–f). Further characterization of this 24-amino acid region by alanine-scanning mutagenesis showed that mutation of Lys505 to alanine (K505A), but not Lys504Ala (K504A), abolished the binding of KBP to Fbxo15 (Supplementary Figs 2b–d and 2f-g). Mutation of Lys505 to arginine partially impaired the binding to Fbxo15 (perhaps due to a compensatory effect of Lys504), but mutations of Lys505 and Lys504 [KBP(KK/RR)] rendered KBP completely unable to interact with Fbxo15 (Supplementary Fig. 2f–g).
Human and mouse KBP share 91.5% identity and 96% similarity. The inability of KBP(KK/RR) to bind endogenous Fbxo15 was confirmed in mESCs that were infected with lentiviruses expressing either wild type mouse KBP or mouse KBP(KK/RR) (the latter generated by mutating Lys500 and Lys501, corresponding to human Lys504 and Lys505, to arginine) (Fig. 1d). Importantly, mouse KBP(KK/RR) displayed a much longer half-life than that of wild type KBP (Fig. 1e).
Fbxo15 appears to interact with the Kif1Bα-free pool of KBP, since it did not co-precipitate Kif1Bα (Supplementary Fig. 1b). Moreover, KBP mutants unable to interact with Fbxo15 still bound Kif1Bα efficiently (Supplementary Figs 2b and 2d). Similarly, the stable KBP(KK/RR) mutant co-precipitated more Kif1Bα from mESCs, compared to the less abundant wild type KBP (Fig. 1d). The latter experiment shows that when free KBP is not degraded through Fbxo15, the fraction of KBP associated with Kif1Bα increases.
We also analyzed the post-translational modifications of immunopurified KBP by mass spectrometry. This analysis revealed that Lys505 is acetylated in vivo (Supplementary Fig. 2h). Next, we used immobilized, synthetic peptides containing the candidate acetylated degron sequence to test its binding to Fbxo15. Whilst the immobilized peptide containing acetylated Lys505 efficiently pulled down Fbxo15 (but not Fbxo1 used as control), the corresponding non-acetylated peptide was unable to bind Fbxo15 (Fig. 1f). Collectively, our results indicate that acetylation of the KBP degron at Lys505 is required for its interaction with Fbxo15. To date, no other SCF substrate degron has been shown to contain an acetylated motif11.
GCN5L1- and TDH-mediated acetylation of KBP is required for its recognition by Fbxo15 in mESCs
To further investigate KBP acetylation in vivo, we generated an acetyl-specific antibody against the acetylated degron motif. This antibody recognized wild type KBP, but not KBP(KK/RR) (Supplementary Fig. 2i), confirming that KBP is acetylated in vivo on Lys505. Notably, depletion of Fbxo15 in mESCs induced an increase in the levels of KBP acetylated on Lys501 (the murine equivalent of Lys505) (Fig. 2a and Supplementary Fig. 3a), providing further support for the degradation of acetyl-KBP via Fbxo15.
To date, two mitochondrial acetyl-transferases have been reported: ACAT1 (AcetylCoA Acetyltransferase 1) and GCN5L1 (General Control Nonrepressed 5-Like 1). ACAT1 is involved in ketone body metabolism and is localized inside the mitochondrial matrix19, whereas GCN5L1 is also localized to the cytoplasm, suggesting that it may be present on the outer mitochondrial membrane20. Moreover, GCN5L1 plays a role in the remodeling and biogenesis of mitochondria and other organelles21, 22. Thus, we tested the hypothesis that GCN5L1 may acetylate KBP. Indeed, silencing of GCN5L1 in mESCs reduced the levels of acetyl-KBP (Lys501), whilst simultaneously increasing the total levels of KBP (Fig. 2a and Supplementary Fig. 3a; compare lanes 1 and 3) and prolonging its half-life (Fig. 2b). Importantly, knockdown of GCN5L1 in mESCs resulted in a dramatic reduction in the binding between KBP and Fbxo15 (Fig. 2c), supporting the hypothesis that GCN5L1 promotes this protein-protein interaction by acetylating KBP.
During mESC self-renewal, the production of acetyl-CoA (Coenzyme A) at the mitochondria requires the activity of TDH (l-Threonine Dehydrogenase)23, and it is assumed that this event is solely required for channeling acetyl-CoA into the tricarboxylic acid cycle and the synthesis of S-adenosylmethionine23, 24. Similar to Fbxo159, levels of TDH are downregulated in primed pluripotent stem cells and upon the induction of differentiation of mESCs23–26. To study the role of TDH in KBP acetylation and stability in mESCs, we depleted its expression by siRNA and observed both less acetylation of KBP on Lys501 and an increase in KBP levels and stability (Fig. 2a–b). Short treatment of mESCs with Qc-1, a pharmacological inhibitor of TDH that impairs the production of acetylCoA in mESCs27, resulted in an increase of both KBP protein level and half-life (Supplementary Fig. 3b–c). Moreover, we found that Qc-1 treatment abrogated the interaction between KBP and Fbxo15 in mESCs (Fig. 2d), suggesting that the activity of TDH and the consequent production of acetylCoA are a requirement for the recognition of KBP by Fbxo15. Importantly, Qc-1 treatment, Fbxo15 knockout, or knockdown of Fbxo15, GCN5L1 or TDH in mESCs did not activate the cell differentiation program, as shown by the unchanged levels of the pluripotency core factors Oct-3/4 and Sox2 (Fig. 1b, Fig. 2a–b, and Supplementary Fig. 3a–d). Accordingly, expression of KBP(KK/RR) did not cause any major transcriptional changes (including transcription of pluripotency core factors), as detected by RNA-sequencing (Supplementary Fig. 4 and Supplementary Table 2).
Fbxo15 and TDH are transcriptionally downregulated when mESCs are induced to differentiate9,23. We confirmed that the reduction of Fbxo15 and TDH protein levels occurs rapidly upon loss of pluripotency, with similar kinetics in cells induced to differentiate with either retinoic acid (RA) or neurobasal medium (NB) (Fig. 2a and Supplementary Figs 3a and 3d). Moreover, under these differentiation conditions, the levels of KBP were similar to the levels observed when Fbxo15, GCN5L1 or TDH were depleted from self-renewing mESCs (Fig. 2a and Supplementary Figs 3a and 3d). Collectively, our results show that the concerted activities of Fbxo15, GCN5L1, and TDH are required to keep KBP levels low during pluripotency, and suggest that the decrease in the levels of Fbxo15 and TDH at the onset of differentiation allows for the accumulation of KBP.
Failure to degrade KBP in mESCs enhances mitochondrial biogenesis in a Kif1Bα–dependent manner
Next, we investigated the biological significance of KBP degradation in self-renewing mESCs, by expressing either wild type KBP or KBP(KK/RR) at near-to-physiologic levels (see Fig. 1d). KBP has been proposed to mediate mitochondrial transport and microtubule dynamics16,14. We found no difference in mitochondrial motility between mESCs expressing wild type KBP or KBP(KK/RR) (Fig. 3a). Microtubules exert an active role in mitochondrial network remodeling28, and the KBP-Kif1Bα complex has been reported to act as a bridge between microtubules and mitochondria15. Accordingly, using 3D reconstructions, we observed that the expression of KBP(KK/RR) significantly enhanced the co-localization between mitochondria and β-tubulin (Fig. 3b). Similar results were obtained when Fbxo15 expression was depleted or TDH was briefly pharmacologically inhibited (Supplementary Fig. 5a–b).
