Abstract
ShigaShield™ is a phage preparation composed of five lytic bacteriophages that specifically target pathogenic Shigella species found in contaminated waters and foods. In this study, we examined the efficacy of various doses (9x105-9x107 PFU/g) of ShigaShield™ in removing experimentally added Shigella on deli meat, smoked salmon, pre-cooked chicken, lettuce, melon and yogurt. The highest dose (2x107 or 9x107 PFU/g) of ShigaShield™ applied to each food type resulted in at least 1 log (90%) reduction of Shigella in all the food types. There was significant (P<0.01) reduction in the Shigella levels in all phage treated foods compared to controls, except for the lowest phage dose (9x105 PFU/g) on melon where reduction was only ca. 45% (0.25 log). The genomes of each component phage in the cocktail were fully sequenced and analyzed, and they were found not to contain any “undesirable genes” including those listed in the US Code for Federal Regulations (40 CFR Ch1). Our data suggest that ShigaShield™ (and similar phage preparations with potent lytic activity against Shigella spp.) may offer a safe and effective approach for reducing the levels of Shigella in various foods that may be contaminated with the bacterium.
Introduction
Shigella is an important cause of morbidity and mortality throughout the world, causing approximately 125 million Shigella infections / year and an estimated 14,000 deaths, mostly among children <5 years of age [1,2]. In the United States, Shigella is the third most common causes of gastroenteritis, with at least 500,000 cases of shigellosis linked diarrheal events in the USA annually [3,4]. General symptoms of shigellosis include bloody watery diarrhea, fever, nausea, and tenesmus (pain in the bowel), while complications include post-infection arthritis, sepsis, seizures (in young children) and hemolytic-uremic syndrome [4]. The two main routes of transmission include (i) through contaminated foods, and (ii) water contaminated with human waste; Shigella are easily transmitted through human contact due to its low infectious dose of 10–200 cells [5]. Shigella have been isolated from almost all food types, including salads (potato, tuna, shrimp, macaroni, or chicken), fresh fruits and vegetables, poultry, milk and dairy products, deli meats, and seafood [6]. In the USA, the main Shigella species isolated from contaminated foods is Shigella sonnei (found in ca. 80% of tested foods during 1998–2008). Internationally, Shigella flexineri (66%) and then Shigella sonnei (18%) are the most common food contaminant Shigella species [6,7]. The “ready-to-eat” foods (i.e., foods that are ready to be eaten, without any additional processing) are of particular concern because of the lack of the heat processing step (e.g., cooking) that could kill Shigella and render those foods safer to eat. Shigella also present a major concern to military personnel and travelers, especially when the deployment and/or travel is in the endemic areas where commercial food sanitation standards are poor and enforcement of those standards is lenient [8]. For example, several studies have shown a considerable loss of person-hours because of traveler’s diarrhea among U.S. military personnel deployed to the Persian Gulf during operations Desert Shield and Desert Storm, well as during peacetime operations [9–12]. The problem is further exacerbated by the increase of Shigella strains that are resistant to many commonly available antibiotics. For example, in 2013, the Centers for Disease Control and Prevention (CDC) declared antibiotic-resistant Shigella an urgent threat in the United States [13]. Also, an increasing prevalence of antibiotic resistance has been recently reported in the USA for the Shigella strains transmitted through sexual contact among men (nearly a quarter of Shigella isolates tested in New York City showed decreased susceptibility or resistance to recommended antibiotics) [14]. The prevalence of antibiotic-resistant Shigella strains also appears to be on the rise in foods; for example, in a recent study where more than 1,600 food samples (seafood, fresh vegetables, and meats) were examined, 89% of the Shigella strains were found to be multidrug resistant [6]. In these settings, novel non-antibiotic approaches are required to reduce the prevalence and levels of Shigella in various foods, which may help reduce the risk of shigellosis in the civilian (including young children) and military (including in US troops stationed in developing countries) populace. Lytic bacteriophages may provide one such relatively novel, environmentally friendly “green” approach.
