Abstract
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fifth most common cause of cancer-related death in Europe with a 5-year survival rate of <5%. Chronic pancreatitis (CP) is a risk factor for PDAC development, but in the majority of cases malignancy is discovered too late for curative treatment. There is at present no reliable diagnostic marker for PDAC available.
Objectives
The aim of the study was to identify single blood-based metabolites or a panel of metabolites discriminating PDAC and CP using liquid chromatography-mass spectrometry (LC-MS).
Methods
A discovery cohort comprising PDAC (n = 44) and CP (n = 23) samples was analyzed by LC-MS followed by univariate (Student’s t test) and multivariate (orthogonal partial least squares-discriminant analysis (OPLS-DA)) statistics. Discriminative metabolite features were subject to raw data examination and identification to ensure high feature quality. Their discriminatory power was then confirmed in an independent validation cohort including PDAC (n = 20) and CP (n = 31) samples.
Results
Glycocholic acid, N-palmitoyl glutamic acid and hexanoylcarnitine were identified as single markers discriminating PDAC and CP by univariate analysis. OPLS-DA resulted in a panel of five metabolites including the aforementioned three metabolites as well as phenylacetylglutamine (PAGN) and chenodeoxyglycocholate.
Conclusion
Using LC-MS-based metabolomics we identified three single metabolites and a five-metabolite panel discriminating PDAC and CP in two independent cohorts. Although further study is needed in larger cohorts, the metabolites identified are potentially of use in PDAC diagnostics.
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-017-1199-6) contains supplementary material, which is available to authorized users.
Keywords: Pancreatic cancer, Pancreatic ductal adenocarcinoma, Pancreatitis, Metabolomics, Metabolism, LC-MS, Biomarkers, Serum, Plasma, Discovery, Validation
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth most common cause of cancer-related death with a 5-year survival rate of <5% (Siegel et al. 2016). In contrary to many other forms of cancer the incidence of PDAC is rising and predicted to become the 2nd most common cause of cancer-related death by 2030 (Rahib et al. 2014). Its lethality can be attributed to late diagnosis, almost complete resistance to conventional chemo- and radiotherapy, and a lack of diagnostic biomarkers. Unlike breast or colorectal cancer, the only treatment with curative intent is surgery, for which < 20% of patients qualify; but even then the chance of survival is very low (Lohr 2006). Therefore, PDAC should be treated as a medical emergency (Lohr 2014). Risk factors for PDAC include chronic pancreatitis (CP) and around 5% of CP patients develop PDAC over time (Pinho et al. 2014; Garrido-Laguna and Hidalgo 2015). Both PDAC and CP are characterized by an excessive and reactive stroma, or desmoplasia, which makes it difficult to distinguish between the two conditions (Kloppel and Adsay 2009). The one FDA-approved biomarker for PDAC, carbohydrate antigen 19-9 (CA 19-9), is only useful for prognosis and detection of disease recurrence (Fong and Winter 2012). Therefore there is an urgent clinical need of a non-invasive diagnostic biomarker for PDAC.
Metabolomics is the untargeted analysis of low molecular weight endogenous compounds, metabolites, present in e.g. a defined biological sample such as tissue or a bodily fluid as blood (Fiehn et al. 2000; Fiehn 2002; Spratlin et al. 2009; Dunn et al. 2011). The most widely used analytical platform for metabolomics providing the largest metabolome coverage is liquid chromatography-mass spectrometry (LC-MS) (Want et al. 2007; Patti et al. 2012; Yin and Xu 2014). Given the established connection between cell metabolism and cancer (Hanahan and Weinberg 2011), LC-MS metabolomics has been applied to identify metabolite markers for several cancers (Spratlin et al. 2009; Nordstrom and Lewensohn 2010; Nicholson et al. 2012), including PDAC (Daemen et al. 2015; Rios Peces et al. 2016). Specifically, there have been previous LC-MS based metabolomics efforts to compare PDAC and CP blood samples (Urayama et al. 2010; Fukutake et al. 2015). There is however still no screening test for PDAC available.
In the present study, we have performed LC-MS based metabolomics on blood samples to compare the metabolic profiles of PDAC and CP in two independent cohorts. Both single discriminative metabolites and a panel of metabolites discriminating PDAC and CP were identified. Our findings have potential clinical relevance for early diagnosis of PDAC among CP patients to improve patient survival.
Materials and methods
Clinical samples
The study comprised a discovery- and a validation cohort (Table 1). All patients had given informed consent in accordance with the Helsinki Declaration (World Med 2013).
Table 1.
Clinical data
| Discovery cohort | Validation cohort | |||
|---|---|---|---|---|
| PDAC | CP | PDAC | CP | |
| Subjects (n) | 44 | 23 | 20 | 31 |
| Male/female ratio | 21/20a | 18/5 | 10/10 | 16/15 |
| Age, median (range) | 67 (30–102)b | 36 (31–93) | 70 (46–80) | 69 (23–87) |
an = 41,
bn = 42
Blood samples for the discovery cohort were collected from German patients with PDAC (n = 44) and patients with CP (n = 23). Serum samples were prepared at the same location using a standardized protocol.
The validation cohort included Swedish PDAC patients (n = 20) and CP patients (n = 31). Plasma samples were supplied by the Karolinska Institutet biobank and had been prepared at different sites following a standardized procedure: whole blood samples were obtained at fasting early in the morning, collected in sodium citrate tubes and immediately placed in −80 °C.
All plasma and serum samples were stored at −80 °C at all times prior to analysis.
Sample preparation
All solvents used below were of HPLC grade (Fisher Chemicals) and the water was Milli-Q (Millipore).
