Abstract
We report a case of severe infection with liver abscess and endophthalmitis caused by a hypervirulent Klebsiella pneumoniae strain in an immunocompetent German male patient without travel history to Asia. Phenotypic and molecular characterization showed high similarity to the reference genome NTUH-K2044 isolated in Asia. The isolate was assigned as ST2398 (clonal complex 66). The findings underline global spread of hypervirulent Klebsiella pneumoniae strains to Europe.
Keywords: Endophthalmitis, hypervirulent, Klebsiella pneumoniae, liver abscess, ST2398
Introduction
Klebsiella pneumoniae is one of the organisms that merits particular attention as a result of rising resistance problems causing high numbers of both healthcare- and community-acquired infections, mainly urinary tract, respiratory tract or bloodstream infections [1]. Apart from antimicrobial resistance—mainly due to β-lactamases like extended-spectrum β-lactamases (ESBL), AmpC β-lactamases or carbapenemases—so-called hypervirulent strains have emerged primarily in Asia, but recent data suggest a worldwide spread [2], [3]. Antimicrobial resistance and hypervirulence are mostly not overlapping [4]. Nonetheless, cases due to hypervirulent carbapenem-resistant K. pneumoniae have been described, particularly in Taiwan and China [5], [6].
To date, there is no consistent definition of hypervirulence in literature. Hypervirulent K. pneumoniae strains typically have a hypermucoviscous phenotype and belong to serotype K1 or K2 [7]. Two genes have previously been associated with invasive infections, the mucoviscosity-associated gene A (magA) and the regulator of mucoid phenotype A (rmpA) [8]. magA is located in the same gene cluster specifying serotype K1 and encodes a particular capsular polymerase. In accordance with the bacterial polysaccharide gene nomenclature (BPGN) scheme, it was suggested to be renamed wzy(KpK1) [9]. The rmpA gene is a plasmid-borne regulator of extracellular polysaccharide synthesis. Hypervirulent strains possess various iron acquisition systems, including enterobactin (Ent); the prototypical catecholate siderophore aerobactin, a hydroxamate siderophore whose receptor is encoded by iutA; and yersiniabactin (ybtS), a phenolate-type siderophore that is structurally distinct from Ent [10]. Kfu, which mediates uptake of ferric iron, seems to be present in many hypervirulent strains [11]. Liver abscess has been associated with demonstration of allS, a gene relevant in allantoin metabolism [12]. Other genes found include fimbrial and nonfimbrial adhesion genes, such as ycfM, KPN and mrkD. The latter is believed to function as the type 3 fimbrial adhesin and to mediate binding to the extracellular matrix [13].
A 61-year-old previously healthy man presented with fever for 3 days, progressive jaundice and a red left eye with pain and lid swelling, as well as reduced vision in his left eye. He denied any travel to Asia but reported a recent journey to Brazil. He was hospitalized with the presumptive diagnosis of an endophthalmitis as a consequence of systemic infection. At admission his vital signs included a body temperature of 38.7°C, heart rate of 140 beats per minute and an elevated blood pressure of 170/100 mm Hg. Scleras and skin were icteric; no rash was observed. No tenderness could be elicited in any quadrant of the abdomen; hepatosplenomegaly was not detected. Neurologic examination was revealed nothing abnormal. The initial laboratory examination revealed mild leucocytosis (white blood cell count 13 800/mL) and thrombocytopenia (platelet count 123 000/mL). Levels of C-reactive protein (CRP) and liver enzymes were markedly elevated (CRP 252 mg/L, alanine aminotransferase 59 U/L, alkaline phosphatase 156 U/L, total bilirubin 5.7 mg/dL). Kidney function was preserved. Clotting factors were in the normal range. Computed tomographic scan revealed a liver abscess in segment II (4.8 × 4.2 cm). Obstructive cholestasis or any other intrabdominal pathology was not present. The patient underwent emergent ultrasound-guided percutaneous catheter drainage of the liver abscess. Intravenous piperacillin/tazobactam 4.5 g three times daily was empirically administered, and intravenous levofloxacin 750 mg once daily was added 3 days later. Cultures from the liver abscess yielded K. pneumoniae. Blood cultures taken at admission remained sterile. To clarify the extent of the disease, a head magnetic resonance imaging (MRI) was done, which permitted us to exclude a brain abscess. Transthoracic echocardiography revealed no signs of endocarditis. Furthermore, the patient underwent colonoscopy to rule out a diverticulitis or colorectal carcinoma as the underlying focus. Cultures from vitreous samples remained negative in culture but were PCR positive. Follow-up MRI of the abdomen 6 days later showed a reduction in abscess cavity size but unfortunately revealed a thrombosis of adjacent left hepatic vein as well as a new abscess cavity located dorsally to the original abscess. The new abscess was interpreted as a complication of the percutaneous abscess drainage. A repeated percutaneous drainage or chirurgical intervention was not indicated. The patient was successfully treated by intravenous antibiotic therapy with piperacillin/tazobactam for 10 days followed by oral levofloxacin for up to 4 weeks. Repeated abdominal ultrasound during follow-up revealed complete resolution of the liver abscess.
