Abstract
Hepatitis C virus (HCV) evolves rapidly in a single host and circulates as a quasispecies wich is a complex mixture of genetically distinct virus’s but closely related namely variants. To identify intra-individual diversity and investigate their functional properties in vitro, it is necessary to define their quasispecies composition and isolate the HCV variants. This is possible using single genome amplification (SGA). This technique, based on serially diluted cDNA to amplify a single cDNA molecule (clonal amplicon), has already been used to determine individual HCV diversity. In these studies, positive PCR reactions from SGA were directly sequenced using Sanger technology. The detection of non-clonal amplicons is necessary for excluding them to facilitate further functional analysis. Here, we compared Next Generation Sequencing (NGS) with De Novo assembly and Sanger sequencing for their ability to distinguish clonal and non-clonal amplicons after SGA on one plasma specimen. All amplicons (n = 42) classified as clonal by NGS were also classified as clonal by Sanger sequencing. No double peaks were seen on electropherograms for non-clonal amplicons with position-specific nucleotide variation below 15% by NGS. Altogether, NGS circumvented many of the difficulties encountered when using Sanger sequencing after SGA and is an appropriate tool to reliability select clonal amplicons for further functional studies.
Introduction
Hepatitis C virus (HCV) is an enveloped positive sense single stranded RNA virus of 9600 bases, which infects 130–150 million people worldwide. Most (70%) HCV infections become chronic and progress toward liver diseases such as cirrhosis and hepatocellular carcinoma[1]. Approximately 500 000 people die each year from hepatitis C-related liver diseases[2].
In vivo, HCV replicates rapidly using a viral RNA polymerase that lacks proofreading activity[3]. The error rate of HCV polymerase has been estimated in vitro to be 10−3 nucleotide substitutions per site per year[4]. This high mutation rate combined with a short generation time (1012 virions produced per day[5]) is at the origin of the quasispecies dynamics of RNA viruses[6]. Seven genotypes have been described, which differ by 30 to 35% in their nucleotide sequence[7]. HCV circulates in infected individuals as a complex mixture of genetically different, but closely related, viral variants[8], [9]. Constraints in the viral genome and protein structure prevent some variants from proliferating[10]. Rapid HCV evolution in a single host favors the emergence of mutants that can escape from specific immunity[11].
Investigating the functional properties of intra-individual HCV variants in vitro requires accurate identification of HCV variants within a quasispecies. Single genome amplification (SGA) can be used in this context. It consists of serially diluting cDNA to amplify single cDNA molecules (clonal amplicon). However, the amplification of two or more cDNA molecules (non-clonal amplicon) cannot be fully ruled out. Although initially developed for the study of HIV quasispecies diversity[12],[13], SGA has also been used to describe early diversification of HCV after transmission events[14],[15],[16], [17],[18]. In those studies, conventional Sanger sequencing technique was performed on positive PCR reactions coming from SGAs. Visual inspection of electropherograms is required for detecting mixed populations. Non-clonal amplicons must be excluded to achieve the most accurate representation of the variant population. This is paramount for performing further functional analyses to study, for example, the transmission of variants from mother to child using in vitro models, such as HCV retroviral pseudoparticles (HCVpps)[19] or infectious hepatitis C virus coming from cell culture (HCVcc) [20]. Another limitation of conventional Sanger sequencing is the necessity to use specific sequencing primers. This represents a major challenge, especially in variable regions such as the HCV E1E2 gene, and could compromise the sequencing of long fragments.
Here, we compared Next Generation Sequencing (NGS) with de novo assembly to the classical Sanger approach with the aim of improving the differentiation between clonal and non-clonal amplicons after SGA.
Materials and methods
Biological material
A plasma specimen was obtained in the year 2000 from a Thai woman infected by the HCV 1b genotype. She was participating in HIV prevention clinical trial that assessed different duration of maternal and infants zidovudine for the prevention of perinatal transmission of HIV[21],[22]. The HCV genotype was determined by NS5B amplification and sequencing.