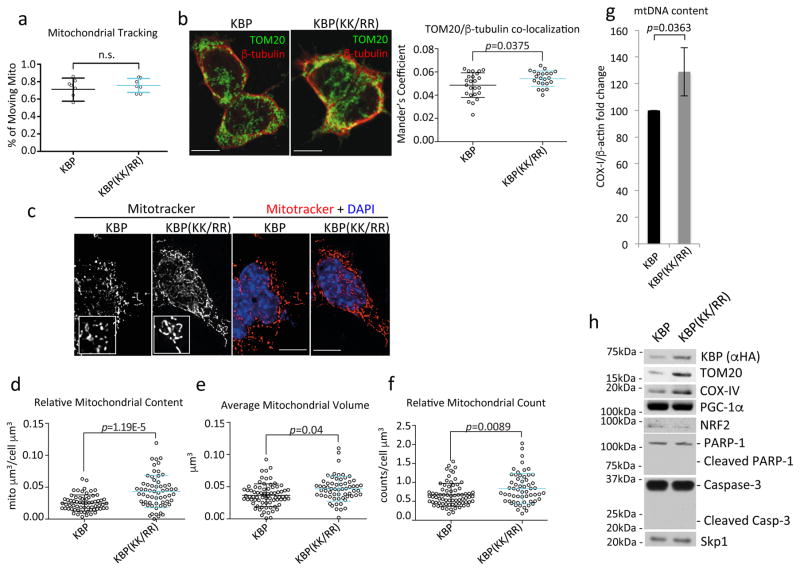
Fig. 3. Expression of a stable mutant of KBP in mESCs induces mitochondrial biogenesis.
a. mESCs stably infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) were transfected with mitochondria-targeted GFP and monitored with live-cell imaging. The graph shows the percentage of moving mitochondria per cell. n=7 independent field acquisitions. p value was calculated by unpaired t-test. Error bars indicate +/−SD. This experiment was performed once.
b. Cells as in (a) were immuno-stained for TOM20 and β-tubulin. Microtubule-mitochondria co-localization was detected by confocal microscopy. Scale bar = 15μm. In the right panel, the Manders’ Overlap Coefficient is shown. n=24 and 22 [KBP and KBP(KK/RR), respectively]. p value was calculated by t-test. Error bars indicate +/−SD.
c–f. Cells as in (a) were stained with Mitotracker and DAPI. 3D images were collected through widefield microscopy and reconstructed by 3D deconvolution. Panel c shows representative maximum intensity projections (MIPs) of the mitochondrial network of KBP and KBP(KK/RR) expressing mESCs. Scale bar = 15 μm. KBP and KBP(KK/RR) expressing mESCs were compared for their relative mitochondrial content (d), average mitochondrial volume (e), and number of mitochondria per cell (f). n=65 and 60 [KBP and KBP(KK/RR), respectively]. p values were calculated by unpaired t-test. Error bars indicate +/−SD.
g. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR). After completing the selection process, total cellular DNA was extracted and mtDNA copy number was evaluated by qPCR normalized using β-actin DNA copy number. mtDNA amount in mESCs expressing wild type KBP was set as 100 and the graph shows the fold change. p value was calculated by unpaired t-test. Error bars indicate +/−SD. n= 4 biologically independent experiments (see Supplementary Table 6).
h. mESCs stably infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) were collected and lysed for immunoblotting as indicated. Unprocessed original scans of blots are shown Supplementary Fig. 8.
Unless otherwise noted, experiments were performed at least three times.
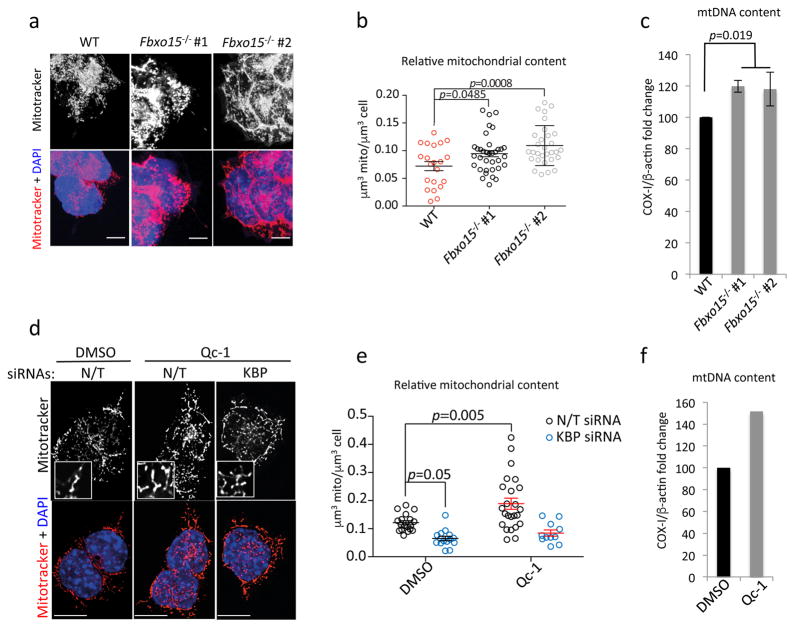
Upon induction of differentiation, mESCs undergo mitochondrial biogenesis. This is required to allow cells to adapt to the metabolic switch in favor of oxidative phosphorylation induced by differentiation3, 5, 29. We confirmed that mESCs expanded their mitochondrial content 24 hours after the induction of differentiation (Supplementary Fig. 5c). This increase occurs before the induction of PGC-1α, a master regulator of mitochondrial biogenesis (Fig. 2a). Significantly, reconstructing three dimensional images by 3D deconvolution, we found that, compared to wild type KBP, expression of KBP(KK/RR) in self-renewing mESCs induced a significant increase in the number of mitochondria per cell, the mitochondrial volume per cell, and the average volume of individual mitochondria (Fig. 3c–f). Moreover, mESCs expressing KBP(KK/RR) displayed an increase in mitochondrial DNA and specific mitochondrial proteins (i.e. TOM20 and COX-IV) compared to mESCs expressing wild type KBP (Fig. 3g–h). Similar results were obtained by knocking out Fbxo15 (Fig. 4a–c), inhibiting TDH in self-renewing mESCs (Fig. 4d–f), or silencing Fbxo15 (Supplementary Fig. 5d–g). The increase in mitochondrial content was dependent on KBP expression, as the knockdown of KBP reverted the effect obtained by silencing Fbxo15 (Supplementary Fig. 5d–e) or inhibiting TDH (Fig. 4d–e).
Fig. 4. Failure to degrade KBP in mESCs induces mitochondrial biogenesis.
a–b. Wild type mESCs (WT) and Fbxo15−/− mESCs (two different clones) were maintained in self-renewing conditions. Cells were stained with Mitotracker and DAPI prior to fixation and then analyzed by confocal microscopy. Panel a shows the MIPs of representative cells (scale bar = 15 μm). Panel b shows the quantification of the relative mitochondrial content. n=21, 35, and 30 cells (WT, Fbxo15−/− #1 and Fbxo15−/− #2, respectively). p value was calculated by one-way ANOVA. Error bars indicate +/−SD.
c. Wild type mESCs (WT) and Fbxo15−/− mESCs (two different clones) were maintained in self-renewing conditions. Total cellular DNA was extracted and mtDNA copy number was evaluated by qPCR normalized using b-actin DNA copy number. mtDNA amount in wild type mESCs was set as 100 and the graph shows the fold change. p value was calculated by one-way ANOVA. Error bars indicate +/−SD. n= 3 biologically independent experiments. (see Supplementary Table 6).
d–e. mESCs were transfected with either a nontargeting (N/T) siRNA or an siRNA to KBP, and then incubated with either DMSO or Qc-1 for 12 hours. Cells were stained with Mitotracker and DAPI prior to fixation and then analyzed by confocal microscopy. Panel d shows the MIPs of representative cells (scale bar = 15 μm). Panel e shows the quantification of average mitochondrial content. n=19, 15, 24, and 11 cells (N/T siRNA+DMSO, KBP siRNA+DMSO, N/T siRNA+Qc-1, and KBP siRNA+Qc-1, respectively). This experiment was performed twice. p value was calculated by two-way ANOVA. Error bars indicate +/−SD.
f. mESCs were treated with DMSO or Qc-1 for 12 hours. Total cellular DNA was extracted and mtDNA copy number was evaluated by qPCR normalized using β-actin DNA copy number. mtDNA amount in mESCs treated with vehicle (DMSO) was set as 100 and the graph shows the fold change. n= 2 biologically independent experiments with three technical replicates each (see Supplementary Table 6).