Bacteriophages are bacterial viruses that are the most abundant biological entities in the world, in one ml of seawater there is an estimated 107 phages, with approximately 1030–1031 in the world [15]. Lytic phages have a potent, highly specific bactericidal activity against their targeted bacterial cells–a feature that enables a targeted killing of specific problem-causing bacteria in various settings, without disturbing the normal–and often beneficial–microflora. Various bacteriophage-based food additives have been approved by the FDA for food safety applications, including (listed in chronological order of receipt of regulatory approval in the USA): ListShield™, Listex P-100™, EcoShield™, SalmoFresh™ and Salmonelex™ [16]. Here, we report the results of studies in which the ability of ShigaShield™ to reduce Shigella levels was evaluated in various foods experimentally contaminated with a S. sonnei strain. Various Shigella phage preparations have been extensively used therapeutically previously to prevent and/or treat shigellosis in humans [17] reviewed by [18]; however, to the best of our knowledge, there is only one previous report of successful use of a Shigella phage preparation in food safety applications [19]. The phage preparation described in this communication (i.e., ShigaShield™) is currently undergoing FDA and USDA review for the GRAS (Generally Recognized As Safe) status (GRN 672).
Materials and methods
Bacteriophage preparation
ShigaShield™ is a bacteriophage “cocktail” essentially identical to the previously-described ShigActive™ preparation [20]. It is composed of 5 lytic phages (mixed in approximately equal concentrations): SHSML-52-1 (ATCC PTA-121241), SHFML-11 (ATCC PTA-121234), SHSML-45 (ATCC PTA-121238), SHFML-26 (ATCC PTA-121236), and SHBML-50-1 (ATCC PTA-121239). The same lot (# 1112I210158) was used in all studies. The phage preparation was supplied in normal saline solution (0.1 M NaCl, pH 6.5–7.5), and was stored refrigerated (2–8°C) until use. An additional 36 strains were examined for their susceptibility to ShigaShield™ employing the same method used on the other Shigella strains in our previous study [20].
Genomic sequencing of component bacteriophages
Each phage included in ShigaShield™ was sequenced, annotated, and the sequences were deposited in GenBank at the National Center for Biotechnology Information (NCBI) (Accession numbers: KX130865.1; KX130864.1; KX130863.1; KX130862.1; and KX130861.1). Briefly, each monophage was sequenced on a MiSeq (Illumina, San Diego, CA), assembled and annotated on the CLC Bio Genomic Workbench software, using default settings (version 7.0.2; CLC Bio, Cambridge, MA) at the BioAnalytical Laboratory of the Institute of Marine and Environmental Technology (IMET). Annotations were confirmed using two additional annotation pipelines: Rapid Annotation Subsystem Technology (RAST, version 4.0) and the Phage Annotations Using Subsystems Technology server (PHAST, version 1.0). Lastly, each genome was scanned for the “undesirable genes” listed in 40 CFR §725.421, and for any potential virulence factors on the Virulence BLAST Interface (VBI) using default parameters.
Food items
The ability of ShigaShield™ to reduce the levels of Shigella in foods was examined in (1) smoked salmon, (2) pre-cooked chicken breast strips, (3) sliced deli corned beef, (4) honeydew melon, pre-cut and packaged, (5) 1.5% vanilla flavored yogurt, and (6) long leaf lettuce packaged in a bag. These foods were selected to encompass a variety of viscosities, carbon/protein levels, and textures. All food items were purchased in local Baltimore grocery stores and were not washed, heated, or otherwise pre-treated prior to the studies.
Bacterial strain used to experimentally contaminate foods
A nalidixic acid resistant strain of Shigella sonnei was selected by serially passaging the Intralytix SH.s43 (original identification University of Maryland-Pakistan Isolate 90) on Luria-Bertani (LB) (Neogen, MI) agar plates supplemented with increasing concentrations of nalidixic acid (Arcos Organics, NJ). The strain underwent ≤8 serial passages before it was determined to be nalidixic acid-resistant at a concentration of 25 μg/ml. After the serial passages, the strain was assigned an Intralytix designation SH.s53. It is susceptible to all phages in ShigaShield™. The strain was stored at –80°C in 70% LB broth/30% glycerol supplemented with 25 μg of nalidixic acid/mL. For each study, a frozen aliquot of SH.s53 was thawed and grown (37 ± 2°C, 16–24 h) in LB broth supplemented with nalidixic acid (25 μg/ml). Overnight growth for this strain corresponds to ca. 2x108 Colony Forming Units per mL (CFU/mL). An overnight culture of SH.s53 was applied to all foods in approximately same concentrations, ranging from ca. 2x103 CFU/g (melon, chicken, beef deli and yogurt) to ca. 3x103 CFU/g (lettuce) to ca. 4x103 CFU/g (salmon). The bacteria was evenly spread across the surfaces of or mixed into each food item using a hockey stick. The bacteria-contaminated samples then rested in room temperature for 60 minutes before ShigaShield™ application.