Viral RNA extraction and cDNA synthesis
Viral RNA was extracted from 400 μL plasma using the Macherey Nagel NucleoSpin Virus kit (Macherey Nagel, Hoerdt, France). The RNA was eluted and immediately reverse-transcribed using random hexamer primers and Superscript III kit (Invitrogen, Life technologies, Courtaboeuf, France) to generate cDNA according to the manufacturer’s protocol.
Single genome amplification of full-length E1E2 glycoproteins
Full-length E1E2 glycoprotein genes were amplified using an SGA approach. A series of cDNA dilutions (1:40, 1:80, 1:100, 1:150, 1:200) was amplified by nested PCR, resulting in a fragment of 2097 base pairs. We selected the dilution giving a maximum of 30% positive PCR reactions (dilution 1:40)[12,17]. Indeed, according to the Poisson distribution law, a majority of wells at this dilution contains a single cDNA molecule. PCR amplification was carried out using high fidelity Platinum Taq PCR SuperMix (Invitrogen Life technologies, Courtaboeuf, France) according to the manufacturer’s protocol. The PCR primers for generating the full-length E1E2 glycoproteins sequences were: first-round sense primer P1bE1E2extsens (5’-ACCAAACGTAACACCAACCGC-3’; position 372 to 392, H77), first-round antisense primer P1bE1E2extantisens (5’-GCTCTGGTGATAAAATATTGTAACCAC-3’; position 2873 to 2899, H77), second-round sense primer P1bE1E2intsens (5’-TGGGTAAGGTCATCGATACCCT-3’; position 697 to 718, H77), and second-round antisense primer P1bE1E2intantisens (5’-CACGATGCAGCCATCTCCCG-3’; position 2775 to 2794, H77). The PCR amplification conditions were: 94°C for 2 min followed by 35 cycles of 94°C for 30 s; 58°C for 30 s; 68°C for 2 min 30 s; before a final extension at 68°C for 10 min. The product of the first PCR was used as a template for the 2nd PCR under the same conditions. The PCR products were separated by microfluidics capillary electrophoresis (LabChip GX–Perkin Elmer). Only PCR products obtained at the dilution giving a maximum of 30% positive PCR reactions are selected.
DNA sequencing using NGS
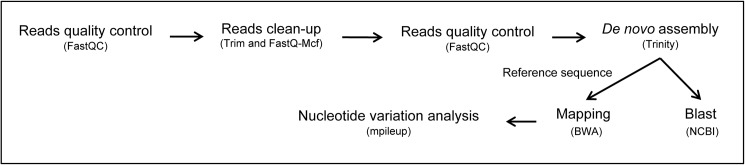
The sequencing library was built using a Nextera XT DNA sample preparation kit (Illumina, San Diego, USA) according to the manufacturer’s protocol. 1 ng of DNA per sample is necessary. This protocol includes: tagmentation of both DNA strands, a PCR amplification step that adds adapters and indexes, and a clean-up step using AMPure XP beads (Agencourt—Beckman Coulter, Roissy, France). Finally, a beads-based normalization of each library according to the manufacturer’s instructions was performed to ensure equal library representation in the pooled sample. Heat-denaturation of the library pool was performed before the sequencing run. Paired-end sequencing of 151 base-pairs was performed on a Miseq (Illumina, San Diego, USA) platform. Illumina sequencer output files matching 151 base-pair sequencing reads were processed using the “Biomina Galaxy platform”[23] after verifying read quality (FastQC algorithm). De novo assembly was performed using the Trinity program. The constructed sequences were verified using the Blast option of the National Center for Biotechnology Information (NCBI). Single reads with a QC score over 30 and a length of over 30 nucleotides were conserved for de novo assembly using the Trinity program. Reads were then mapped using the “Burrows-Wheeler Aligner” (BWA) to the reference sequence given by the de novo assembly. Nucleotide analysis was performed position by position using the mpileup program (Fig 1). Positions with a sequencing depth of over 100X were retained for further analysis. Non-clonal samples (ie amplicons containing multiple templates) were identified by determining nucleotide heterogeneity, position by position.