Materials and Methods
Susceptibility testing
Minimum inhibitory concentrations (MICs) for various antibiotics were obtained by broth microdilution antimicrobial susceptibility testing with the Micronaut system (Merlin Diagnostika, Bornheim-Hersel, Germany) according to standard procedures ISO 20776-1:2006. Results were interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (Breakpoint tables for interpretation of MICs and zone diameters, version 6.0, 2016, http://www.eucast.org). MIC values of relevant β-lactam agents are shown in Table 1. Phenotypic detection of ESBL production could be excluded with the combination disc test as recommended by EUCAST using cefotaxime, ceftazidime and cefepime with or without clavulanate. AmpC production was excluded by cefoxitin/cloxacillin disc test.
Table 1.
Summary of relevant characteristics of K2 Klebsiella pneumoniae isolate KP_FR_2016 sequence type 2398 (clonal complex 66)
| Characteristic | Value |
|---|---|
| MICs for selected β-lactams determined by broth microdilution (Micronaut system) (mg/L) | |
| Ampicillin/sulbactam | 2 |
| Amoxicillin/clavulanate | 4 |
| Piperacillin/tazobactam | 1 |
| Ceftazidime | 0.25 |
| Cefotaxime | 0.25 |
| Cefepime | 0.25 |
| Meropenem | 0.5 |
| Ertapenem | 0.125 |
| Levofloxacin | 0.06 |
| Resistance profile | Negative for ESBL, AmpC, carbapenemase |
| Virulence-associated features | |
| Hypermucoviscosity (defined by positive string test) [14] | Positive |
| Cps genotype | K2 |
| Virulence gene profiles | |
| magA gene | Not detected |
| rmpA gene | Present |
| Iron acquisition | |
| Aerobactin (iutA gene) | Present |
| kfu gene | Not detected |
| entB gene | Present |
| ybtS gene | Present |
| Allantoin metabolism | |
| allS gene | Not detected |
| Adhesins | |
| mrkD gene | Present |
ESBL, extended-spectrum β-lactamase; MIC, minimum inhibitory concentration.
Determination of virulence factors
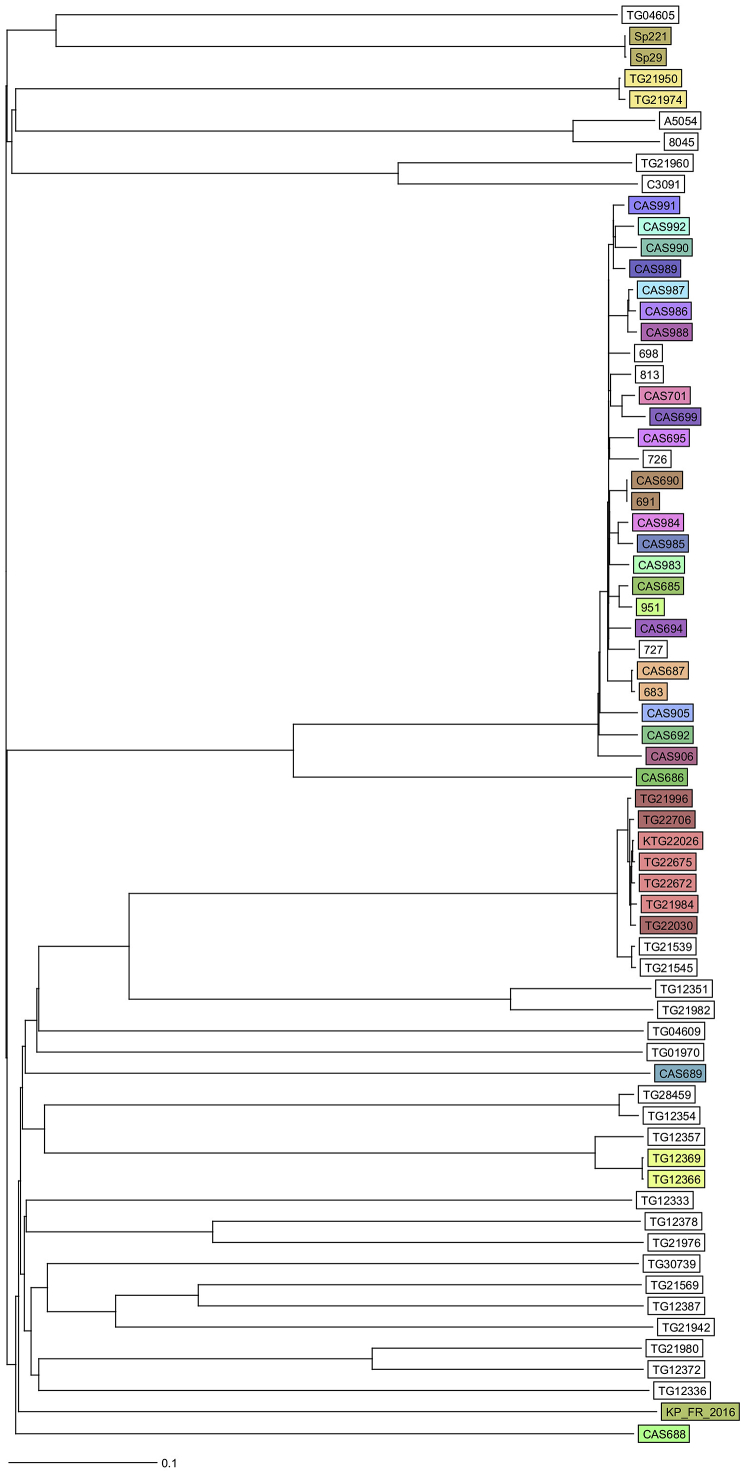