Plasma and serum samples were thawed on ice and 50 µl aliquots were mixed with 150 µl MeOH for protein precipitation. Samples were then centrifuged for 15 min at 15800×g. The supernatant was transferred to a new tube and then evaporated to complete dryness in a vacuum concentrator. Prior to LC-MS analysis, samples were reconstituted in 50 µl MeOH:H2O 1:1 and the two isotopically labelled internal standards phenylalanine (D5) and palmitic acid (D4) (Cambridge Isotope Laboratories) were added at a final concentration of 5 µM.
LC-MS analysis
Metabolites were separated by reversed phase liquid chromatography and detected by electrospray ionization (ESI) mass spectrometry in positive mode. All samples were run in randomized order, randomized by the random number function in Excel (Microsoft, USA).
For the discovery cohort, the analytical platform consisted of a 1200 HPLC-system (Agilent) connected on-line to an LTQ Orbitrap Velos equipped with a HESI probe (Thermo Scientific). 5 µl of each sample were injected onto a Kinetex C18 150 × 2.1 mm column, 2.6 µ, 100 Å (Phenomenex) and the following mobile phases were used at a flow rate of 0.4 ml/min: H2O with 0.1% formic acid (A) and 3:1 acetonitrile:isopropanol with 0.1% formic acid (B). The linear gradient was 0 min, 5% B; 1 min, 5% B; 21 min, 95% B; 26 min, 95% B; 26.5 min, 5% B; 33 min, 5% B. MS data was collected in centroid mode between m/z 80-1200 at an Orbitrap resolution of 60,000 using the following ESI settings: Cap Temp, 350; Sheath Gas Flow, 45; Aux Gas Flow, 15; Source Voltage, 3 kV; S-Lens RF Level, 60%.
For the validation cohort, the analytical platform consisted of a 1290 UHPLC-system (Agilent) connected on-line to a 6550 Q-ToF equipped with a JetStream source (Agilent). 5 µl of each sample were injected onto a Kinetex C18 100 × 2.1 mm column, 2.6 µ, 100 Å (Phenomenex) and the following mobile phases were used at a flow rate of 0.5 ml/min: H2O with 0.1% formic acid (A) and 3:1 acetonitrile:isopropanol with 0.1% formic acid (B). The linear gradient was 0 min, 5% B; 7 min, 95% B; 9 min, 95% B; 9.2 min, 5% B; 11 min, 5% B. MS data was collected in centroid mode between m/z 70-1700 using the following ESI settings: Gas Temp, 300; Gas Flow, 8; Nebulizer Pressure, 40; Sheet Gas Temp, 350; Sheet Gas Flow, 11; Vcap, 4000; Fragmentor, 100; Skimmer1, 45; OctapoleRFPeak, 750. The analysis procedure has also been described previously (Staubert et al. 2015; Lindahl et al. 2016).
The peak heights for the internal standards were continuously monitored during analysis to ensure MS signal stability (Supplementary table 1). In the validation cohort the signal stability was monitored by injection of a pooled QC sample for every sixth sample during the LC-MS analysis. Examples of extracted ion chromatograms are illustrated in Supplementary Fig. 1.
Targeted MS/MS data of a pooled sample was also collected for identification purposes of discriminative features in both cohorts using the ESI settings described above (Supplementary Fig. 2). Samples were analyzed on two different instruments for reasons of instrument availability at the time of receiving the different cohorts to the laboratory.
Discovery cohort data preprocessing
Discovery raw data files were converted to .cdf format using the file converter tool in Xcalibur version 2.2 (Thermo Scientific). Peak detection, integration and alignment were performed with the open-source software XCMS version 1.30.3 (Smith et al. 2006). The following XCMS settings were used: For peak detection and integration with the centWave algorithm, ppm = 10, snthr = 5, peakwidth = c(5.15), mzdiff = −0.01, prefilter = c(6,100), fitgauss = TRUE; for alignment with Obiwarp, distFunc = “cor”, profStep = 1, grouping parameters bw = 1, mzwid = 0.005, minfrac = 0.5. Isotopes and adducts were not removed.
Sample-wise median normalization (Trezzi et al. 2015) was used to correct for technical variation in e.g. sample preparation and MS signal intensity. Next, a two-step peak filtering approach was applied. First, peaks with an intensity level ≥ 60,000 were retained. Second, only peaks present in at least 75% of samples in at least one group were used for the following statistical analyses.
Univariate analysis
Both univariate and multivariate statistical analysis were applied in parallel (Fig. 1).