Fig 1. Workflow of analysis on Galaxy platform.
15 to 30% of position had some variability due to the NGS technique. We subjected a known clonal sample from an RNA virus to the deep sequencing to distinguish variability due to background from that due to non-clonal differences. Thus, we could determine that a clonal sample contained a nucleotide variation of less than 5% or 50 nucleotides at each position. Non-clonal samples were defined as those having a nucleotide variation of more than 10% or 100 nucleotides at one or several positions. Ambiguous samples had a nucleotide variation of between 5 and 10% or 50 and 100 nucleotides at one or several positions.
DNA sequencing using Sanger method
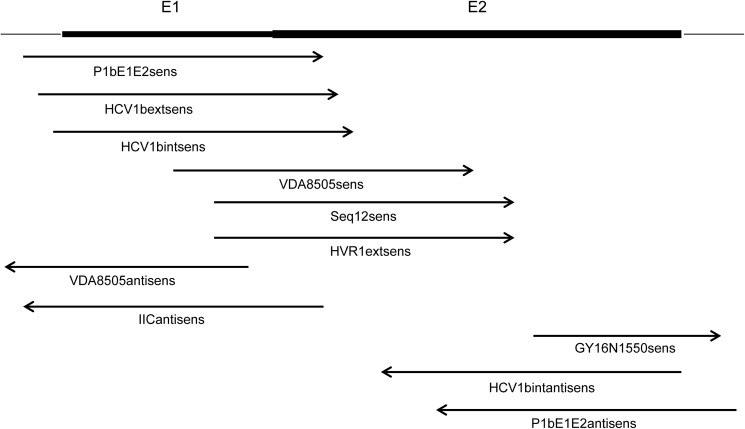
The selected samples from SGA were also sequenced using BigDye terminator chemistry, on an ABI 3130xl capillary sequencer (Applied Biosystems—Life Science Technologies, Carlsbad, USA). According to the manufacturer’s instructions, 10 ng of DNA was used for each sequencing reaction. The sequencing primers were designed using vector NTI® (Thermo Fisher Scientific, Courtaboeuf, France) based on the consensus sequences obtained following the Miseq de novo assembly (Fig 2).
Fig 2. Overlapping primers for Sanger sequencing.
During a preliminary analysis (performed with Sequencing Analysis software™), the threshold for second peak height was set at 25% of the main peak. A secondary step of analysis was needed for the final validation of second peaks (performed with CLC Main WorkbenchTM). It was based on visual inspection of electropherograms especially in homopolymer regions. Background signal as well as alignments of electropherograms obtained with different primers, enable to validate or reject second peak.
Non-clonal samples were identified by identifying double peaks on electropherograms position by position.
Primer for Sanger sequencing
The primers used for Sanger sequencing are shown in Table 1.
Table 1. Primers for Sanger sequencing.
| Primer | Location* | Sequence (5'-3') | Samples† |
|---|---|---|---|
| P1bE1E2sens | 697–718 | TGGGTAAGGTCATCGATACCCT | All except c14, c16, c17 |
| HCV1bextsens | 821–841 | CGGCGTGAACTATGCAACAGG | All except c3, c14, c15, c16, c17, c37 |
| HCV1bintsens | 843–873 | CACCATGGGTTGCTCTTTCTCTATCTTCC | c6, c12, c20, c27, c37, c42 |
| VDA8505sens | 1178–1196 | CCTGTGCGGGTCGTTTTT | All except c14, c16, c17 |
| seq12sens | 1290–1309 | CGCATGGCTTGGGATATGAT | c34, c36 |
| HVR1extsens | 1290–1310 | CGCATGGCGTGGGACATGATG | c15 |
| VDA8505antisens | 1435–1454 | AACCTTAGCCCAGTTCCCCG | All except c14, c16, c17 |
| IICantisens | 1605–1620 | ACCCAGTGCTATTCAT | All except c14, c15, c16, c17, c37, c42 |
| GY16N1550sens | 2247–2266 | GTGGAGCACAGGTTCGAAGC | All except c14, c16, c17, c37, c41 |
| HCV1bintantisens | 2559–2584 | AATCAGGCCTCAGCCTAGGCTATCTG | c12, c20, c27, c37, c42 |
| P1bE1E2antisens | 2775–2794 | CACGATGCAGCCATCTCCCG | All except c14, c16, c17 |
* Location indicate nucleotide postion relative to the H77 genome
† Samples sequencing with each primer
Nucleotide sequence accession numbers
The clonal sequences contigs and the reads from the NGS sequencing were deposited in GenBank under the accession numbers KX358443 to KX358470.