The isolate underwent further phenotypic and molecular tests for a series of virulence factors. Hypermucoviscosity was defined by a positive string test, which monitored the formation of a viscous string that is greater than 5 mm in length by stretching bacterial colonies on an agar plate as previously described [14]. DNA was extracted from pure cultures with the Ultraclean Microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA). The DNA library was prepared using the Nextera XT library preparation kit with the Nextera XT index kit (Illumina, San Diego, CA, USA). Subsequently, the library was sequenced on a MiSeq sequencer using the MiSeq reagent kit generating 250 bp paired-end reads (Illumina, San Diego, CA, USA) aiming at a coverage of at least 60-fold. Quality trimming of reads was performed with CLC Genomics Workbench 9.0.1 (Qiagen, Hilden, Germany) using a minimum Phred (Q) score of 28. De novo assembly was performed using CLC Genomics Workbench 7.0.4 (Qiagen) with optimal word sizes based on the maximum N50 value. For the assembled genome, the coverage (mean depth) was 85, the number of contigs was also 85, the N50 was 164 351, the maximum contig length was 368 426 nt and the total genome size was 5 459 317 nt. The multilocus sequence typing (MLST) sequence type (ST) was extracted from the assembled genome using Seqsphere+ version 3.0 (Ridom, Muenster, Germany) and appeared to be a new ST that was subsequently submitted to the MLST database (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). A clonal complex analysis was performed by eBURST (http://eburst.mlst.net/). Genes relating to virulence were detected using the mapping unit of CLC Genomics Workbench to map reads and/or by blasting assembled genomes to a pseudomolecule generated by concatenating a set of K. pneumoniae genes. Finally, the genetic similarity of our isolate with other K. pneumoniae strains [15] was determined by a gene-by-gene comparison using Seqsphere+ version 3.5.0 (Fig. 1).
Fig. 1.
Ridom Seqsphere+ neighbour-joining tree for 68 samples including Klebsiella pneumoniae strain KP_FR_2016 [1]. Tree is based on 2358 columns, pairwise, ignoring missing values. Distance is based on columns from K. pneumoniae sensu lato cgMLST scheme provided by Seqsphere+.
Study accession number
Generated raw reads were submitted to the European Nucleotide Archive (ENA) of the European Bioinformatics Institute (EBI) under the study accession number PRJEB19331.