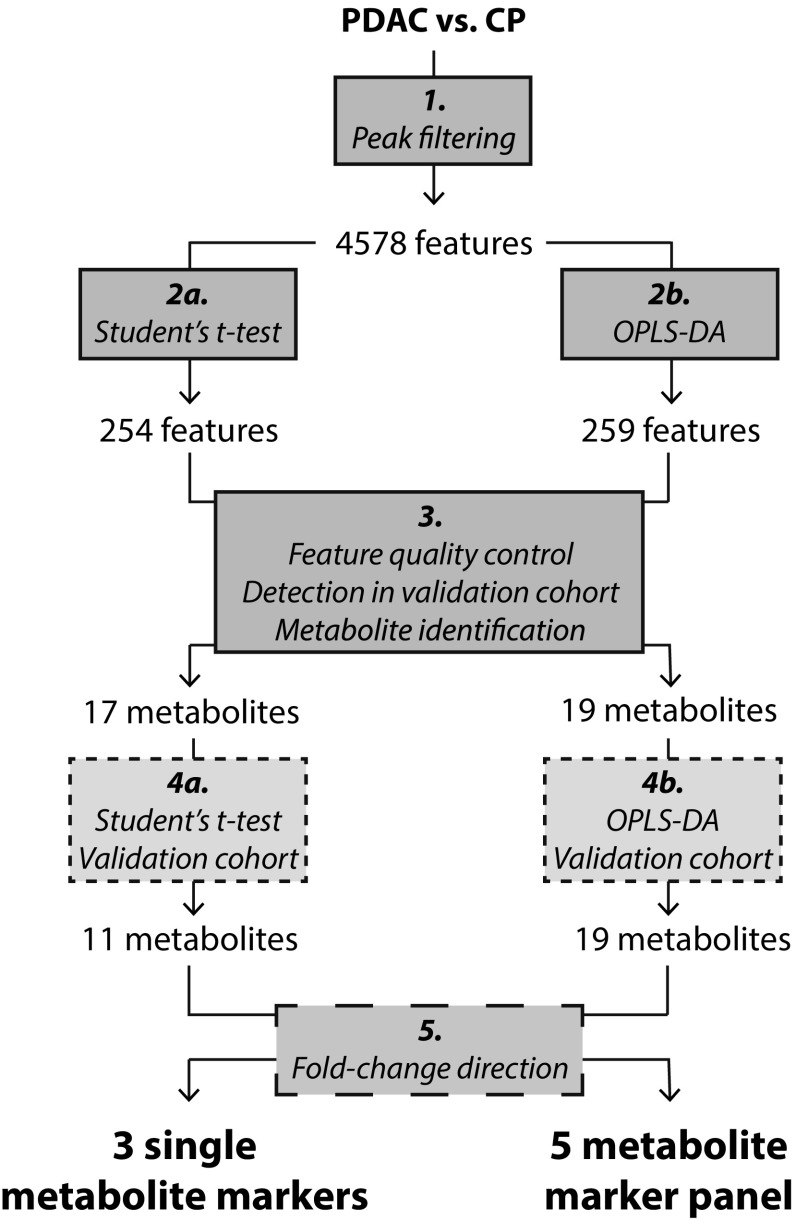
Fig. 1.
Schematic overview of data analysis workflow. Univariate (left) and multivariate (right) analyses comparing PDAC and CP were performed in parallel, starting with the abundance filtered discovery cohort data set (step 1). The large number of metabolite features (n = 4578) is due to superfluous features e.g. adducts and isotopes. Following statistical analysis of the discovery cohort (step 2a and b), selected metabolite features were examined manually in the raw data and low quality features (e.g. adducts, isotopes, low intensity peaks) were excluded (step 3). High quality features that were not present in the validation cohort were excluded as well, together with features that could not be identified. The discriminative capacity of the remaining identified features, now referred to as metabolites, was confirmed in the validation cohort by statistical analysis (step 4a and b). Finally, metabolites that were regulated in opposite directions in the two cohorts were excluded (step 5), resulting in three single metabolite markers and a panel of five metabolite markers
Univariate analysis was performed in the software GraphPad Prism version 6.07 (GraphPad), where PDAC and CP were compared using a Student’s t test with Welch’s correction. In the discovery cohort, metabolite features with p-value > 0.05 were excluded in order to limit the number of features to be examined in the raw data.
Box plots and bar graphs were created in GraphPad Prism.
Multivariate analysis
For multivariate analysis, data sets were imported into the software SIMCA version 14 (Umetrics). Unit variance scaling was applied to give metabolite features with low and high variation between samples equal importance, and log transformation was applied to shift data towards Gaussian distribution. Principal component analysis (PCA) was initially applied to all variables extracted using XCMS (Supplementary Fig. 3). Subsequently was orthogonal partial least squares-discriminant analysis (OPLS-DA) (Bylesjo et al. 2006) with default SIMCA settings was used to identify metabolite feature patterns discriminating PDAC and CP. The features with the highest discriminatory power between classes were selected based on the variable importance for the projection (VIP) plot. Features significant with a 95% confidence interval, based on the cross- validation, were selected for further analysis. For the discovery cohort, the feature selection was done in the same way with the additional criterion that the VIP-values should be >1.5.
For refined OPLS-DA models, i.e. those built on selected metabolite features only, cross-validation settings were changed from the default 1/7 to 1/5 of samples constituting the internal prediction set. The purpose was to minimize the bias of the refined model towards the selected metabolite features.
All score- and loading scatter plots were created in SIMCA.
Feature quality control
The discriminative metabolite features selected in the discovery cohort had to meet several criteria to ensure feature quality, regardless of the method of statistical analysis. First, features were manually examined in the raw data. Isotopes, adducts and poor quality features, e.g. low intensity peaks were excluded following manual inspection of peak shape. This removed features judged by the software to be chromatographic peaks, but which they human eye rapidly can assign to be column bleed or similar artefacts. Second, for the univariate part of the analysis workflow, features were re-integrated in the raw data using the Qual and Quan Browsers in Xcalibur version 2.2 (Thermo Scientific) and their statistical significance was confirmed with a Student´s t test (p < 0.05). Third, ion chromatograms of the selected features were extracted from the validation cohort raw data files using the software MassHunter Qual version B.06.00 (Agilent); discovery cohort features that were either not detected or of low quality in the validation cohort were excluded. Fourth, unidentifiable metabolite features were excluded.
Peak areas of the identified metabolites that met quality control criteria were integrated in the validation cohort raw data files using the “Agile” setting in MassHunter Qual. The median normalized peak area values were used for further uni- and multivariate analyses to confirm the results from the discovery cohort.
Metabolite identification
Accurate mass measurements were subject to database searches in the public databases METLIN (Smith et al. 2005) and Human Metabolome Database (Wishart et al. 2013) as well as an in-house library comprising 384 synthetic standards. Database hits were then confirmed by retention time match (in-house library only) and MS/MS spectral match from pooled samples (Supplementary Fig. 2). In a few cases additional synthetic standard compounds were acquired and analyzed on the same platform to confirm compound identity.