Importantly, Qc-1 treatment of mESCs stably transduced with wild type KBP induced both an accumulation of exogenous KBP (Supplementary Fig. 5h) [in agreement with what is observed with endogenous KBP (Supplementary Fig. 3b–c)] and an increase in mitochondrial number similar to what is observed in mESCs that were induced to differentiate (Supplementary Fig. 5i–j). In contrast, in mESCs expressing KBP(KK/RR), Qc-1 treatment failed to increase the already high levels of KBP(KK/RR) (Supplementary Fig. 5h). Moreover, KBP(KK/RR) expressing mESCs displayed high mitochondrial content regardless of their Qc-1 treatment or differentiation status (Supplementary Fig. 5i–j).
The stabilization of KBP did not affect the rate of fission and fusion events (Supplementary Fig. 6), suggesting that the observed increase in mitochondrial mass is due to de novo biogenesis.
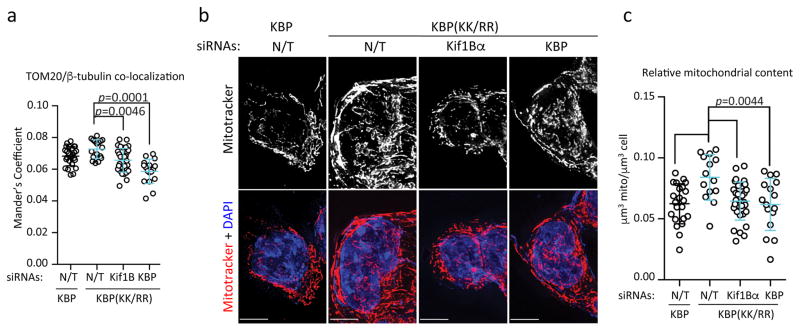
As KBP stabilization results in its enhanced interaction with Kif1Bα (Fig. 1d) and increased mitochondria-microtubule association (Fig. 3a and Supplementary Fig. 5a–b), we silenced Kif1Bα, which connects KBP to microtubules, in mESCs expressing KBP(KK/RR). Notably, depletion of Kif1Bα reversed the enhancement of mitochondria-microtubule co-localization (Fig. 5a) and the increased mitochondrial biogenesis induced by KBP(KK/RR) (Fig. 5b–c), showing that the effects of KBP on mitochondria depend on Kif1Bα.
Fig. 5. The mitochondrial biogenesis induced by KBP stabilization depends on Kif1Bα.
a. Following infection with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR), mESCs were transfected with a nontargeting (N/T) siRNA or siRNAs to Kif1Bα or KBP, and immunostained for TOM20 and α-tubulin. Microtubule-mitochondria co-localization was detected by confocal microscopy and the Manders’ Overlap Coefficient is shown. n=26, 17, 28, and 17 cells [KBP+N/T siRNA, and KBP(KK/RR)+N/T, or Kif1Bα or KBP siRNA, respectively]. p value was calculated by one-way ANOVA. Error bars indicate +/−SD.
b–c. Cells treated as in (a) were stained with Mitotracker and DAPI. 3D images were collected through widefield microscopy and reconstructed by 3D deconvolution. Panel b shows representative MIPs of the mitochondrial network of KBP and KBP(KK/RR)-expressing mESCs, with or without depletion of Kif1Bα or KBP (scale bar = 15μm). Panel c shows quantification of average mitochondrial content per μm3. n=22, 14, 31, and 16 cells [KBP+N/T siRNA, KBP(KK/RR)+N/T, or Kif1Bα or KBP siRNA, respectively]. p value was calculated by one-way ANOVA. Error bars indicate +/−SD.
KBP degradation preserves the fitness of mESCs
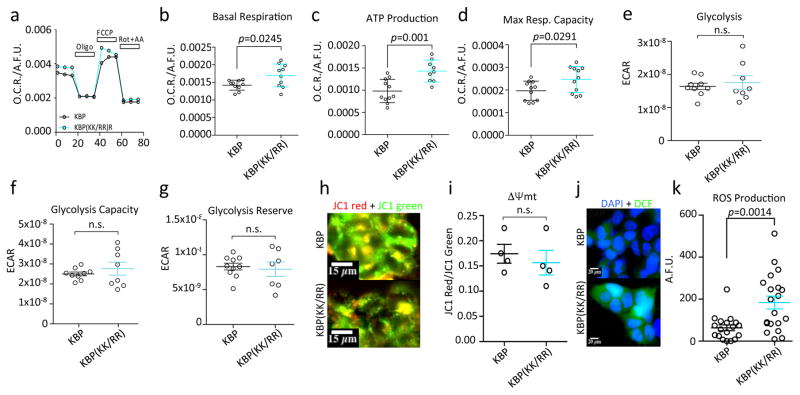
To evaluate the performance of mitochondria in KBP(KK/RR) expressing mESCs, we measured critical cellular bioenergetic parameters. We found that the increase in mitochondrial mass was accompanied by an increase in respiration and ATP production, without affecting glycolysis (Fig. 6a–g). We also measured the differential membrane potential (Δψmt) at the mitochondria, and found that wild type KBP and KBP(KK/RR) expressing mESCs did not differ in Δψmt (Fig. 6h–i) and, therefore, had comparable individual mitochondrion activity. Taken together, these data show that KBP(KK/RR) expressing mESCs perform increased mitochondrial respiration due to an overall higher mitochondrial mass and not a metabolic switch. Importantly, KBP stabilization promoted production of ROS (Fig. 6j–k).
Fig. 6. Effects of KBP accumulation on mitochondrial respiration.
a–d. mESCs infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) were analyzed for their mitochondrial respiratory capacity. Panel a shows cell representative kinetics of oxygen consumption rate (OCR). Panel b compares cellular basal respiration. Panel c reports ATP synthase-dependent mitochondrial OCR. Panel d shows the maximal respiratory capacity. p values were calculated by unpaired t-test. Error bars: S.E.M.; n=10 independent cell culture dishes.
e–g. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR). Cells underwent a real-time analysis of glycolytic through the measurement of the extracellular acidification rate after the induction of acute metabolic stress. p value was calculated by unpaired t-test. Error bars = S.E.M; n=8 independent cell culture dishes.
h–i. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and the Δψmt was measured using the ratiometric Δψmt sensitive dye JC1. Panel h shows representative images. p value was calculated by unpaired t-test. Error bars = S.E.M; n = 4 independent cell culture dishes (see Supplementary Table 6). Scale bars = 15 μm.
j–k. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and analyzed for ROS production using the peroxide sensitive dye dichlorofluorescein (DCF). Panel j shows representative images. p value was calculated by unpaired t-test. Error bars = S.E.M. n = 18 and 20 independent cell culture dishes [KBP and KBP(KK/RR), respectively]. Scale bars = 20 μm.