ShigaShield™ application
Immediately prior to use, ShigaShield™ was removed from refrigerated storage and diluted in clean tap water as necessary. ShigaShield™ and controls were applied using a Basic Spray Gun Model #250 (Badger Air-Brush Co., Franklin Park, IL) to evenly spray the treatment onto all food surfaces, except the yogurt, where the ShigaShield™ or water treatments were mixed into the food. Each food item, except lettuce, was treated with three levels of ShigaShield™ or water. All treatments were 0.9mL per 100g of food, the same volume of water was applied for the control. Three concentrations (in Plaque Forming Units, or PFU) of ShigaShield™ (1x1010 PFU/mL, 1x109 PFU/mL, or 1x108 PFU/mL) were used to obtain a final concentration of ca. 9x107 PFU/g, 9x106 PFU/g, or 9x105 PFU/g on the foods, respectively. Lettuce was treated with two levels of ShigaShield™ or PBS, applied at 1mL per 100g. Two concentrations of ShigaShield™ (2x109 PFU/mL or 2x108 PFU/mL) were used to obtain a final concentration of ca. 2 x107 PFU/g or 2x106 PFU/g on the lettuce, respectively.
General design of efficacy studies in foods
After treatment, food samples were incubated at room temperature for 5 minutes. Three replicates of 25 g sample sizes from each food type and experimental group were placed into sterile bags, and 225 mL of sterile peptone water (Becton, Dickinson and Co., MD) was added. The bags were hand mushed briefly and stomached for a minimum of 30 seconds. The number of viable Shigella in the samples was determined by plating 0.5 mL aliquots of the stomached food/peptone water mixture onto separate MacConkey plates (Becton, Dickinson and Co., MD) supplemented with nalidixic acid (25 mg/mL). The plates were incubated (35 ± 2°C, 24±2 hr), and the final bacterial amounts (CFU/g) were calculated after counting the colonies as follows:
Evaluation of results and statistical analysis
The efficacy of ShigaShield™ in reducing the levels of Shigella in various foods was determined by comparing the levels of Shigella recovered from the foods treated with phage preparation vs. controls. Statistical analysis was performed using the Analysis of Variance (ANOVA) test for each food type independently. Tukey- Kramer multiple comparison post-hoc tests were done to determine which phage concentrations were significantly different when significance was determined via ANOVA (P<0.01). All statistical analysis was performed using version 3.05 of GraphPad InStat and version 4.0 of GraphPad Prism (GraphPad Software, San Diego, CA; www.graphpad.com).
Results and discussion
Phage host range
In a previous study by Mai and colleagues, this phage cocktail was shown to lyse 62 of 65 Shigella strains representing all four known species of Shigella: S. flexneri, S. dysenteriae, S. sonnei and S. boydii [20] Since that publication, the cocktail was tested for its activity against an additional 36 multidrug-resistant Shigella strains obtained from the Centers for Disease Control and Prevention (CDC). This additional collection of Shigella included 5 S. boydii, 4 S. dysenteriae, 8 S. flexneri, and 19 S. sonnei strains (See Table 1). All new strains were resistant to at least 3 of the antibiotics tested by the National Antimicrobial Resistance Monitoring System (NARMS) (Nancy Strockbine, personal communication). All new multidrug-resistant Shigella strains were susceptible to the ShigaShield™ phage preparation when tested in the standard concentration of ca. 1x109 PFU/mL. When these data are combined with the previous data reported by Mai and colleagues [20], the ShigaShield™ phage cocktail lysed 98 (97%) of the 101 Shigella strains in our collection. [20].
Table 1. List of new Shigella isolates tested for susceptibility to ShigaShield™.
