Abstract
We report the principal characteristics of ‘Anaerococcus mediterraneensis’ strain Marseille P2765, a new member of the Anaerococcus genus. Strain Marseille P2765 was isolated in a vaginal sample of a 26-year-old patient with bacterial vaginosis.
Keywords: Anaerococcus mediterraneensis, bacterial vaginosis, culturomics, human microbiota, vaginal flora
We are currently studying the human microbiota by culturomics in our laboratory in Marseille, France [1]. As part of this study, we isolated in the vaginal flora of a 26-year-old French woman with bacterial vaginosis [2] a bacterium which could not be identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) performed with a MicroFlex spectrometer (Bruker Daltonics, Leipzig, Germany) [3]. The agreement number of the National Ethics Committee of the IFR48 (Marseille, France) for this study is 09-022. The patient provided written consent.
First, the vaginal sample was preincubated at 37°C for 21 days in a blood culture bottle (BD Diagnostics, Le Pont-de-Claix, France) enriched with 4 mL rumen that was filter-sterilized through a 0.2 μm pore filter (Thermo Fisher Scientific, Villebon-sur-Yvette, France) and 3 mL of sheep’s blood (bioMérieux, Marcy l’Etoile, France). After 21 days of preincubation, the sample was inoculated on Schaedler agar enriched with sheep’s blood and vitamin K (BD Diagnostics) and incubated for 7 days in anaerobic conditions at 37°C. On sheep’s blood agar (bioMérieux), colonies were white with a mean diameter of 2 mm. Bacterial cells were Gram-positive cocci. Catalase activity was positive; oxidase activity was negative.
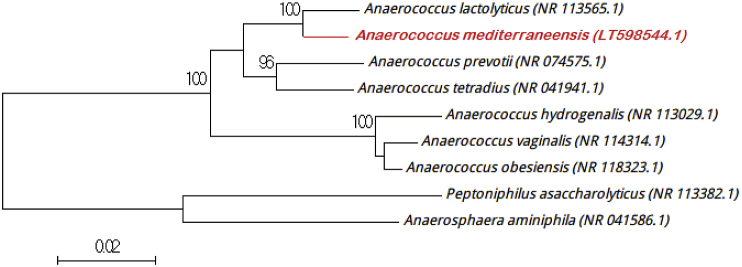
The 16S rRNA gene was amplified and sequenced using the universal primers (fD1 and rp2) and a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France), as described elsewhere [4]. Strain Marseille P2765 exhibited a 97.2% sequence identity with Anaerococcus lactolyticus strain JCM 8140 (GenBank accession no. NR_113565.1), the phylogenetically closest validated species (Fig. 1). This degree of similarity was lower than the 98.7% threshold to define a new species [5], and we propose that strain Marseille P2765 be considered representative of a new species within the Anaerococcus genus in the phylum Firmicutes. The Anaerococcus genus was created by Ezaki et al. [6]. This genus is one of the three genera obtained after the subdivision of the Peptostreptococcus genus [6]. Bacterial species from the Anaerococcus genus have been already reported from diverse human clinical specimens [6]. Anaerococcus lactolyticus, the phylogenetically closest validated species, was first isolated from vaginal discharges [6] like strain Marseille P2765T.
Fig. 1.
Phylogenetic tree showing phylogenetic position of ‘Anaerococcus mediterraneensis’ strain Marseille 2765T relative to close species in genus Anaerococcus. GenBank accession numbers are indicated after name. Sequences were aligned using Muscle v3.8.31 with default parameters, and phylogenetic inferences were obtained using neighbour-joining method with 500 bootstrap replicates within MEGA6 software. Only bootstrap values >95% are shown. Scale bar represents 2% nucleotide sequence divergence.
Because strain Marseille P2765 is more than 2.8% divergent in the 16S rRNA gene sequence with its closest phylogenetic neighbour [7], we propose that it may be the representative strain of a novel species named ‘Anaerococcus mediterraneensis’ (me.di.ter.ra.ne.en′sis, L. masc. adj., mediterraneensis, ‘of Mediterraneum,’ the Latin name of the Mediterranean Sea by which Marseille, where strain P2765 was isolated, is located). Strain Marseille P2765T is the type strain of the new species ‘Anaerococcus mediterraneensis’ sp. nov.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of ‘Anaerococcus mediterraneensis’ is available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in European Molecular Biology Laboratory–European Bioinformatics Institute under accession number LN598544.1.
Deposit in a culture collection
The type isolate of ‘Anaerococcus mediterraneensis’ was deposited in the collection Deutsche Sammlung von Mikroorganismen (DSM 103343) and the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P2765.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menard J.P., Fenollar F., Henry M., Bretelle F., Raoult D. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis. 2008;20:33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 6.Ezaki T., Kawamura Y., Li N., Li Z.Y., Zhao L., Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–1528. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- 7.Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K.H. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]