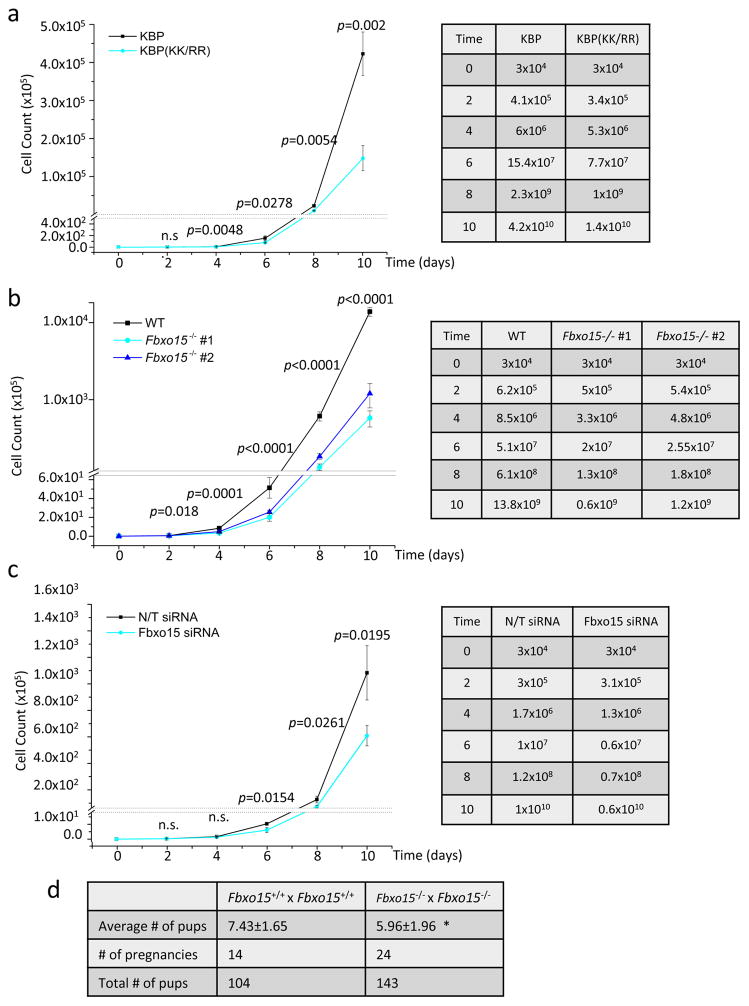
Based on the latter results, we assessed whether impairing the degradation of KBP affected cell growth rate. We found that KBP(KK/RR) expressing mESCs proliferated significantly slower than wild type KBP expressing mESCs (Fig. 7a).
Fig. 7. In vivo and in vitro effects of KBP stabilization and loss of Fbxo15 on mESC growth.
a. mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and plated at a density of 3×104 cells/well. Every 48 hours, cells were trypsinized, counted, and re-plated at a 1:10 dilution. No changes in viability and cell death were detected in the two groups by Annexin-V/PI analysis [0.25% +/− 0.127 for wild type KBP and 0.29% +/− 0.255 for KBP(KK/RR); n=2 biologically independent experiments (see Supplementary Table 6)]. Right panel summarizes the average of each cell count. p value was calculated by unpaired t-test. Error bars indicate +/−SD. n=3 biologically independent experiments (see Supplementary Table 6).
b. Wild type mESCs (WT) and Fbxo15−/− mESCs (two different clones) were plated at a density of 3×104 cells/well. Every 48 hours, cells were trypsinized, counted, and re-plated at a 1:10 dilution. Cell counts were performed using a Vi-CELL, an automated cell counter that simultaneously assesses cell viability. No changes in viability were detected in the three groups. Right panel summarizes the average of each cell count. p value was calculated by one-way ANOVA. Error bars indicate +/−SD. n=3 biologically independent experiments (see Supplementary Table 6).
c. mESCs were transfected with either a nontargeting (N/T) siRNA or an siRNA to Fbxo15 and plated at a density of 3×104 cells/well. Every 48 hours, cells were trypsinized, counted, and re-plated at a 1:10 dilution with a further round of siRNA transfection. Cell counts were performed using a Vi-CELL, an automated cell counter that simultaneously assesses cell viability. No changes in viability were detected in the two groups. Right panel summarizes the average of each cell count. p value was calculated by unpaired t-test. Error bars indicate +/−SD. n=3 biologically independent experiments (see Supplementary Table 6).
d. Fbxo15+/+ and Fbxo15−/− mice were mated and the average offspring per litter was compared. n=14 and 24 pregnancies. *= p=0.0242, calculated with unpaired t-test (see Supplementary Table 6).
The observation that Fbxo15 knockout and Fbxo15 silencing significantly reduced the proliferation of mESCs (Fig. 7b–c) corroborated our finding. This reduction in the growth rates of Fbxo15 is compatible with the observation that the average litter size of Fbxo15−/− mice is significantly smaller than that of Fbxo15+/+ mice (Fig. 7d)
KBP accumulation is necessary for mitochondrial biogenesis during differentiation
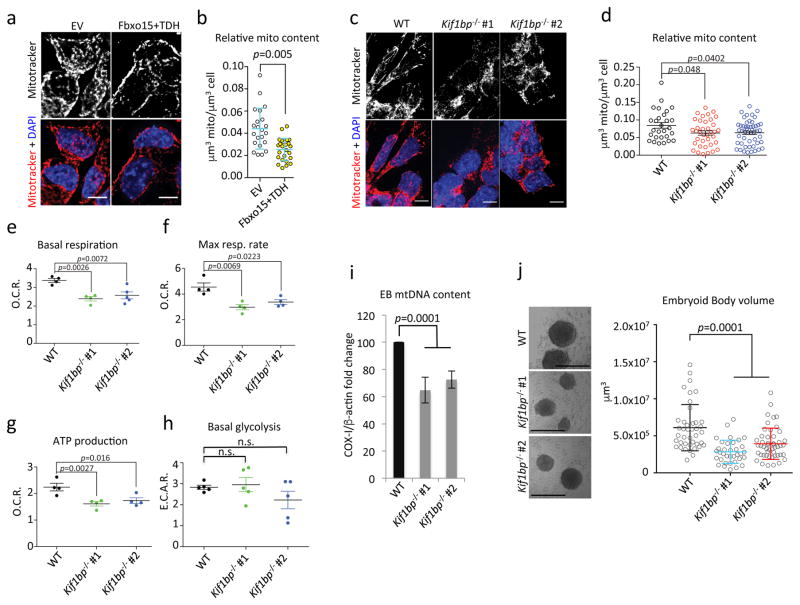
Altogether, the experiments in Fig. 3c–f, Fig. 4a–b, Fig. 4d–e and Supplementary Fig. 5 demonstrate that the defects in KBP degradation in naïve mESCs result in the acquisition of a mitochondrial content that is typically observed at the beginning of differentiation. Conversely, when Fbxo15 and TDH were overexpressed in mESCs undergoing differentiation, a statistically significant reduction of mitochondrial content was observed (Fig. 8a–b). This result suggests that Fbxo15 and TDH are downregulated upon the initiation of differentiation to allow the accumulation of KBP and the expansion of the mitochondrial network. To further substantiate this hypothesis, we eliminated both alleles of Kif1bp (the gene encoding KBP) in mESCs employing a CRISPR/Cas9-dependent strategy (Supplementary Fig. 7a–b). We found that in 2 out 2 clones, the absence of KBP induced a defect in both respiration and differentiation. Specifically, we observed that, compared to wild type cells, KBP-null cells induced to differentiate displayed reduced mitochondrial mass, mitochondrial DNA, oxygen consumption, and ATP production, without a compensatory increase in basal glycolysis (Fig. 8c–i). Notably, the absence of KBP induced a reduction in the volume of embryoid bodies and a change in the transcriptional signature (Fig. 8j and Supplementary Tables 3 and 4), likely due to an energetic disadvantage.