Ethics statement
The parent studies received initial and annual approvals by the Thai Ministry of Public Health Ethics Committees and other relevant Ethics committees.
All participants or their guardians provided their informed consent to participate in the parent studies. Their consent includes explicitly the use of the blood/plasma samples in other future studies.
Results
NGS and Sanger sequencing
Forty-two positive PCR products issued from SGA fulfilled the criteria of PCR selection process (ie PCR products obtained at the dilution giving a maximum of 30% positive PCR reactions). were purified for sequencing.
We first sequenced the HCV E1E2 amplicons derived from SGA by NGS. In total, 42 PCR products (samples), identified c1 to c42, were successfully sequenced and analyzed. The sequencing depth varied between samples, but was between 100x (our threshold) at the end of fragments and 6545x in the middle of the fragments. We classified 28 samples as clonal, 11 as non-clonal and three as ambiguous.
The same 42 samples were also sequenced by Sanger technology using 6 to 11 primers based on the consensus sequences as described before. Samples c14, c16, and c17 could not be sequenced because of an insufficient quantity of DNA. After analysis of the electropherograms of the remaining 39 samples, we classified 32 samples as clonal and 7 as non-clonal.
Assignment of clonal and non-clonal samples: Comparison between NGS and Sanger sequencing
We compared the assembled sequences obtained with NGS and those obtained with Sanger sequencing (Table 2). All 28 samples classified as clonal by NGS were also classified as clonal after direct Sanger sequencing. Of the three samples classified as ambiguous by NGS, two were considered to be clonal after direct Sanger sequencing, and the remaining one non-clonal. Among the 11 samples classified as non-clonal by NGS, six were also classified as non-clonal after direct Sanger sequencing and two was re-assigned to the clonal category; three samples could not be sequenced. These discrepancies in the classification of samples were probably due to the depth of the NGS sequencing at each position for the nucleotide variation analysis. The sequencing depth for all positions was at least 100X and nucleotide variations at each position are clearly indicated by the software. This is less subjective than interpreting electropherograms following direct Sanger sequencing.
Table 2. Comparaison of clonality sample using two different sequencing technology.
| Sample | MiSeq sequencing | Sanger sequencing | Agreement |
|---|---|---|---|
| c1 | C | C | ● |
| c2 | A (0*; 2†) | C | ○ |
| c3 | NC (1; 1) | NC (1‡; 0§) | ● |
| c4 | NC (3; 0) | A (0; 3) | ○ |
| c5 | NC (3; 2) | NC (4; 0) | ● |
| c6 | C | C | ● |
| c7 | A (0; 1) | C | ● |
| c8 | C | C | ● |
| c9 | C | C | ● |
| c10 | C | C | ● |
| c11 | C | C | ● |
| c12 | A (0; 3) | NC (2; 0) | ○ |
| c13 | C | C | ● |
| c14 | NC (2; 0) | /// | ◊ |
| c15 | NC (2; 2) | A (0; 1) | ○ |
| c16 | NC (1; 0) | /// | ◊ |
| c17 | NC (7; 0) | /// | ◊ |
| c18 | C | C | ● |
| c19 | C | C | ● |
| c20 | C | C | ● |
| c21 | C | C | ● |
| c22 | C | C | ● |
| c23 | C | C | ● |
| c24 | C | C | ● |
| c25 | C | C | ● |
| c26 | NC (1; 0) | NC (1; 0) | ● |
| c27 | C | C | ● |
| c28 | C | C | ● |
| c29 | C | C | ● |
| c30 | C | C | ● |
| c31 | NC (1; 0) | C | ○ |
| c32 | C | C | ● |
| c33 | C | C | ● |
| c34 | C | C | ● |
| c35 | C | C | ● |
| c36 | C | C | ● |
| c37 | C | C | ● |
| c38 | NC (1; 0) | C | ○ |
| c39 | C | C | ● |
| c40 | C | C | ● |
| c41 | NC (2; 0) | NC (2; 0) | ● |
| c42 | C | C | ● |
* number of non clonal position with MiSeq sequencing analysis.