| Original ID | Intralytix ID | Provider | Shigella Serotype |
|---|---|---|---|
| 2013C-3160 | SH.f71 | CDC, Atlanta GA | flexneri |
| 2013C-3473 | SH.s72 | CDC, Atlanta GA | sonnei |
| 2013C-3606 | SH.f73 | CDC, Atlanta GA | flexneri |
| 2013C-3787 | SH.f74 | CDC, Atlanta GA | flexneri |
| 2013C-4189 | SH.f75 | CDC, Atlanta GA | flexneri |
| 2014C-3799 | SH.s76 | CDC, Atlanta GA | sonnei |
| 2015C-3053 | SH.s77 | CDC, Atlanta GA | sonnei |
| 2015C-3237 | SH.s78 | CDC, Atlanta GA | sonnei |
| 2015C-3288 | SH.s79 | CDC, Atlanta GA | sonnei |
| 2015C-3306 | SH.s80 | CDC, Atlanta GA | sonnei |
| 2015C-3349 | SH.s81 | CDC, Atlanta GA | sonnei |
| 2015C-3626 | SH.s82 | CDC, Atlanta GA | sonnei |
| 2015C-3627 | SH.s83 | CDC, Atlanta GA | sonnei |
| 2015C-3802 | SH.s84 | CDC, Atlanta GA | sonnei |
| 2015C-3811 | SH.s85 | CDC, Atlanta GA | sonnei |
| 2015C-4077 | SH.s86 | CDC, Atlanta GA | sonnei |
| 2015C-4287 | SH.s87 | CDC, Atlanta GA | sonnei |
| 2015C-4463 | SH.s88 | CDC, Atlanta GA | sonnei |
| 2015C-4465 | SH.s89 | CDC, Atlanta GA | sonnei |
| 2015C-4836 | SH.f90 | CDC, Atlanta GA | flexneri |
| 2015C-5216 | SH.s91 | CDC, Atlanta GA | sonnei |
| 2016C-3073 | SH.f92 | CDC, Atlanta GA | flexneri |
| 2016C-3082 | SH.s93 | CDC, Atlanta GA | sonnei |
| 2016C-3328 | SH.s94 | CDC, Atlanta GA | sonnei |
| 2016C-3355 | SH.f95 | CDC, Atlanta GA | flexneri |
| 2016C-3375 | SH.f96 | CDC, Atlanta GA | flexneri |
| 2013AM-2809 | SH.d97 | CDC, Atlanta GA | dysenteriae |
| 2014AM-1029 | SH.b98 | CDC, Atlanta GA | boydii |
| AM11413 | SH.d99 | CDC, Atlanta GA | dysenteriae |
| AM17886 | SH.d100 | CDC, Atlanta GA | dysenteriae |
| AM22438 | SH.b101 | CDC, Atlanta GA | boydii |
| AM25896 | SH.d102 | CDC, Atlanta GA | dysenteriae |
| AM38301 | SH.b103 | CDC, Atlanta GA | boydii |
| AM41657 | SH.b104 | CDC, Atlanta GA | boydii |
| AM49802 | SH.b105 | CDC, Atlanta GA | boydii |
All strain listed are susceptible to ShigaShield™. These strains are in addition to the 62 strains previously analyzed for susceptibility [20]
Phage genome analysis
Each phage genome in ShigaShield™ was fully sequenced and annotated to determine whether there were any potentially "undesirable" genes (e.g., virulent and/or toxic genes) present. No toxin, virulence, repressor genes, integrases, recombinases nor any bacterial gene listed in the US Code for Federal Regulations (40 CFR §725.421) were detected. The absence of these "undesirable" genes has important safety implications. In this context, the physiological safety of ShigaShield™ (under the name ShigActive™) has been previously demonstrated in mice that were administered oral doses at both the recommend upper limit dosage (ca. 1x 108 PFU/g) and a 10-fold higher dosage (1x109 PFU/g) [20]. In that study, no physiological signs of toxicity were detected even with the highest dose of ShigActive™. Moreover, the metagenomic analysis showed that oral administration of bacteriophage preparation (in contrast to an antibiotic) elicited no changes in the overall microbiome of the mice (i.e., non-targeted bacteria were not affected) [20].
The genome analyses data presented in this manuscript provide further supporting evidence of the safety of these bacteriophages from the genomic composition standpoint. Namely, the genomic data suggest that the component bacteriophages are lytic phages with no "undesirable" genes in their genomes (and thus no evidence of any potentially dangerous transducing ability), are safe, and well suited for biocontrol applications.