Fig. 8. Ectopic expression of Fbxo15 and TDH reduces the mitochondrial content in differentiating mESCs, and Kif1bp knockout impairs mitochondrial biogenesis and differentiation.
a–b. mESCs were transfected with either empty (EV) or Fbxo15 and TDH expressing vectors, and induced to differentiate for 24 hours. Panel a shows the MIPs of representative cells (scale bar = 15 μm). Panel b shows the average mitochondrial content. n=22 and 24 cells (EV and Fbxo15+TDH, respectively). This experiment was performed twice. p value was calculated by unpaired t-test. Error bars indicate +/−SD.
c–d. Wild type (WT) and Kif1bp−/− mESCs (two different clones) were induced to differentiate with retinoic acid for 48 hours. Cells were stained with Mitotracker and DAPI prior to fixation and then analyzed by confocal microscopy. Panel c shows the MIPs of representative cells (scale bar = 15 μm). Panel d shows the average mitochondrial content. n=28, 38, and 52 cells (WT, Kif1bp−/− #1, and Kif1bp−/− #2, respectively). p value was calculated by one-way ANOVA. Error bars indicate +/−SD.
e–h. Cells treated as in (c) were analysed for their mitochondrial respiratory capacity. Panel e compares cellular basal respiration. Panel f shows the maximal respiratory capacity. Panel g reports ATP synthase-dependent mitochondrial OCR. Panel h shows glycolytic capacity. p value was calculated by one-way ANOVA. Bars: S.E.M.; n=4 or 5 independent cell culture dishes (see Supplementary Table 6).
i. Cells as in (c) were induced to generate embyoid bodies (EBs). After 4 days, total cellular DNA was extracted and mtDNA copy number was evaluated by qPCR normalized using β-actin DNA copy number. mtDNA amount in wild type mESCs was set as 100 and the graph shows the fold change. p value was calculated by one-way ANOVA. Error bars indicate +/−SD. n= 4 independent cell culture dishes (see Supplementary Table 6).
j. EBs were generated as in i and volume was calculated. Left panel shows representative images (scale bar = 400 μm). n=42, 35, and 49 embryoid bodies (WT, Kif1bp−/− #1, and Kif1bp−/− #2, respectively). p value was calculated by one-way ANOVA. Error bars indicate +/−SD.
Discussion
In this study, we have described a regulatory network that controls mitochondrial biogenesis through the proteolysis of KBP in mESCs. Defects in KBP degradation do not affect the expression of the pluripotency core factors, supporting a model in which the observed effects on mitochondrial biogenesis are due to a mechanism that acts locally at the mitochondria, independently of transcriptional changes. Since stabilized KBP binds Kif1Bα and promotes the association of mitochondria to microtubules, we speculate that this association provides a microtubule track along which mitochondria grow. In addition, the motor activity of Kif1Bα may increase the mechanical forces applied to mitochondria, which, in turn, may promote the growth of these organelles through a mechanism that is reminiscent of that acting at the level of the muscle fiber in which the application of force or mechanical tension is responsible for increasing the number of sarcomeres. In support of this proposed mechanism, Kif1Bα silencing reverts the phenotypes induced by the stabilization of KBP. A role of the microtubule network in mitochondrial biogenesis was previously proposed30 and our findings integrate and expand those observations.
Importantly, we found that the mitochondrial-specific production of acetylCoA by TDH is required for GCN5L1 to acetylate KBP, which allows its recognition by Fbxo15 (see model in Supplementary Fig. 7c). In this context, mitochondrial outsourced acetylCoA, rather than being a mere substrate for protein acetylation, could be considered analogous to an intracellular messenger that is locally sensed by the GCN5L1-Fbxo15 machinery. In self-renewing naïve mESCs, this machinery controls KBP levels and acts as a gatekeeper of the mitochondria steady state, thus, reducing ROS production and preserving the optimal fitness of mESCs. When naïve mESCs are induced to differentiate, levels of Fbxo15 and TDH decrease, acetylation of KBP on Lysine 501 is inhibited, and KBP levels increase (Supplementary Fig. 7d).
In vivo, around the time of implantation, derivative cells of the inner cell mass - the embryonic counterpart of cultured naïve mESCs - enter a so-called primed pluripotent state, which is characterized by profound transcriptional rearrangements that include the downregulation of Fbxo15 and TDH26. At this developmental stage, pluripotent cells display a more expanded mitochondrial network31, in agreement with our model; yet, their oxidative phosphorylation is inhibited, likely due to a downregulation of key components of the respiratory chain31, 32.
Loss of Fbxo15 in mice does not induce major defects (Ref. 9 and http://www.mousephenotype.org/data/genes/MGI:1354755#section-associations). This could be due to compensatory events occurring in the Fbxo15−/− embryo and/or the fact that the time window of naïve pluripotency in which Fbxo15 is expressed is very limited (~1 day) 33, and animals are fine once they have passed this bottleneck. This latter hypothesis is in agreement with the observed cell proliferation defects of Fbxo15−/− mESCs, as well as with the reduced litter size of Fbxo15−/− mice compared to Fbxo15+/+ animals. Follow up work might clarify whether and how loss of Fbxo15 plays a role in early development.
In conclusion, we propose that KBP degradation is necessary to keep mitochondrial biogenesis in check during self-renewal. In contrast, the accumulation of KBP occurring upon the differentiation of mESCs is functional to their increase in mitochondrial mass, which allows proper differentiation.
Methods
Cell Culture
mESCs were kindly provided by Dr. Iannis Aifantis and maintained on gelatin coated tissue culture plates with LIF/2i as described in34. ES cells were trypsinized and passaged every 48h and periodically checked for the expression of pluripotency markers. mESCs differentiation was induced either by LIF/2i withdrawal and supplementation with 10 μM retinoic acid (RA) on gelatin-coated plates, or by seeding cells on non-adherent petri dishes as in34. Where indicated, Qc-1 was administered at the concentration of 10 μM for 12 hours. HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. HEK293T cells were not authenticated, as they were not used in experiments designed for the functional characterization of KBP degradation. mESCs were not authenticated. Cells were periodically screened for Mycoplasma contamination. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample.
Transient transfections
siRNA duplexes (see Supplementary Table 5 for sequence details) were transfected into mESCs using RNAiMAX reagent (Life Technologies) according to the manufacturer’s instructions. To achieve satisfactory RNAi-mediated depletion, siRNA duplexes were transfected over 2 or 3 rounds. Plasmids were transfected into mESCs using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. HEK293T cells were always transduced with vectors expressing human gene sequences, while mESCs were transfected with their mouse counterparts. HEK293T cells were transfected using the calcium phosphate method, as described35.