† number of ambiguous postion with MiSeq sequencing analysis.
‡ number of non clonal position with Sanger sequencing analysis.
§ number of ambiguous postion with Sanger sequencing analysis.
● MiSeq and Sanger are in agreement.
○ MiSeq and Sanger are in desagreement.
◊Failure of Sanger sequencing.
To check our results, 6 amplicons (positive after SGA) were selected to be sequenced a second time with the two techniques. This relates to all samples in disagreement between NGS and Sanger sequencing (c2, c7, c31 and c38) and two in agreement (c12 and c41).
The sequencing results (NGS and Sanger sequencing) are similar to our previous results (data not shown).
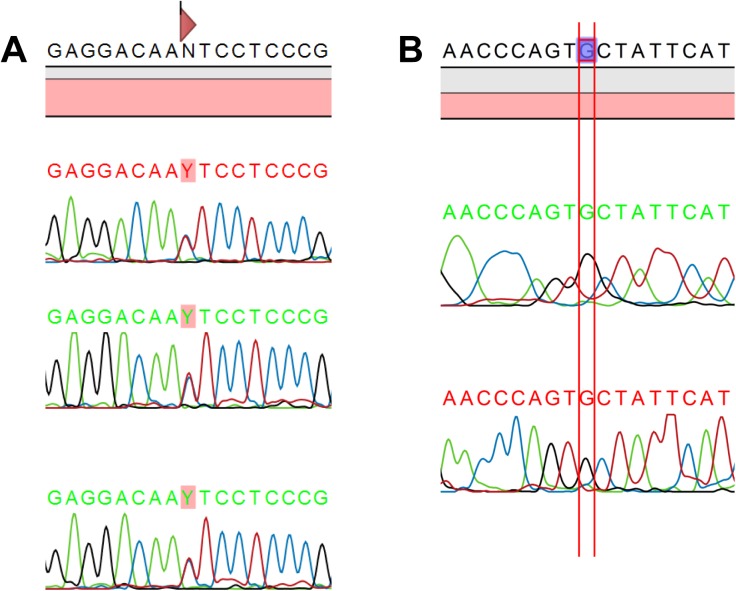
We compared the percentage of nucleotide variation observed after NGS analysis at each non-clonal and ambiguous position with the electropherogram profiles obtained by Sanger sequencing. Double peaks were clearly visible on electropherograms for position-specific nucleotide variation of above 35% by NGS. Double peaks were less visible and mixed bases could be easily missed for 15–20% nucleotide variation by NGS, and no double peaks were visible on the electropherograms for a nucleotide variation of less than 15% by NGS (Table 3). Two examples of electropherograms are presented in Fig 3.