For the discovery cohort, samples were also fractionated by LC and relevant fractions were then analyzed by direct infusion to acquire MSn structural information (van der Hooft et al. 2012).
Results
Univariate analysis identifies three single metabolites discriminating PDAC and CP
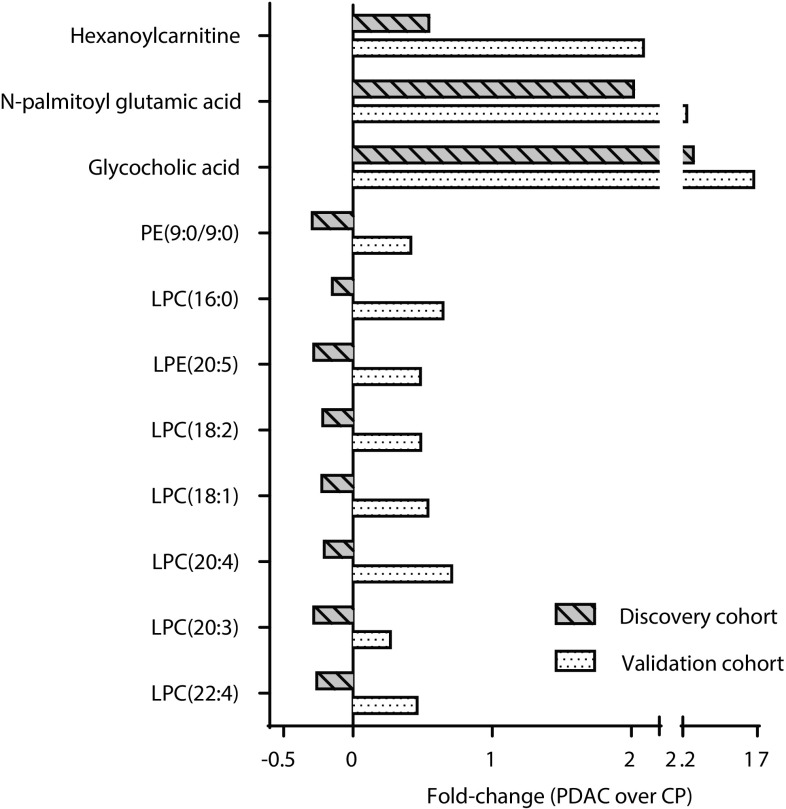
After abundance peak filtering, the discovery cohort data set contained 4578 metabolite features including isotopes, adducts and low quality features (Fig. 1). 254 of these features had a p-value < 0.05 as determined by Student’s t test with Welch’s correction. After (i) raw data examination, (ii) re-integration and confirmation of statistical significance, (iii) detection in validation cohort and (iv) feature identification, 17 metabolites remained. 11 of these metabolites had a p-value < 0.05 (Student’s t test) in the validation cohort as well. Fold-change calculations showed that the eight metabolites with the lowest intensity were down-regulated in PDAC compared to CP in the discovery cohort, as opposed to up-regulated in PDAC compared to CP in the validation cohort (Fig. 2). These eight metabolites were phospholipids. After exclusion of the phospholipids, the three metabolites glycocholic acid, hexanoylcarnitine and N-palmitoyl glutamic acid remained as single discriminative markers for PDAC compared to CP (Fig. 3). In the discovery cohort, applying an FDR cut-off of 5% to the 254 metabolites did not yield any significant metabolites. However the manual inspection and re-integration together with the validation in an independent patient cohort strengthens the findings.
Fig. 2.
Fold-change comparison between cohorts. Following confirmation of metabolite significance in the validation cohort (Fig. 1, step 4a and b), fold-change calculations revealed that eight metabolites were regulated in opposite directions in the discovery- and validation cohorts. All eight metabolites were phospholipids that were down-regulated in PDAC compared to CP in the discovery cohort, but up-regulated in PDAC compared to CP in the validation cohort. Consequently all phospholipids were excluded as potential single metabolite markers
Fig. 3.
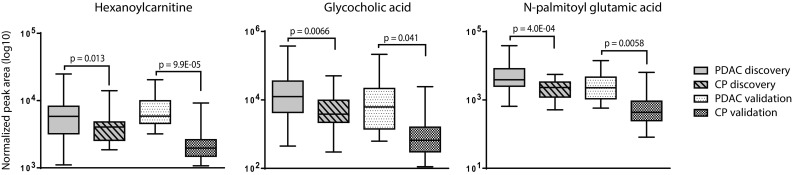
Single metabolite markers discriminating PDAC and CP. After identification of significant features in the discovery cohort (n = 67), feature quality control and confirmation of significance in the validation cohort (n = 51), three single metabolite markers for PDAC compared to CP remained. All three were up-regulated in PDAC. Statistical test: Welch’s unequal variances t test. Box plot settings: Range, minimum to maximum value; line at median
Multivariate analysis identifies a panel of five metabolites discriminating PDAC and CP
259 of the 4578 metabolite features in the discovery cohort (Fig. 1, step 2b) were selected as potential markers for PDAC as determined by the VIP-plot in the initial OPLS-DA model (Fig. 4a). 19 metabolites remained after feature quality control, detection in validation cohort and metabolite identification. These 19 metabolites were significant in the validation cohort OPLS-DA model (Fig. 4b). A comparison of the loading scatter plots for the discovery- and validation cohort OPLS-DA models showed that a group of 14 metabolites was regulated in opposite directions in the two cohorts (Fig. 4c, d). As in the univariate analysis, all 14 were phospholipids that were down-regulated in PDAC compared to CP in the discovery cohort but up-regulated in PDAC in the validation cohort.