Efficacy of ShigaShield™ on foods experimentally contaminated with Shigella
Lettuce
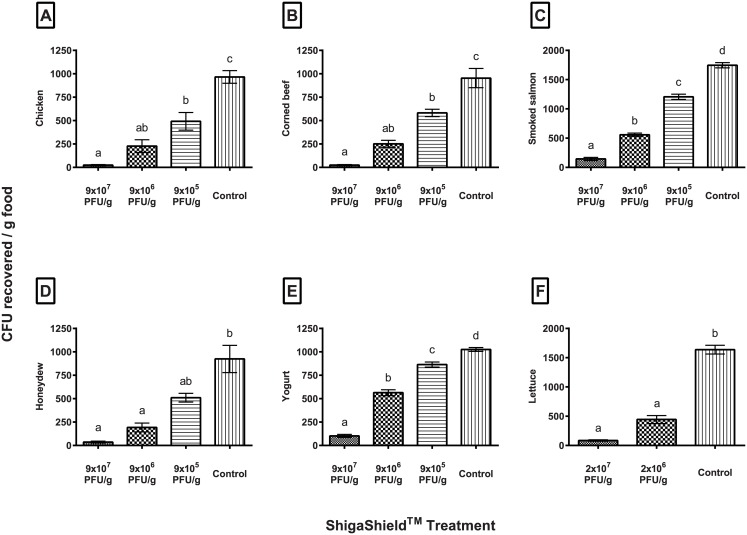
Contaminated lettuce has been the cause of several major foodborne Shigella outbreaks [21]. In this study, lettuce was the first food type tested with ShigaShield™. Because lettuce was the preliminary testing food, some of the methods were slightly different compared to the other foods examined. For example, PBS instead of water was used as a control, and different ShigaShield™ doses (i.e., the highest dose on lettuce was 2x107 vs. 9x107 PFU/g in the other studies). Even with the comparatively lower phage dosages in lettuce vs. the other studies, the higher phage application (2x107 PFU/g) still reduced Shigella levels by ca.1.3 log (95%) (Fig 1-F, Table 2). The lower dosage tested (2x106 PFU/g) also performed well, with 73% reduction or 0.6 log reduction in Shigella. Both doses were significantly different from the PBS controls (P<0.001) (Fig 1-F, Table 2), but not significantly different from each other, indicating that the lower and upper doses worked statistically similarly. This suggests that slight variations in application rates (e.g., due to human error or minor modifications in treatment protocols among various food producers) are not likely to significantly alter the efficacy.
Fig 1. Effect of ShigaShield on the Shigella levels in various foods.
Mean CFU recovered per gram of food (±SEM); for each food, means with different letters are significantly different (P<0.01). P-values are based on Tukey-Kramer multiple comparison post-hoc tests.
Table 2. Reduction in the Shigella levels in various foods treated with bacteriophages vs. water- or PBS-treated controls.
| Food | Group (phage concentration) | % Reduction±SEM | Log Reduction±SEM |
|---|---|---|---|
| Lettuce | A* | 95±0.73 | 1.3±0.06 |
| B* | 73±4.13 | 0.6±0.07 | |
| D* | 0±4.58 | 0±0.02 | |
| Salmon | A | 92±2.73 | 1.098±0.08 |
| B | 68±2.73 | 0.50±0.02 | |
| C | 31±4.49 | 0.16±0.02 | |
| D | 0±4.49 | 0±0.01 | |
| Chicken | A | 98±0.62 | 1.6±0.10 |
| B | 76±6.94 | 0.7±0.15 | |
| C | 49±9.73 | 0.3±0.08 | |
| D | 0±6.94 | 0±0.03 | |
| Corned beef | A | 97±0.63 | 1.6±0.10 |
| B | 74±3.93 | 0.6±0.11 | |
| C | 39±4.12 | 0.2±0.10 | |
| D | 0±10.73 | 0±0.049 | |
| Yogurt | A | 90±1.55 | 1.01±0.07 |
| B | 45±3.09 | 0.26±0.03 | |
| C | 16±2.68 | 0.07±0.01 | |
| D | 0±2.026 | 0±0.01 | |
| Melon | A | 96±1.95 | 1.44±0.14 |
| B | 79±8.78 | 0.7±0.12 | |
| C | 45±8.78 | 0.25±0.04 | |
| D | 0±27.30 | 0±0.07 |
Mean and Standard Error of Mean are shown (SEM). Group A* = 2x107 PFU/g; Group B* = 2x106 PFU/g; Group D* = PBS control; Group A = 9x107 PFU/g; Group B = 9x106 PFU/g; Group C = 9x105 PFU/g; Group D = water control.
Smoked salmon
After treatment regimens and doses were established using lettuce, additional foods were treated with ShigaShield™, starting with smoked salmon. Treatment with all doses of ShigaShield™ resulted in statistically significant reduction in the Shigella levels compared to water controls (ANOVA P<0.0001; post-hoc tests = P<0.001 for all) (Table 2). As expected, the lowest dose was less effective at reducing Shigella contamination compared to the higher doses (31% vs 68% and 92%). There was ≥ 1 log reduction in the Shigella levels with 9x107 PFU/g in all the tested replicates (Fig 1-C, Table 2). These data are in general agreement with the reduction of Listeria monocytogenes observed in smoked salmon samples treated with L. monocytogenes-specific phage preparation ListShield™ [22].