Purification, MudPIT, and PTM Analyses
HEK293T cells were transfected with constructs encoding Strep-FLAG-tagged human Fbxo15, Strep-FLAG-tagged human Fbxo21, or Strep-FLAG-tagged human KBP(K504G) (the latter at the C-terminus). Forty-eight hours after transfection, cells were collected and lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 0.5% NP-40, plus protease and phosphatase inhibitors). Proteins were immunopurified with Streptavidin beads (IBA) and, after extensive washing, eluted by competition with biotin. The eluate was then subjected to a second immunopurification with anti-FLAG M2 agarose beads (Sigma), prior to elution by competition with a 3x-FLAG peptide (Sigma). The final eluate was then precipitated with TCA. TCA-precipitated proteins were urea-denatured, reduced, alkylated and digested with endoproteinase Lys-C (Roche) and modified trypsin (Roche), as described in36, 37. Peptide mixtures were loaded onto 100 μm fused silica microcapillary columns packed with 5-μm C18 reverse phase (Aqua, Phenomenex), strong cation exchange particles (Partisphere SCX, Whatman), and reverse phase38. Loaded microcapillary columns were placed in-line with a Quaternary Agilent 1100 series HPLC pump and a LTQ linear ion trap mass spectrometer equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Fully automated 10-step MudPIT runs were carried out on the electrosprayed peptides, as described in37. Tandem mass (MS/MS) spectra were interpreted using SEQUEST39 against a database of 61430 sequences, consisting of 30552 human proteins (downloaded from NCBI on 2008-03-04), 177 usual contaminants and, to estimate false discovery rates, 30712 randomized amino acid sequences derived from each non-redundant protein entry. Peptide/spectrum matches were sorted and selected using DTASelect40 with the following criteria set: spectra/peptide matches were only retained if they had a DeltCn of at least 0.08 and a minimum XCorr of 1.8 for singly-, 2.0 for doubly-, and 3.0 for triply-charged spectra. In addition, peptides had to be fully-tryptic and at least 7 amino acids long. Combining all runs, proteins had to be detected by at least 2 such peptides, or 1 peptide with 2 independent spectra. Under these criteria the final FDRs at the protein and spectral levels were 1.6% and 0.13%±0.05, respectively. Peptide hits from multiple runs were compared using CONTRAST40. To estimate relative protein levels, Normalized Spectral Abundance Factors (NSAFs) were calculated for each detected protein, as described in37, 41, 42.
Biochemical Methods
Extract preparation, immunoprecipitation, and immunoblotting have been previously described43, 44.
CRISPR genome editing
To generate Kif1bp and Fbxo15 knockout mESCs, optimal gRNA target sequences closest to the genomic target sites were designed using the Benchling CRISPR Genome Engineering tool. Kif1b and Fbxo15 gRNA target sequences (see Supplementary Table 5) were cloned into the pSpCas9(BB)-2A-GFP (PX458), a gift from Feng Zhang (Addgene plasmid # 48138)45. mESCs were seeded into 10cm dishes at a ~70% confluency, and transfected with 5μg of the appropriate gRNA-containing PX458 plasmid, using Lipofectamine 3000 (Life Technologies). The transfection was performed according to the manufacturer’s recommended protocol, using a 2:1 ratio of Lipofectamine:DNA. Two days after transfection, GFP positive cells were sorted using the Beckman Coulter MoFlo XDP cell sorter (100 μm nozzle), and 15,000 cells were plated on a 15cm dish. 8–10 days later, single mESC clones were picked, trypsinized in 0.25% Trypsin-EDTA for 5 minutes, and plated into individual wells of a 96-well plate for genotyping. Genomic DNA was collected using QuickExtract (Epicentre). Genotyping PCRs were performed with MangoTaq DNA Polymerase (Bioline), using primers surrounding the genomic target site. The resulting PCR products were purified and sequenced to determine the presence of an insertion or deletion event. To further validate the mutational status of candidate clones, the PCR products were subjected to TOPO-TA Cloning (Invitrogen), and sequenced in order to distinguish the amplified products of distinct alleles. Fifty bacterial colonies for each TOPO-TA cloning reaction were sequenced and aligned to the corresponding wild-type template in Benchling. Clones positive to insertion or deletion events were validated also by western blot.
Plasmids
Mouse KBP cDNA was amplified by PCR using a cDNA library generated from mESCs and then subcloned into pLenti-III-UbC (ABM). Mouse Fbxo15 and TDH were amplified by PCR using a cDNA library generated from mESCs and then subcloned into pHAGE2-EF1α46. Human Fbxo15, Fbxo21 and KBP cDNAs were amplified by PCR using a cDNA library generated from HEK293T cells. cDNAs were then subcloned into pcDNA 3.1 (Life Technologies). A plasmid expressing a mitochondrial-targeted GFP (mtGFP) under the MSCV promoter was purchased from ABM. Human and mouse KBP mutants were generated by either using the QuikChange Site-directed Mutagenesis kit (Stratagene) or by standard PCR procedures. All cDNAs were sequenced.
Stable infections
Lentiviruses were generated in HEK293T cells and concentrated as in46. Infections were carried as in47. 24 hours after the infection, mESCs were selected with puromycin (1.5 μg/mL).
Antibodies
The anti-acetyl-KBP antibody was generated immunizing rabbits with the peptide C-DSHIVKacKINNLNKSALKYYQLFLDSL after its conjugation with KLH. The following rabbit polyclonal antibodies were used: Fbxo15 (M-291; Santa Cruz Biotechnology; 1:500 diluition), KBP (kindly provided by Rainer Lammers, Tubingen, Germany; 1:5000 diluition), Skp1 (H-163; Santa Cruz Biotechnology; 1:1000 diluition), Oct-4 (#PA5-27438, Thermo Fisher; 1:2000 diluition), Sox2 (#48-1400, Thermo Fisher; 1:2000 diluition), GCN5L1 (kindly provided by Michael Sack, NIH; 1:1000 diluition), PGC-1α (ST1204, Calbiochem; 1:1000), Caspase-3 (#9662, Cell Signaling Technologies; 1:1000 diluition), NRF-2 (#12721, Cell Signaling Technologies; 1:1000 diluition), PARP-1 (#9542, Cell Signaling Technologies; 1:1000 diluition), HA (A190-108A, Bethyl; 1:5000 diluition) and FLAG (Sigma; 1:5000 diluition). The following mouse monoclonal antibodies were used: TOM20 (#612278, BD; 1:2000 diluition), α-tubulin (T5168; Sigma; 1:5000 diluition), and COX-IV (4D11-B3-E8, Cell Signaling Technologies; 1:2000 diluition). The following goat polyclonal antibodies were used: TDH (D-14; Santa Cruz Biotechnology; 1:1000 diluition) and Kif1Bα (L-20; Santa Cruz Biotechnology; 1:1000 diluition).
Immunofluorescence microscopy
For indirect immunofluorescence staining, mESCs were grown on StemXVivo matrix (RD Systems) coated glass coverslips and then fixed with paraformaldehyde 4% for 10 minutes at room temperature. The cells were permeabilized with PBS/0.05% Triton X-100 for 10 minutes and blocked for 1 hr in PBS/0.05% Triton X-100 containing 3% BSA prior to incubation with primary antibodies. Secondary antibodies were Alexa Fluor conjugated (Life Technologies). DAPI was used to counterstain DNA. Slides were mounted with ProLong Gold Antifade Mountant (Life Technologies). MitoTracker Red (Life Technologies) was incubated 30 minutes prior to fixation. Images were acquired at the Nyquist rate in x,y and z axes by a LSM 800 equipped with Zen software (Zeiss), or by a motorized custom epifluorescence microscope (Crisel Instruments). Three dimensional co-localizations between mitochondria and microtubules were quantified according to the Manders method using a custom-made Fiji macro as in48. To evaluate the extent of spontaneous mitophagy, cells were incubated with Mitotracker (Life Technologies) for 30′ in growth medium, washed in PBS and incubated with Lysotracker (Life Technologies) for 30′ in PBS +15% serum. Live cells were scanned with the Cytation 5 plate reader (Biotek). Co-localization coefficients were calculated in ImageJ using the JACoP plugin.