Fig. 4.
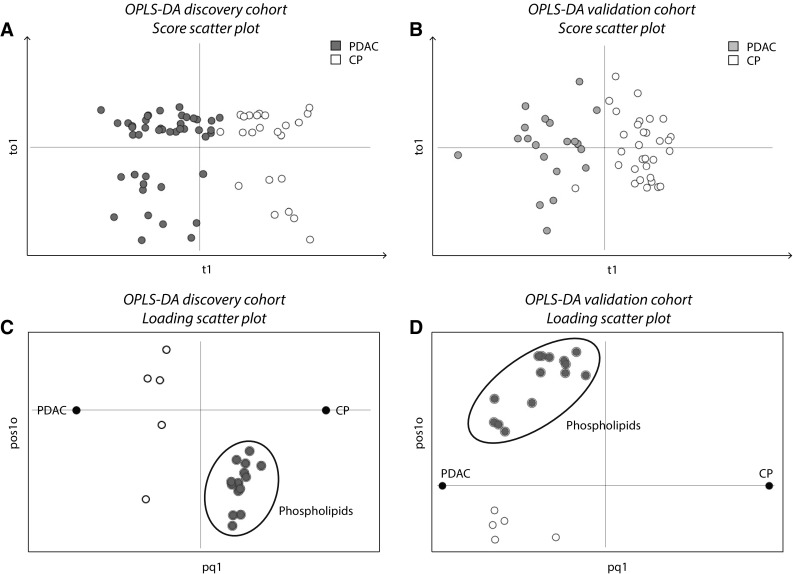
Reversed metabolite fold-change direction in discovery- and validation cohorts. a Score scatter plot for the initial OPLS-DA model of 4578 metabolite features in the discovery cohort (Fig. 1, step 2b). Blue dots represent PDAC samples (scores, n = 44); white dots represent CP samples (n = 23); t1 on the x-axis, first component; to1 on the y-axis, first of two orthogonal components. Model parameters: CV-groups, 7; R2X(cum) 0.39; Q2(cum) 0.22; CV-ANOVA p = 0.02. The model was used to select 259 features for further analysis. b Score scatter plot for the OPLS-DA model in the validation cohort based on the 19 metabolites remaining from the discovery cohort (Fig. 1, step 4b). Orange dots represent PDAC samples (n = 20); white dots represent CP samples (n = 31); to1 on the y-axis, first (and only) orthogonal component. Model parameters: CV-groups, 7; R2X(cum) 0.648; Q2(cum) 0.699; CV-ANOVA p = 1.7E-11. c Loading (metabolite) scatter plot for the discovery cohort OPLS-DA model. The phospholipids are situated to the right of the vertical line, close to the dummy CP variable, indicating that they are down-regulated in PDAC compared to CP. d Loading scatter plot for the validation cohort OPLS-DA model. The phospholipids are situated to the left of the vertical line, close to the dummy PDAC variable; hence they are up-regulated in PDAC compared to CP. Based on this reversal of their fold-change directions, the 14 phospholipids were excluded from the final metabolite marker panel
After exclusion of the phospholipids the five metabolites N-palmitoyl glutamic acid, glycocholic acid, hexanoylcarnitine, chenodeoxyglycocholate and PAGN remained. They were used to build a refined validation cohort OPLS-DA model (Fig. 5a, b) to evaluate their discriminatory power in the absence of phospholipids. The predictive ability of the refined model as indicated by the Q2(cum) value was 0.513, i.e. approximately 50% of samples were correctly classified, and the R2X(cum) value was 0.736. The statistical significance of the model as indicated by cross-validated ANOVA was p = 8.2E−07.
Fig. 5.
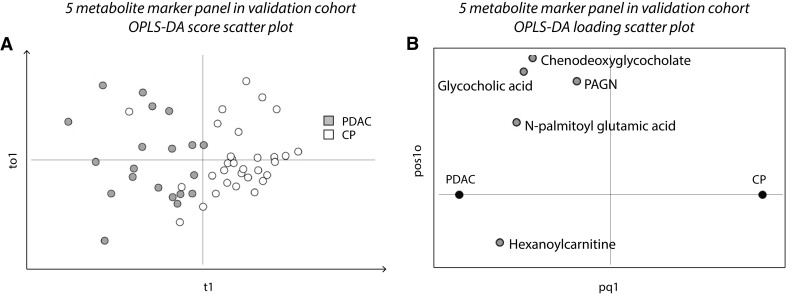
A marker panel of five metabolites discriminates PDAC and CP. a Score scatter plot for the refined OPLS-DA model of the five discriminative metabolites with consistent fold-change directions in the validation cohort (Fig. 1, step 4b and 5). Orange dots represent PDAC samples (n = 20); white dots represent CP samples (n = 31); t1 on the x-axis, first component; to1 on the y-axis, first (and only) orthogonal component. Model parameters: CV-groups, 5; R2X(cum) 0.736; Q2(cum) 0.513; CV-ANOVA p = 8.2E-07. b Corresponding loading scatter plot. All five metabolites in the marker panel show increased levels in PDAC compared to CP
Discussion
The PDAC incidence rate is expected to increase worldwide over the next years (Rahib et al. 2014), making the need for improved diagnostic tools even more acute. One group at risk of PDAC development is CP patients (Pinho et al. 2014) who would clearly benefit from the discovery of novel markers for PDAC diagnosis.
Any preclinical biomarker study at the discovery stage should include an inflammatory control of the target organ to increase the chance of identifying disease-specific markers (Chechlinska et al. 2010; Lindahl et al. 2016). Here, CP serves as an inflammatory control of the pancreas for PDAC-specific marker discovery. In addition, CP is a risk factor for PDAC development and the two diseases share several inflammatory parameters. It is therefore imperative to identify PDAC-unique markers specific for the discrimination of PDAC and CP.