Precooked chicken breast strips
When ShigaShield™ was applied onto precooked chicken breast strips at the dose of 9x107 PFU/g, it significantly reduced Shigella levels by 98% (1.6 log) compared to the water control (P<0.001). This reduction is somewhat lower than that reported previously for other Shigella phages applied onto pre-cooked chicken samples [19]. In that study, phages were found to reduce the levels of Shigella by ca. 2.0 log after 48 hours. Two factors may be responsible for this difference, first, in our study, the contact time for phages was limited to 5 minutes vs. 48 h in the study by Zhang and colleagues [19], and longer incubation time in theory may increase phage treatment efficacy. Secondly, and perhaps more importantly, they [19] used a larger concentration of phages compared to our study (3x108 PFU/g in their study vs. max. of 9x107 PFU/g in our study). The efficacy of phage treatment is concentration-dependent, and using more phage is likely to yield a better reduction of the targeted bacterium’s level [23,24]. Our studies further support this idea. Namely, we observed less reduction in the Shigella levels with the lowest dose of ShigaShield™ (9x105 PFU/g) compared to the highest dose (P<0.01) (Fig 1-A, Table 2). The difference between various doses fades away when the doses differ by less than 1 log. For example, the highest dose of 9x107 reduced the targeted bacteria by 98% (1.6 logs), which is not significantly different (P>0.01) from the medium dose of 9x106, which resulted in ~76% reduction (0.7 log). As noted earlier, this range of dose variation vs. efficacy may be important to ensure the efficacy of the phage applications in the real-life commercial settings, when slight variations in application rates (e.g., due to minor deviations in treatment protocols among various food producers) may be encountered.
Corned beef deli meat
ShigaShield™ application was also effective in reducing the levels of Shigella in corned beef samples, at all concentrations examined (p<0.001). Among the different doses, the highest dosage resulted in 97% (1.6 log) reduction of Shigella compared to 74% (0.6 log) resulting from the next dose of 9x106 PFU/g (Fig 1-B, Table 2). All three treatments had significantly reduced CFU/g compared to water controls (P<0.001 for the concentrations 9x107, 9x106 PFU/g and P<0.01 for group 9x105 PFU/g -Table 2). There was no significance (P>0.01) between group 9x107 and 9x106, or 9x106 and 9x105, however 9x107 and 9x105 PFU/g were significantly different (P<0.001) (Fig 1-C). The lowest dose group 9x105 PFU/g had 39% reduction compared to water controls and 0.2 log reduction.
Yogurt
ShigaShield™ reduced the levels of Shigella in yogurt by ca. 1 log (90%) when the highest dose (9x107 PFU/g) was used. There was a significant difference among the various treatment groups (P< 0.001), and all treatments were statistically different from each other (P<0.001) and from water controls (group 9x107 and 9x106, P<0.001, group 9x105 PFU/g, P<0.01- Table 2, Fig 1-E). Interestingly, the efficacy of phage treatment declined more rapidly in yogurt compared to the other foods when lower concentrations of phage were used for treatment. For example, we observed only a 0.07 log (16%) reduction in the Shigella levels for the lowest dose (9x105 PFU/g) of ShigaShield™ vs., for example, reduction of 0.2 log (39%) in corned beef samples with the similar low dose. Various factors may have contributed to this outcome. For example, in the yogurt experiment, the same PFU/g was applied as other foods, but phages were mixed inside the yogurt with a greater overall surface area compared to the other foods, which have likely resulted in fewer contacts between phages and their targeted bacterial cells. Another (or additional) possibility is that some yogurt ingredient(s) inhibited phages or provided additional layer of protection for the targeted bacterial cells so that larger phage concentrations were required to effectively lyse the bacteria. Additional studies will be required to determine the underlying mechanisms, but our studies show that with the proper phage concentration, ShigaShield™ can provide a significant reduction in Shigella levels even in such complex food matrices as is yogurt.
Honeydew melon
ShigaShield™ application significantly reduced the levels of Shigella in honeydew melon samples at all concentrations examined. The two higher doses (9x107 PFU/g and 9x106 PFU/g) both resulted in significant reduction in the Shigella levels compared to water controls (P<0.001), ranging from 0.7 to 1.44 log reductions, respectively (Table 2, Fig 1-D). While the lowest dose of 9x105 did not significantly reduce Shigella, there was still a respectable 45% (0.25 log) reduction of Shigella compared to the water controls (Table 2, Fig 1-D). There was no significant difference among the 9x106 PFU/g dose and either the higher or lower dosage; the middle range dose reduced Shigella by 79% (0.7 log) (Fig 1-D).