Table 3. Comparaison of non clonal position with two sequencing techniques according to the percentage of MiSeq nucleotide variation.
| MiSeq sequencing | Sanger sequencing | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Location* | Depth | A | C | G | T | % variation† | Double peak | Primers‡ |
| c41 | 1028 | 1759 | 1 | 819 | 0 | 878 | 48,3 | "+" | 3 |
| c5 | 1380 | 3085 | 1821 | 0 | 1264 | 0 | 41 | "+" | 3 |
| c5 | 916 | 1937 | 1147 | 0 | 790 | 0 | 40,8 | "+" | 3 |
| c5 | 2048 | 3202 | 0 | 1992 | 0 | 1210 | 37,8 | "+" | 2 |
| c3 | 1431 | 2523 | 910 | 0 | 1612 | 1 | 36,1 | "+" | 2 |
| c26 | 1973 | 1803 | 626 | 0 | 1177 | 0 | 34,7 | "+" | 2 |
| c4 | 1707 | 2745 | 2217 | 0 | 528 | 0 | 19,2 | "+/-" | 2 |
| c4 | 1670 | 2302 | 0 | 421 | 0 | 1881 | 18,3 | "+/-" | 2 |
| c15 | 905 | 1479 | 1237 | 0 | 0 | 242 | 16,4 | "+/-" | 2 |
| c4 | 2168 | 2630 | 400 | 0 | 2230 | 0 | 15,2 | "+/-" | 2 |
| c31 | 2379 | 2387 | 2069 | 0 | 318 | 0 | 13,3 | "-" | 2 |
| c38 | 2012 | 1571 | 0 | 1406 | 0 | 165 | 10,5 | "-" | 2 |
| c15 | 1615 | 1343 | 0 | 1236 | 0 | 107 | 8 | "-" | 1 |
| c41 | 2479 | 2952 | 0 | 2726 | 0 | 226 | 7,7 | "-" | 1 |
* Location indicate nucleotide postion relative to the H77 genome.
† % nucleotide variation at this postion after MiSeq analysis.
‡ Number of nucleotide read at this position according to the numbers of primers used.
"+" Double peak clearly visible on electrophoregram at this position.
"+/-" Double peak no clearly visible on electrophoregram at this position.
"-" No double peak on electrophoregram at this position.
Fig 3. Examples of superimposed electropherograms.
(A) in agreement with NGS (sample c41; 48,3% nucleotide variation; double peak seen on electropherograms) (B) in disagreement with NGS (sample c38, 10,5% nucleotide variation; absence of double peak on electropherograms).
For nucleotide variations below 7.4% (i.e. at ambiguous positions), the electropherogram profiles were very heterogeneous with double peaks visible in some but not in others (Table 4).
Table 4. Comparaison of ambiguous position with two sequencing techniques according to the percentage of MiSeq nucleotide variation.
| MiSeq sequencing | Sanger sequencing | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Location* | Depth | A | C | G | T | % variation† | Double peak | Primers‡ |
| c15 | 1008 | 1114 | 1032 | 0 | 82 | 0 | 7,4 | "-" | 1 |
| c12 | 2082 | 1736 | 0 | 0 | 80 | 1656 | 4,6 | "+" | 2 |
| c2 | 826 | 1615 | 0 | 68 | 0 | 1547 | 4,2 | "-" | 3 |
| c15 | 1322 | 1681 | 1617 | 0 | 64 | 0 | 3,8 | "-" | 1 |
| c12 | 1858 | 1723 | 0 | 62 | 0 | 1661 | 3,6 | "-" | 1 |
| c12 | 2291 | 2104 | 70 | 0 | 2034 | 0 | 3,3 | "+" | 1 |
| c5 | 1274 | 1856 | 0 | 1798 | 0 | 58 | 3,1 | "+" | 4 |
| c3 | 2191 | 3735 | 3636 | 0 | 99 | 0 | 2,7 | "-" | 2 |
| c5 | 1811 | 3209 | 3129 | 0 | 80 | 0 | 2,5 | "-" | 2 |
| c10 | 1603 | 2531 | 2472 | 0 | 59 | 0 | 2,3 | "-" | 2 |
| c2 | 2558 | 3174 | 72 | 0 | 3102 | 0 | 2,3 | "-" | 3 |
* Location indicate nucleotide postion relative to the H77 genome.
† % nucleotide variation at this postion after MiSeq analysis.
‡ Number of nucleotide read at this position according to the numbers of primers used.
"+" Double peak clearly visible on electrophoregram at this position.