Apoptosis assay
Annexin-V/PI staining was performed as previously described49. Two-dimensional flow cytometry was performed to detect Alexa-488-conjugated Annexin-V and PI using a LSRII flow-cytometer (BD Biosciences) and FlowJo software (Tree Star Inc., Stanford).
Analysis of mitochondrial morphology
Three dimensional images were reconstructed by 3D deconvolution using experimentally measured PSF and the 3D iterative deconvolution plug-in for Fiji (available at: http://fiji.sc/Iterative_Deconvolve_3D). After reconstruction, cells were manually demarcated using the β-tubulin signal which defines the cytosol limit. For each cell, mitochondria and nuclei were segmented automatically using the moment algorithm. Then object amount and size were obtained using the 3D object counter plug-in for Fiji (available at: http://fiji.sc/3D_Objects_Counter).
Quantification of mitochondrial DNA content
Total DNA was extracted from cells using DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instruction and used as template for qPCR, using primers for COX-I (to assess the copies of mtDNA) and to β-actin (to normalize the mtDNA content per cell) (see Supplementary Table 5 for primer sequences). The reaction was performed with a Roche480 thermal cycler, by using the Absolute Blue QPCR SYBR Green Mix (Thermo Scientific).
Mitochondrial Tracking and mitochondrial dynamics analyses
mESCs were stained with Mitotracker Red according to the manufacturer’s instructions, and then imaged with a Nikon Swept Field confocal equipped with CFI Plan Apo VC60XH objective (n.a. 1.4) and an Andor DU885 EM-CCD camera. Coverslips were placed in a chamber with controlled temperature, CO2 and humidity; z-stacks were acquired by 73 planes with 0.3 μm distance, to allow acquisition of the whole cell. Images were acquired every 3 seconds for a total of 30 seconds. Mitochondrial movement was tracked through the Imaris software (Bitplane). Out of the total amount of mitochondria, the proportion of moving mitochondria was calculated. To assess the occurrence of events of mitochondrial fission and fusion, we applied an algorithm to detect a fusion event as an increase in the object volume larger then the smallest object detectable; inversely, a fission event was considered as a reduction in object volume larger then the smallest object detectable.
XF bioenergetic analysis
OCRs in mESCs were measured using the Seahorse XF96 bioanalyzer (Seahorse Biosciences) according to the manufacturer’s protocols. Cells were seeded in a XF96 microplate at a density of 15,000 cells per well and allowed to attach. The following day or 48 hours after the induction of differentiation, the medium was exchanged, where indicated, with 175 μl unbuffered XF assay media at pH 7.4 (Seahorse Biosciences) supplemented with 5.5 mM glucose (Sigma), 1 mM sodium pyruvate and 1 mM glutamine, or with 175 μl unbuffered XF assay media at pH 7.4 (Seahorse Biosciences) without glucose, sodium pyruvate or glutamine. Then, the microplate was placed in a 37°C non-CO2 incubator for 60 min. Respiration was measured in four blocks of three for 3 min each. The first block measured the basal respiration rate. Next, 1 μm oligomycin (Seahorse Biosciences) was added to inhibit complex V, and the second block was measured. Then, 1 μm FCCP (Seahorse Biosciences) was added to uncouple respiration and the third block was measured. Finally, 1 μm antimycin A (Seahorse Biosciences) and 1 μm rotenone (Seahorse Biosciences) were added to inhibit complex III and the last measurements were performed. Immediately after finishing the measurements, the cells were washed with PBS, fixed in 4% paraformaldehyde and stained with 1 μM DAPI. After staining, the plate was scanned on an automated Olympus Scan^R workstation and total DAPI content measured. An empty well was used as background reference. For each well, the DAPI content (subtracted of the background) was used to normalize OCR values on cell content. Glycolysis capacity and reserve were measured in real-time with the Seahorse Bioanalyzer using the XF Glycolysis Stress Test Kit (Seahorse Biosciences), according to the manufacturer’s recommendations. Briefly, cells were seeded in a XF96 microplate at a density of 15,000 cells per well and allowed to attach. The following day, the extracellular acidification rate (ECAR) was measured after triggering acute stress with the kit buffers.
Intracellular ROS measurement
mESCs infected with lentiviruses expressing wild type KBP and KBP(KK/RR) were seeded in a 4-chamber 35 mm glass bottom petri dish (Cellvis) coated with 0.1% gelatin 24h before the experiment. Then, the cells were stained with 1 μM 2′-7′dichlorodihydrofluorescein diacetate (H2DCFDA, ThermoFisher) supplemented with 1 μM Hoechst and 0.02% pluronic acid (Sigma-Aldrich) in Krebs ringer buffer supplemented with 1 g/L glucose and 1 mM CaCl2 (complete KRB) for 30 min at 37°C. After staining, the cells were washed with complete KRB and then measurements were acquired with an automated Olympus Scan^R workstation (Olympus). Fluorescence intensity in the FITC channel was measured for each cluster of cells.
Measurement of mitochondrial membrane potential
mESCs infected with lentiviruses expressing wild type KBP and KBP(KK/RR) were seeded in a 4-chamber 35 mm glass bottom petri dish (Cellvis) coated with 0.1% gelatin 24h before the experiment. Then, the cells were stained with JC1 (ThermoFisher) 0.2 μM in complete KRB for 30 min at 37°C. After staining, the cells were washed with complete KRB and then measurements were acquired with an automated Olympus Scan^R workstation (Olympus). Integrated fluorescence intensity in the FITC and Rhodamine channels were measured for each cluster of cells. The ratio between Rhodamine and FITC channel intensity was used as index of mitochondrial membrane potential extent.
RNA Extraction and Next-Generation Sequencing Analysis
Libraries for RNA-seq were prepared according to manufacturer’s instructions (Illumina). Briefly, total RNA was extracted from cultured cells using the RNeasy Plus Mini Kit (QIAGEN). Poly(A) RNA was isolated using Dynabeads Oligo(dT)25 (Invitrogen) and used as input for library construction utilizing the dUTP method as described50. Barcodes were used for sample multiplexing. RNA libraries were sequenced on an Illumina HiSeq. RNA-seq data were mapped to mm10 (Genome Reference Consortium GRCm38) with Bowtie using parameters -v2 -m40. Sequence reads were assigned to genes using DEGseq (R package) and the ENSEMBL annotation. Differential gene expression was normalized using DESeq2 (R package) and the resulting values were used to generate an MA plot (i.e. a Bland-Altman plot of log2 fold changes versus the mean of normalized counts).
Mice
Fbxo15−/− mice were originally generated by the Yamanaka laboratory (Kyoto University). Fbxo15+/+ and Fbxo15−/− embryos were obtained from RIKEN BRC and backcrossed into a C57BL6 background for 5–6 generations. Both male and female mice in fertile age were used for breeding. For breeding experiments, randomization was not applicable and the operator was not blinded. All animal experiments were performed in accordance with protocols approved by the Tokushima University Institutional Animal Care and Use Committee.