In the present study, we have compared the metabolic profiles of PDAC and CP in blood in two independent cohorts using untargeted LC-MS. The fact that both cohorts did not consist of either serum or plasma is a drawback of the study. However, that they represent completely independent patients strengthens the validity of the proposed biomarkers. Previous reports comparing the metabolite profiles of serum and plasma samples have found good correlation between the two sample matrices (Denery et al. 2011; Yu et al. 2011; Ishikawa et al. 2014). We have identified three single discriminative metabolites as well as a five-metabolite panel of markers, since a marker panel can increase specificity and sensitivity compared to single discriminative metabolites (Wang et al. 2011; Wingren et al. 2012). All five metabolites identified here can be easily measured in a clinical MS-lab.
PDAC and CP blood samples have been compared by LC-MS metabolomics analysis in two previous studies (Urayama et al. 2010; Fukutake et al. 2015). One of these targeted free amino acids in plasma. The advantage of a targeted approach is that sample preparation and analysis is optimized for a specific class of metabolites, but at the cost of covering only a limited part of the metabolome. Untargeted LC-MS on the other hand has the largest potential to identify novel metabolites due to increased metabolome coverage (Patti et al. 2012). In the second previous study comparing PDAC and CP, untargeted LC-MS was applied to plasma samples. Contrary to the present study, clinical data on e.g. disease stage and smoking status was available, which is important to avoid known confounding factors; however, the sample size (n = 10) was very limited which makes the results less generalizable. Other studies have applied different analytical metabolomics platforms to discriminate PDAC and CP in bodily fluids. In a study using gas chromatography-mass spectrometry, a majority of the serum discriminative metabolites were amino acids (Kobayashi et al. 2013), but included also some metabolites that would not have been particularly well captured in our LC-MS set-up such as some sugar species including, arabinose, ribulose and 1,5-Anhydro-D-glucitol and some organic acids such as uric acid, nonanoic acid and caprylic acid. In the mentioned study, CP samples were however included in the validation cohort only. Amino acids were also connected to future risk of pancreatic cancer in a metabolomics study applying LC-MS to prediagnostic plasma (Mayers et al. 2014). In the present study a number of amino acids were found dysregulated in PDAC compared to CP in the validation cohort only (data not shown), but lacking confirmation in a second cohort these results were excluded. Further, two studies have used nuclear magnetic resonance to study plasma and urine samples, respectively, but have not validated their results in a second cohort (Zhang et al. 2012) or had only three CP samples out of a total of 25 samples included in the benign control group (Davis et al. 2013).
In the present study, the levels of a number of phospholipids were altered in PDAC compared to CP in both cohorts. Phospholipids were also found dysregulated in previous studies (Urayama et al. 2010; Ritchie et al. 2013; Sakai et al. 2016). However, we excluded all phospholipids as potential markers since they were down-regulated in PDAC in the discovery cohort as opposed to up-regulated in the validation cohort (Figs. 2, 4c, d). A possible explanation for these results is that different blood sample matrices, i.e. serum and plasma, were used in the two cohorts. Serum and plasma samples from the same individual are known to display different concentration levels for some metabolites, including phospholipids, due to the differences in sample handling (Yu et al. 2011). Noteworthy, as systemic phospholipid levels are altered by inflammatory responses in general, they are unlikely candidates for disease specific markers (Lindahl et al. 2016).
As mentioned above, the two bile acids glycocholic acid and chenodeoxyglycocholate were significantly increased in PDAC compared to CP in the present study (Figs. 3, 5b). A possible explanation for increased levels of circulating bile acids is tumor growth into the bile duct. Another theory includes bile acid reflux into the pancreas, leading to pancreatitis and eventually malignant cell transformation; a direct carcinogenic effect of bile acids is also possible (Feng and Chen 2016). Bile acid levels in prediagnostic plasma have also been connected to future risk of pancreatic cancer (Mayers et al. 2014), supporting our results and reasoning.
Two of the discriminative metabolite markers identified here, N-palmitoyl glutamic acid and hexanoylcarnitine (Figs. 3, 5b), are fatty acid conjugates with an amino acid and carnitine, respectively. N-acyl amino acids were only recently discovered as a novel group of compounds and little is known about their function (Hanus et al. 2014). However, a recent study in mice showed that certain N-acyl amino acids regulate cell metabolism through mitochondrial uncoupling (Long et al. 2016). Acylcarnitines in turn are vital for the transport of fatty acids into the mitochondrial matrix (Indiveri et al. 2011). Short-chain acylcarnitines including hexanoylcarnitine were previously found increased in serum in type 2 diabetes compared to normal glucose tolerance (Mai et al. 2013); both CP and PDAC may cause diabetes (Pinho et al. 2014).
The levels of the nitrogenous metabolite PAGN were also increased in PDAC compared to CP in the present study (Fig. 5b). PAGN is a product of gut microbiota-host co-metabolism found in urine and blood (Barrios et al. 2015), and there are indications that the gut microbiota has an important role in carcinogenesis (Schwabe and Jobin 2013).
In conclusion, this work has identified five metabolites capable of discriminating PDAC and CP in blood either as single or as a panel of markers. Findings were validated in a separate patient cohort. To determine the potential clinical benefit of these markers, further evaluation in larger clinical studies is needed. Nevertheless, we believe that these metabolites are potentially useful for early diagnosis of PDAC in CP patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
A.N. acknowledges The Swedish Foundation for Strategic Research (FFL4) and the Jane and Dan Olsson Foundation for funding. ML acknowledges ALF and Richard Julin Stiftelse for financial support and RH acknowledges Cancerfonden (CAN2008/982, CAN2013/780) for financial support. The Swedish Metabolomics Centre is acknowledged for support with LC-MS analysis.