Conclusion
Our studies continue to support the idea that lytic bacteriophages can be used to effectively reduce the levels of various foodborne bacteria in various foods, thus rendering those foods safer for human consumption. Although major foodborne outbreaks of Shigella infections are relatively rare in the United States, Shigella spp. do cause ca. 500,000 cases of illness in the USA annually [3,4,25]. The problem of shigellosis was recently highlighted by an outbreak in Flint, Michigan during September—October 2016, where the population was afraid to use water for handwashing due to the concerns of the water being contaminated with lead [26]. Although the role of contaminated foods in the outbreak has not been firmly established, it is likely that they played at least some—and possibly major—role in spreading the disease. Another, more direct example of foodborne shigellosis outbreak is the 406-person antibiotic resistant S. sonnei outbreak in 2000, which was linked to ready to eat dip [27]. ShigaShield™ and similar phage preparations lytic for Shigella may help reduce the incidence or severity of such outbreaks. Another important area of potential application is the use of phages to improve the safety of foods for the US military and/or travelers; e.g., for treating fresh fruits and vegetables in high risk Shigella locations overseas, where US troops are stationed but where the local sanitation standards and/or quality of water are not optimal, thus creating an increased risk of Shigella contamination.
The use of phage preparations for food safety applications has been gradually gaining traction in the United States, where several phage preparations targeting major foodborne pathogens like L. monocytogenes, E. coli O157:H7, and Salmonella are currently on the market. These preparations offer safe and effective intervention modalities for removing their targeted bacteria from various foods, without altering organoleptic qualities of those foods and their normal microflora / nutritional characteristics [16]. After ShigaShield™ becomes commercially available, it will be another addition to that growing family of all natural lytic phage preparations that can serve as an important additional tool for reducing the levels of Shigella in our foods and making them safer to eat.
Acknowledgments
We thank Karen Kotloff (University of Maryland, Baltimore, MD), Afsar Ali and J. Glenn Morris (University of Florida, Gainesville, FL), and Nancy Strockbine (CDC, Atlanta, GA) for sharing Shigella strains.
Data Availability
All phage genomes are available from the NCBI database (accession numbers KX130865.1; KX130864.1; KX130863.1; KX130862.1; and KX130861.1).
Funding Statement
Our studies were supported, in part, by a Small Business Innovation Research (SBIR) award W911NF-11-C-0074 and Contract No. W911QY-09-C-0079 from the United States Army (to A.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bardhan P, Faruque ASG, Naheed A, Sack DA. Decrease in shigellosis-related deaths without shigella spp.- specific interventions, Asia. Emerg Infect Dis. 2010;16(11):1718–23. 10.3201/eid1611.090934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. Global burden of Shigella infections: Implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–66. [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED. Laboratory-confirmed shigellosis in the United States, 1989–2002: epidemiologic trends and patterns. Clin Infect Dis. 2004;38(10):1372–7. 10.1086/386326 [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). Shigella-Shigellosis [Internet]. The Center for Disease Control and Prevention. https://www.cdc.gov/shigella/general-information.html
- 5.DuPont Herbert L; Levine Myron M; Hornick Richard B; Formal SB. Inoculum Size in Shigellosis and Implications for Expected Mode of Transmission Author (s): Herbert L. DuPont, Myron M. Levine, Richard B. Hornick and Samuel B. Formal Reviewed work (s): Published by: Oxford University Press Stable URL: http:/. J Infect Dis. 1989;159(6):1126–8. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed AM, Shimamoto T. Molecular characterization of multidrug-resistant Shigella spp. of food origin. Int J Food Microbiol [Internet].2015;194:78–82. Available from: 10.1016/j.ijfoodmicro.2014.11.013 [DOI] [PubMed] [Google Scholar]
- 7.Nygren BL, Schilling KA, Blanton EM, Silk BJ, Cole DJ, Mintz ED, et al. HHS Public Access. Epidemiol Infect. 2013;141(2):233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani S, Wierzba T, Walker RI. Status of vaccine research and development for Shigella. Vaccine [Internet]. 2016;34(26):2887–94. Available from: 10.1016/j.vaccine.2016.02.075 [DOI] [PubMed] [Google Scholar]
- 9.Quin NE. The impact of diseases on military operations in the Persian Gulf.e. Mil Med. 1982;147(9):728 [PubMed] [Google Scholar]