Statistics and reproducibility
All data were collected and analyzed by Prism 6 (Graphpad). Sample sizes and reproducibility for each figure are denoted in the figure legends. Unless otherwise noted, data are representative of at least three biologically independent experiments. For mouse experiments, no statistical method was used to predetermine sample size. Furthermore, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. Two-group datasets were analyzed by student’s unpaired T-test. For three- or four-group analysis, one-way or two-way ANOVA was used. All graphs show mean values. Error bars indicate +/− S.D. or S.E.M., as indicated in figure legends. Raw data from independent replicate experiments with n<5 can be found in the Statistics Source Data file (Supplementary Table 6).
Data availability
The next-generation sequencing data that support the findings of this study in Supplementary Fig. 4 and in Supplementary Tables 2, 3 and 4 have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE84861. The MudPIT data that support the findings of this study have been provided in Supplementary Table 1. Statistics source data have been provided as Supplementary Table 6 and include data from Fig 3g, 4c, 4f, 6i, 7a–d, 8e–g, 8i, and Supplementary Figure 5f. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
The authors thank C. Haynes, J.K. Pagan, and T. Pozzan for critical reading of the manuscript; I. Aifantis, R. Lammers, M.N. Sacks, and F. Zhang for reagents. M.P. is grateful to T.M. Thor and T.B. Balduur for continuous support. V.D. dedicates this work to Federica, Giorgia and Adriano. P.P. is grateful to Camilla degli Scrovegni for providing continuous support. This work was funded by grants from the National Institutes of Health (R37-CA076584, R01-GM057587, and R21-CA161108) and New York State Health Department (NYSTEM-N11G-255) to M.P.; the American-Italian Cancer Foundation and the National Institutes of Health (5T32CA009161-40) to V.D.; and The Italian Ministry of Health and the Italian Ministry of Education, University and Research (COFIN n. 20129JLHSY_002, FIRB n. RBAP11FXBC_002, and Futuro in Ricerca n. RBFR10EGVP_001), Telethon (GGP15219/B), the Italian Cystic Fibrosis Research Foundation (19/2014) and the Italian Association for Cancer Research (IG-18624) and local funds from the University of Ferrara and the
Italian Ministry of Health to P.P. The authors also thank the NYU Genome Technology Center (partially funded by the Perlmutter Cancer Center Support Grant P30CA016087) for expert library preparation and sequencing for RNA-seq, and the NYU Applied Bioinformatics Laboratories for providing bioinformatics support and helping with the analysis and interpretation of the RNA-seq data using computing resources at the NYU High Performance Computing Facility.. A.S., L.F. and M.P.W. are supported by the Stowers Institute for Medical Research. M.P. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Contributions
V.D. conceived the project, planned and performed most experiments, and co-wrote the manuscript. M.P. directed and coordinated the study, oversaw the results, and co-wrote the manuscript. M.B. and P.P. performed analyses of mitochondrial morphology and physiology. D.Si. generated the CRISPR knockout mESC clones and helped with biochemical experiments and with RNA-seq data analysis. D.Sa. helped with some biochemical experiments. Y.K. performed the Fbxo15+/+ and Fbxo15−/− mouse breeding. M.S. provided reagents and advice. A.S., L.F. and M.P.W. performed the mass spectrometry analyses of the purifications performed by V.D. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 2.Giorgi C, et al. Mitochondrial Ca(2+) and apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X, et al. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci. 2012;125:5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehman J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med (Berl) 2010;88:981–986. doi: 10.1007/s00109-010-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanet A, Arnould T, Najimi M, Renard P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 8.Spiegelman BM. Transcriptional control of energy homeostasis through the PGC1 coactivators. Novartis Found Symp. 2007;286:3–6. discusssion 6–12, 162–163, 196–203. [PubMed] [Google Scholar]
- 9.Tokuzawa Y, et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol. 2003;23:2699–2708. doi: 10.1128/MCB.23.8.2699-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen BB, et al. E3 ligase subunit Fbxo15 and PINK1 kinase regulate cardiolipin synthase 1 stability and mitochondrial function in pneumonia. Cell Rep. 2014;7:476–487. doi: 10.1016/j.celrep.2014.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks AS, et al. Homozygous nonsense mutations in KIAA1279 are associated with malformations of the central and enteric nervous systems. Am J Hum Genet. 2005;77:120–126. doi: 10.1086/431244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyons DA, Naylor SG, Mercurio S, Dominguez C, Talbot WS. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development. 2008;135:599–608. doi: 10.1242/dev.012377. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 16.Wozniak MJ, Melzer M, Dorner C, Haring HU, Lammers R. The novel protein KBP regulates mitochondria localization by interaction with a kinesin-like protein. BMC Cell Biol. 2005;6:35. doi: 10.1186/1471-2121-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kevenaar JT, et al. Kinesin-Binding Protein Controls Microtubule Dynamics and Cargo Trafficking by Regulating Kinesin Motor Activity. Curr Biol. 2016;26:849–861. doi: 10.1016/j.cub.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 18.Drerup CM, Lusk S, Nechiporuk A. Kif1B Interacts with KBP to Promote Axon Elongation by Localizing a Microtubule Regulator to Growth Cones. J Neurosci. 2016;36:7014–7026. doi: 10.1523/JNEUROSCI.0054-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Still AJ, et al. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webster BR, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott I, et al. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci U S A. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prowse AB, et al. Analysis of mitochondrial function and localisation during human embryonic stem cell differentiation in vitro. PLoS One. 2012;7:e52214. doi: 10.1371/journal.pone.0052214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karbowski M, et al. Opposite effects of microtubule-stabilizing and microtubule-destabilizing drugs on biogenesis of mitochondria in mammalian cells. J Cell Sci. 2001;114:281–291. doi: 10.1242/jcs.114.2.281. [DOI] [PubMed] [Google Scholar]
- 31.Zhou W, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbognin E, Betto RM, Soriano ME, Smith AG, Martello G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016;35:618–634. doi: 10.15252/embj.201592629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauklin S, Pedersen RA, Vallier L. Mouse pluripotent stem cells at a glance. J Cell Sci. 2011;124:3727–3732. doi: 10.1242/jcs.074120. [DOI] [PubMed] [Google Scholar]
- 34.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young LM, et al. TIMELESS Forms a Complex with PARP1 Distinct from Its Complex with TIPIN and Plays a Role in the DNA Damage Response. Cell Rep. 2015;13:451–459. doi: 10.1016/j.celrep.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 37.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacCoss MJ, et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci U S A. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paoletti AC, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zybailov B, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 43.D’Angiolella V, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuchay S, et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–480. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagan JK, et al. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat Cell Biol. 2015;17:31–43. doi: 10.1038/ncb3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giorgi C, et al. p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner. Proc Natl Acad Sci U S A. 2015;112:1779–1784. doi: 10.1073/pnas.1410723112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dankert JF, et al. Cyclin F-Mediated Degradation of SLBP Limits H2A.X Accumulation and Apoptosis upon Genotoxic Stress in G2. Mol Cell. 2016;64:507–519. doi: 10.1016/j.molcel.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The next-generation sequencing data that support the findings of this study in Supplementary Fig. 4 and in Supplementary Tables 2, 3 and 4 have been deposited in the Gene Expression Omnibus (GEO) database under the accession code GSE84861. The MudPIT data that support the findings of this study have been provided in Supplementary Table 1. Statistics source data have been provided as Supplementary Table 6 and include data from Fig 3g, 4c, 4f, 6i, 7a–d, 8e–g, 8i, and Supplementary Figure 5f. All other data supporting the findings of this study are available from the corresponding author on reasonable request.