Abbreviations
- RPLC-MS
Reversed phase liquid chromatography coupled to mass spectrometry
- PDAC
Pancreatic ductal adenocarcinoma
- CP
Chronic pancreatitis
- OPLS-DA
Orthogonal partial least squares discriminant analysis
Conflict of interest
The authors declare that they have no conflict of interests in relation to the work described.
Ethical approval
This study was approved by regional etic committee in Gothenburg, Sweden and by Die ethic-kommission-II, Fakultät für klinische Medizin Mannheim der Ruprecht-Karls-Universität Heidelberg, Germany, and was conducted in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Blood samples were collected after informed consent from all patients.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11306-017-1199-6) contains supplementary material, which is available to authorized users.
References
- Barrios C, Beaumont M, Pallister T, Villar J, Goodrich JK, Clark A, Pascual J, Ley RE, Spector TD, Bell JT, Menni C. Gut-microbiota-metabolite axis in early renal function decline. Plos ONE. 2015;10(8):e0134311. doi: 10.1371/journal.pone.0134311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylesjo M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics. 2006;20:341–351. doi: 10.1002/cem.1006. [DOI] [Google Scholar]
- Chechlinska M, Kowalewska M, Nowak R. Systemic inflammation as a confounding factor in cancer biomarker discovery and validation. Nature Reviews Cancer. 2010;10:2–13. doi: 10.1038/nrc2782. [DOI] [PubMed] [Google Scholar]
- Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, Hong R, Moffat J, Gao M, Boudreau A, Mroue R, Corson L, O’Brien T, Qing J, Sampath D, Merchant M, Yauch R, Manning G, Settleman J, Hatzivassiliou G, Evangelista M. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4410–E4417. doi: 10.1073/pnas.1501605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis VW, Schiller DE, Eurich D, Bathe OF, Sawyer MB. Pancreatic ductal adenocarcinoma is associated with a distinct urinary metabolomic signature. Annals of Surgical Oncology. 2013;20:S415–S423. doi: 10.1245/s10434-012-2686-7. [DOI] [PubMed] [Google Scholar]
- Denery JR, Nunes AA, Dickerson TJ. Characterization of differences between blood sample matrices in untargeted metabolomics. Analytical Chemistry. 2011;83:1040–1047. doi: 10.1021/ac102806p. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R, Human Serum Metabolome C. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Feng HY, Chen YC. Role of bile acids in carcinogenesis of pancreatic cancer: An old topic with new perspective. World Journal of Gastroenterology. 2016;22:7463–7477. doi: 10.3748/wjg.v22.i33.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant molecular biology. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fong ZV, Winter JM. Biomarkers in pancreatic cancer diagnostic, prognostic, and predictive. Cancer Journal. 2012;18:530–538. doi: 10.1097/PPO.0b013e31827654ea. [DOI] [PubMed] [Google Scholar]
- Fukutake N, Ueno M, Hiraoka N, Shimada K, Shiraishi K, Saruki N, Ito T, Yamakado M, Ono N, Imaizumi A, Kikuchi S, Yamamoto H, Katayama K. A novel multivariate index for pancreatic cancer detection based on the plasma free amino acid profile. Plos ONE. 2015;10(7):e0132223. doi: 10.1371/journal.pone.0132223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nature Reviews. Clinical Oncology. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanus L, Shohami E, Bab I, Mechoulam R. N-Acyl amino acids and their impact on biological processes. BioFactors. 2014;40:381–388. doi: 10.1002/biof.1166. [DOI] [PubMed] [Google Scholar]
- Indiveri C, Iacobazzi V, Tonazzi A, Giangregorio N, Infantino V, Convertini P, Console L, Palmieri F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Molecular Aspects of Medicine. 2011;32:223–233. doi: 10.1016/j.mam.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Maekawa K, Saito K, Senoo Y, Urata M, Murayama M, Tajima Y, Kumagai Y, Saito Y. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS ONE. 2014;9:e91806. doi: 10.1371/journal.pone.0091806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppel G, Adsay NV. Chronic pancreatitis and the differential diagnosis versus pancreatic cancer. archives of pathology & laboratory. Medicine. 2009;133:382–387. doi: 10.5858/133.3.382. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishiumi S, Ikeda A, Yoshie T, Sakai A, Matsubara A, Izumi Y, Tsumura H, Tsuda M, Nishisaki H, Hayashi N, Kawano S, Fujiwara Y, Minami H, Takenawa T, Azuma T, Yoshida M. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiology Biomarkers & Prevention. 2013;22:571–579. doi: 10.1158/1055-9965.EPI-12-1033. [DOI] [PubMed] [Google Scholar]
- Lindahl A, Forshed J, Nordstrom A. Overlap in serum metabolic profiles between non-related diseases: Implications for LC-MS metabolomics biomarker discovery. Biochemical and Biophysical Research Communications. 2016;478:1472–1477. doi: 10.1016/j.bbrc.2016.08.155. [DOI] [PubMed] [Google Scholar]
- Lohr JM. Personal view Pancreatic cancer should be treated as a medical emergency. Bmj-British Medical Journal. 2014;349:g5261. doi: 10.1136/bmj.g5261. [DOI] [PubMed] [Google Scholar]