Discussion
Biotype profiling of different K. pneumoniae clones revealed several clusters [16], of which our isolate belonged to clonal complex 66. A recent analysis conducted by Holt et al. [1] showed that ST23 and ST65 strains are dominant among hypervirulent K. pneumoniae strains. Our isolate is very close to ST65 and was assigned to ST2398. Many of the characteristic virulence genes could be detected in our isolate by whole genome sequencing and PCR (Table 1). It is possible that the number of hypervirulent cases would be indeed much higher as currently estimated if systematic investigations were done more often. Interestingly, our patient did not show any risk factors like underlying diabetes mellitus (HbA1c 6.1%) or immunodeficiency. He tested negative for HIV. An underlying hepatobiliary disease was not found. His travel history was positive for Brazil, where cases of infections due to K1 serotype K. pneumoniae have been described [17]. In Europe several cases have also been described, with many cases in France [18], [19]. When performing a Medline search using the terms ‘hypervirulent,’ ‘Klebsiella pneumoniae’ and ‘Germany,’ we found no case of severe infection published using these keywords. Currently investigations to determine a predisposition for infections by hypervirulent strains are ongoing [20].
Our case illustrates worldwide occurrence of hypervirulent strains. Adequate infection control and antimicrobial stewardship measures must be in place to contain further spread.
Conflict of Interest
None declared.
References
- 1.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keynan Y., Rubinstein E. The changing face of Klebsiella pneumoniae infections in the community. Int J Antimicrob Agents. 2007;30:385–389. doi: 10.1016/j.ijantimicag.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Li W., Sun G., Yu Y., Li N., Chen M., Jin R. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58:225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 4.Hennequin C., Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2016;35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Sun J., Mi C., Li W., Zhao S., Wang Q. First report of two rapid-onset fatal infections caused by a newly emerging hypervirulent K. pneumonia ST86 strain of serotype K2 in China. Front Microbiol. 2015;6:721. doi: 10.3389/fmicb.2015.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Zeng J., Liu W., Zhao F., Hu Z., Zhao C. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71:553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Lee H.C., Chuang Y.C., Yu W.L., Lee N.Y., Chang C.M., Ko N.Y. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med. 2006;259:606–614. doi: 10.1111/j.1365-2796.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 8.Yu W.L., Ko W.C., Cheng K.C., Lee H.C., Ke D.S., Lee C.C. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42:1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 9.Yeh K.M., Lin J.C., Yin F.Y., Fung C.P., Hung H.C., Siu L.K. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis. 2010;201:1259–1267. doi: 10.1086/606010. [DOI] [PubMed] [Google Scholar]
- 10.Bachman M.A., Oyler J.E., Burns S.H., Caza M., Lepine F., Dozois C.M. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh P.F., Lin T.L., Lee C.Z., Tsai S.F., Wang J.T. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2008;197:1717–1727. doi: 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 12.Chou H.C., Lee C.Z., Ma L.C., Fang C.T., Chang S.C., Wang J.T. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun. 2004;72:3783–3792. doi: 10.1128/IAI.72.7.3783-3792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jagnow J., Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149:2397–2405. doi: 10.1099/mic.0.26434-0. [DOI] [PubMed] [Google Scholar]
- 14.Shon A.S., Bajwa R.P., Russo T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Struve C., Roe C.C., Stegger M., Stahlhut S.G., Hansen D.S., Engelthaler D.M. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brisse S., Fevre C., Passet V., Issenhuth-Jeanjean S., Tournebize R., Diancourt L. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutinho R.L., Visconde M.F., Descio F.J., Nicoletti A.G., Pinto F.C., Silva A.C. Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: a case report of hypervirulent ST23. Mem Inst Oswaldo Cruz. 2014;109:970–971. doi: 10.1590/0074-0276140196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surgers L., Boyd A., Girard P.M., Arlet G., Decre D. ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22:1687–1688. doi: 10.3201/eid2209.160681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentzien M., Rosman J., Decre D., Brenkle K., Mendes-Martins L., Mateu P. Seven hypervirulent ST380 Klebsiella pneumoniae septic localizations. Med Mal Infect. 2017 doi: 10.1016/j.medmal.2016.10.002. In press. [DOI] [PubMed] [Google Scholar]
- 20.Lee I.R., Molton J.S., Wyres K.L., Gorrie C., Wong J., Hoh C.H. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep. 2016;6:29316. doi: 10.1038/srep29316. [DOI] [PMC free article] [PubMed] [Google Scholar]