"+/-" Double peak no clearly visible on electrophoregram at this position.
"-" No double peak on electrophoregram at this position.
Discussion
Exploration of the in vitro functional properties of HCV variants circulating in a single host requires accurate determination of the HCV quasispecies sequences. SGA followed by Sanger sequencing has been used for this purpose by several groups. However, direct sequencing has several limitations that we aimed to circumvent using NGS. Bioinformatics analysis after NGS sequencing is clearly more complex than Sanger sequencing but our work was facilitated by the possibility to design a convenient workflow using the Galaxy Biomina platform.
We were able to entirely sequence envelope genes of all 42 HCV samples (approximately 2000 nt) without any failures using the Illumina NGS technology (Table 2). Only 1 ng of DNA per sample was necessary for NGS whereas a total amount of 60 ng was necessary for Sanger sequencing: 10 n g for each of the six primers (480 Sanger sequencing PCR reactions were necessary for the 42 samples). Finally, three sequences could not be obtained (Table 1), because of the limited quantity of the amplified DNA.
NGS Sequencing has obvious advantages because it spares DNA and enables further plasmid construction for functional in vitro analyses. The use of non-specific sequencing primers for Illumina NGS sequencing provides a clear benefit. DNA is simultaneously fragmented and tagged with sequencing adapters. The HCV E1/E2 gene exhibits a high rate of genetic polymorphism, making it challenging to choose a reference sequence and design specific primers. In NGS analysis, no reference sequence is required. The reads are assembled as contigs and the coverage quality of the de novo consensus sequence depends only on the size and continuity of the contigs. This de novo consensus sequence can then be used to choose adequate primers for Sanger sequencing. This strategy has clearly reduced the sequencing failures encountered using Sanger sequencing, but may have introduced a potential bias in our comparison.
All samples classified as clonal after NGS were also classified as clonal after Sanger sequencing. According to the Poisson distribution, cDNA dilutions that yield PCR products in no more than 30% of wells should contain one amplifiable cDNA template per positive PCR more than 80% of the time[13]. In our study, only 67% of samples contained a unique genome. This difference could be explained by the nested PCR performed during the SGA step, which could be responsible for artefactual mutations caused by the DNA polymerase, despite our choice of a high fidelity proofreading enzyme. Here, we applied stringent criteria for the selection of clonal samples. The proportion of samples containing one amplifiable cDNA template would have increased from 67 to 74% if we had included the ambiguous samples.
Some positions classified as ambiguous after NGS analysis, showed unexpected double peaks in the Sanger electropherograms profiles (Table 4). Neither of the two techniques was able to place these samples in a defined category. Samples classified as ambiguous should be rejected for functional analysis.
In conclusion, NGS circumvented many difficulties encountered with Sanger sequencing after SGA. NGS does not require the design of specific sequencing primers and spares valuable DNA. In addition, for our purpose, NGS (around 46$ per sample) is more economical than Sanger sequencing (about 77$ per sample).
Finally, this work also demonstrates that NGS of long viral RNA fragments allows more reliable selection of clonal samples obtained by SGA, which is crucial for the study of their functional properties using in vitro models.
Acknowledgments
We thank Sylviane Marouillat (UMR INSERM U930, Université François Rabelais, Tours) and the PST ASB platform (Université François Rabelais, Tours, France) for their helpful technical assistance. We thank also Dr. François Darrouzain for helpful discussion on methodological aspects.
Data Availability
The sequences of clonal samples are available from the Genbank database (accession numbers KX358443 to KX358470). The raw data issued from deep sequencing are available from the SRA NCBI database (accession numbers SRR5351778 to SRR5351819).