- Lohr M. Is it possible to survive pancreatic cancer? Nature clinical practice gastroenterology &. Hepatology. 2006;3:236–237. doi: 10.1038/ncpgasthep0469. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Bateman LA, Lin H, Kamenecka T, Lokurkar IA, Lou J, Rao RR, Chang MR, Jedrychowski MP, Paulo JA, Gygi SP, Griffin PR, Nomura DK, Spiegelman BM. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell. 2016;166:424–435. doi: 10.1016/j.cell.2016.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai M, Tonjes A, Kovacs P, Stumvoll M, Fiedler GM, Leichtle AB. Serum levels of acylcarnitines are altered in prediabetic conditions. Plos ONE. 2013;8(12):e82459. doi: 10.1371/journal.pone.0082459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, Yuan C, Bao Y, Townsend MK, Tworoger SS, Davidson SM, Papagiannakopoulos T, Yang A, Dayton TL, Ogino S, Stampfer MJ, Giovannucci EL, Qian ZR, Rubinson DA, Ma J, Sesso HD, Gaziano JM, Cochrane BB, Liu SM, Wactawski-Wende J, Manson JE, Pollak MN, Kimmelman AC, Souza A, Pierce K, Wang TJ, Gerszten RE, Fuchs CS, Vander Heiden MG, Wolpin BM. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature Medicine. 2014;20:1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–392. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- Nordstrom A, Lewensohn R. Metabolomics: Moving to the clinic. Journal of Neuroimmune Pharmacology. 2010;5:4–17. doi: 10.1007/s11481-009-9156-4. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews. Molecular Cell Biology. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: A path to pancreatic cancer. Cancer Letters. 2014;345:203–209. doi: 10.1016/j.canlet.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Research. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- Rios Peces S, Diaz Navarro C, Marquez Lopez C, Caba O, Jimenez-Luna C, Melguizo C, Prados JC, Genilloud O, Vicente Perez F, Perez Del Palacio J. Untargeted LC-HRMS-based metabolomics for searching new biomarkers of pancreatic ductal adenocarcinoma: A pilot study. Journal of biomolecular screening: the official journal of the Society for Biomolecular Screening. 2016 doi: 10.1177/1087057116671490. [DOI] [PubMed] [Google Scholar]
- Ritchie SA, Akita H, Takemasa I, Eguchi H, Pastural E, Nagano H, Monden M, Doki Y, Mori M, Jin W, Sajobi TT, Jayasinghe D, Chitou B, Yamazaki Y, White T, Goodenowe DB. Metabolic system alterations in pancreatic cancer patient serum: potential for early detection. Bmc Cancer. 2013;13(1):416. doi: 10.1186/1471-2407-13-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Suzuki M, Kobayashi T, Nishiumi S, Yamanaka K, Hirata Y, Nakagawa T, Azuma T, Yoshida M. Pancreatic cancer screening using a multiplatform human serum metabolomics system. Biomarkers in Medicine. 2016;10:577–586. doi: 10.2217/bmm-2016-0020. [DOI] [PubMed] [Google Scholar]
- Schwabe RF, Jobin C. The microbiome and cancer. Nature Reviews Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca-a Cancer Journal for Clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: A metabolite mass spectral database. Therapeutic drug monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:431–440. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubert C, Bhuiyan H, Lindahl A, Broom OJ, Zhu YF, Islam S, Linnarsson S, Lehtio J, Nordstrom A. Rewired metabolism in drug-resistant leukemia cells a metabolic switch hallmarked by reduced dependence on exogenous glutamine. Journal of Biological Chemistry. 2015;290:8348–8359. doi: 10.1074/jbc.M114.618769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trezzi JP, Vlassis N, Hiller K. The role of metabolomics in the study of cancer biomarkers and in the development of diagnostic tools. Advances in Experimental Medicine and Biology. 2015;867:41–57. doi: 10.1007/978-94-017-7215-0_4. [DOI] [PubMed] [Google Scholar]
- Urayama S, Zou W, Brooks K, Tolstikov V. Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rapid Communications in Mass Spectrometry. 2010;24:613–620. doi: 10.1002/rcm.4420. [DOI] [PubMed] [Google Scholar]
- van der Hooft JJJ, Vervoort J, Bino RJ, de Vos RCH. Spectral trees as a robust annotation tool in LC-MS based metabolomics. Metabolomics. 2012;8:691–703. doi: 10.1007/s11306-011-0363-7. [DOI] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17:448–483. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. Journal of Proteome Research. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- Wingren C, Sandstrom A, Segersvard R, Carlsson A, Andersson R, Lohr M, Borrebaeck CAK. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Research. 2012;72:2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu YF, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia JG, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0-The human metabolome database in 2013. Nucleic Acids Research. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Med A. World medical association declaration of helsinki ethical principles for medical research involving human subjects. Jama-Journal of the American Medical Association. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- Yin P, Xu G. Current state-of-the-art of nontargeted metabolomics based on liquid chromatography-mass spectrometry with special emphasis in clinical applications. Journal of chromatography. A. 2014;1374:1–13. doi: 10.1016/j.chroma.2014.11.050. [DOI] [PubMed] [Google Scholar]
- Yu ZH, Kastenmuller G, He Y, Belcredi P, Moller G, Prehn C, Mendes J, Wahl S, Roemisch-Margl W, Ceglarek U, Polonikov A, Dahmen N, Prokisch H, Xie L, Li YX, Wichmann HE, Peters A, Kronenberg F, Suhre K, Adamski J, Illig T, Wang-Sattler R. Differences between human plasma and serum metabolite profiles. Plos ONE. 2011 doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jin HF, Guo XG, Yang Z, Zhao L, Tang SH, Mo P, Wu KC, Nie YZ, Pan YL, Fan DM. Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by H-1 nuclear magnetic resonance-based metabonomic profiles. Clinical Biochemistry. 2012;45:1064–1069. doi: 10.1016/j.clinbiochem.2012.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.