- 10.Hyams KC, Hanson K, Wignall FS, Division E, Medical N. The Impact of Infectious Diseases on the Health of U. S. Troops Deployed to the Persian Gulf during Operations Desert Shield and Desert Storm Author (s): Kenneth C. Hyams, Kevin Hanson, F. Stephen Wignall, Joel Escamilla, Edward C. Oldfield and. 2017;20(6):1497–504. [DOI] [PubMed] [Google Scholar]
- 11.DEMAIO J, BAILEY L, HALL K, BOYD R. A major outbreak of foodborne gstroenteritis among air force personnel during operation desert storm. Mil Med [Internet]. [cited 2017 Jan 10];158(3):161–4. Available from: http://cat.inist.fr/?aModele=afficheN&cpsidt=4794050 [PubMed] [Google Scholar]
- 12.Riddle MS, Smoak BL, Thornton SA, Bresee JS, Faix DJ, Putnam SD. Epidemic infectious gastrointestinal illness aboard U.S. Navy ships deployed to the Middle East during peacetime operations—2000-2001. BMC Gastroenterol [Internet]. 2006;6:9 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1456974&tool=pmcentrez&rendertype=abstract 10.1186/1471-230X-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.CDC. Antibiotic resistance threats in the United States, 2013. Current [Internet]. 2013;114. http://www.cdc.gov/drugresistance/threat-report-2013/index.html
- 14.Bowen A, Grass J, Bicknese A, Campbell D, Hurd J, Kirkcaldy RD. Elevated risk for antimicrobial drug-resistant Shigella infection among men who have sex with men, United States, 2011–2015. Emerg Infect Dis. 2016;22(9):1613–6. 10.3201/eid2209.160624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11(11):2771–4. 10.1111/j.1462-2920.2009.02101.x [DOI] [PubMed] [Google Scholar]
- 16.Woolston J, Sulakvelidze A. Bacteriophages and Food Safety. eLS [Internet]. 2015;1–13. http://doi.wiley.com/10.1002/9780470015902.a0025962 [Google Scholar]
- 17.Mulczyk M. and Slopek S. Use of a new phage preparation in prophylaxis and treatment of shigellosis. Acta Microbiol Acad Sci Hung. 1973;21(12):115. [PubMed] [Google Scholar]
- 18.Goodridge LD, Bisha B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage [Internet]. 2011;1(3):130–7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3225777/ 10.4161/bact.1.3.17629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Wang R, Bao H. PROCESSING, PRODUCTS, AND FOOD SAFETY Phage inactivation of foodborne Shigella on ready-to-eat spiced chicken. Poult Sci [Internet]. 2013;92:211–7. Available from: http://ps.oxfordjournals.org/content/92/1/211.full.pdf+html 10.3382/ps.2011-02037 [DOI] [PubMed] [Google Scholar]
- 20.Mai V, Ukhanova M, Reinhard MK, Li M, Sulakvelidze A. Bacteriophage administration significantly reduces Shigella colonization and shedding by Shigella-challenged mice without deleterious side effects and distortions in the gut microbiota. Bacteriophage [Internet]. 2015;5(4):e1088124 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4745833%7B&%7Dtool=pmcentrez%7B&%7Drendertype=abstract 10.1080/21597081.2015.1088124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapperud G, Rorvik LM, Hasseltvedt V, Hoiby EA, Iversen BG, Staveland K, et al. Outbreak of Shigella sonnei infection traced to imported iceberg lettuce. J Clin Microbiol. 1995;33(3):609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera MN, Abuladze T, Li M, Woolston J, Sulakvelidze A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol [Internet]. 2015;52:42–8. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0740002015001173 10.1016/j.fm.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 23.GREER GG. Effects of Phage Concentration, Bacterial Density, and Temperature on Phage Control of Beef Spoilage. J Food Sci [Internet]. 1988. July [cited 2017 Jan 11];53(4):1226–7. Available from: http://doi.wiley.com/10.1111/j.1365-2621.1988.tb13570.x [Google Scholar]
- 24.Worley-Morse TO, Zhang L, Gunsch CK. The long-term effects of phage concentration on the inhibition of planktonic bacterial cultures. Environ Sci Process Impacts [Internet]. 2013;16(1):81–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24301469 [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention (CDC). Foodborne Outbreak Online Database (FOOD Tool) [Internet]. The Center for Disease Control and Prevention; https://wwwn.cdc.gov/foodborneoutbreaks/
- 26.Ganim S. Amid water crisis, Flint faces a Shigellosis outbreak. CNN [Internet]. 2016;1 http://www.cnn.com/2016/10/03/health/flint-water-shigellosis-outbreak/ [Google Scholar]
- 27.Kimura AC, Johnson K, Palumbo MS, Hopkins J, Boase JC, Reporter R, et al. Multistate shigellosis outbreak and commercially prepared food, United States. Emerg Infect Dis. 2004;10(6):1147–9. 10.3201/eid1006.030599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All phage genomes are available from the NCBI database (accession numbers KX130865.1; KX130864.1; KX130863.1; KX130862.1; and KX130861.1).