Funding Statement
This work was supported from the Region Centre Val de Loire, "VIROTRANSAC" Grant from: convention 2014 00094545 / operation AE 2014-1850 (http://www.regioncentre-valdeloire.fr/accueil/les-services-en-ligne/appels-a-projets/recherche-et-innovation.html). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ascione A, Tartaglione MT, Giuseppe Di Costanzo G. Natural history of chronic hepatitis C virus infection. Dig Liver Dis. 2007;39: S4–S7. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Hepatitis C. In: Hepatitis C. WHO fact sheet 164 [Internet]. 2015. Available: http://www.who.int/mediacentre/factsheets/fs164/en/
- 3.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122: 281–288. [DOI] [PubMed] [Google Scholar]
- 4.Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A. 1991;88: 3392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-therapy. Science (80-). 1998;282: 103 [DOI] [PubMed] [Google Scholar]
- 6.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76: 159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology. 2014;59: 318–327. 10.1002/hep.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81: 1631–1648. 10.1099/0022-1317-81-7-1631 [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro RM, Li H, Wang S, Stoddard MB, Learn GH, Korber BT, et al. Quantifying the Diversification of Hepatitis C Virus (HCV) during Primary Infection: Estimates of the In Vivo Mutation Rate. PLoS Pathog. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Lello FA, Culasso ACA, Campos RH. Inter and intrapatient evolution of hepatitis C virus. Ann Hepatol. 2015;14: 442–449. [PubMed] [Google Scholar]
- 11.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C Virus Continuously Escapes From Neutralizing Antibody and T-Cell Responses During Chronic Infection In Vivo. Gastroenterology. 2007;132: 667–678. 10.1053/j.gastro.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 12.Keele BF, Giorgi EE, Salazar-gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. PNAS. 2008;105: 7552–7557. 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salazar-gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering Human Immunodeficiency Virus Type 1 Transmission and Early Envelope Diversification by Single-Genome Amplification and Sequencing. J Virol. 2008;82: 3952–3970. 10.1128/JVI.02660-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Stoddard M, Wang S, Giorgi EE, Blair L, Learn G, et al. Single Genome Sequencing of Hepatitis C Virus in Donor-Recipient Pairs Distinguishes Modes and Models of Virus Transmission and Early Diversification. J Virol. 2015;90: JVI.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arienzo VD, Moreau A, Alteroche LD, Gissot V, Blanchard E, Roch E, et al. Sequence and Functionnal Analysis of the Envelope Glycoproteins of Hepatitis C Virus Variants Selectively Transmitted to a New Host. J Virol. 2013;87: 13609–13618. 10.1128/JVI.02119-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferns RB, Tarr AW, Hue S, Urbanowicz RA, McClure CP, Gilson R, et al. Hepatitis C virus quasispecies and pseudotype analysis from acute infection to chronicity in HIV-1 co-infected individuals. Virology. Elsevier; 2016;492: 93–97. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, et al. Elucidation of Hepatitis C Virus Transmission and Early Diversification by Single Genome Sequencing. PLoS Pathog. 2012;8: 31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoddard MB, Li H, Wang S, Saeed M, Andrus L, Ding W, et al. Identification, Molecular Cloning, and Analysis of Full-Lenght Hepatitis C Virus Transmitted / Founder Genotypes 1, 3, and 4. MBio. 2015;6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartosch B, Dubuisson J, Cosset F-L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197: 633–642. 10.1084/jem.20021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakita T, Pietschmann T, Kato T, Date T, Zhao Z, Murthy K, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2010;11: 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lallemant M, Jourdain G, Le Coeur S. Numb Er 14 a Trial of Shortened Zidovudine Regimens To Prevent Mother-To-Child Transmission Of Human Immunodeficiency Virus Type 1. N Engl J Med. 2000;343. [DOI] [PubMed] [Google Scholar]
- 22.Ngo-Giang-Huong N, Jourdain G, Sirirungsi W, Decker L, Khamduang W, Le C??ur S, et al. Human immunodeficiency virus-hepatitis C virus co-infection in pregnant women and perinatal transmission to infants in Thailand. Int J Infect Dis. 2010;14: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44: gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences of clonal samples are available from the Genbank database (accession numbers KX358443 to KX358470). The raw data issued from deep sequencing are available from the SRA NCBI database (accession numbers SRR5351778 to SRR5351819).