Abstract
Scientific discoveries and technological advancements are inseparable but not always take place in a coherent chronological manner. In the next, we will provide a seemingly unconnected and serendipitous series of scientific facts that, in the whole, converged to unveil DNA and its duplication. We will not cover here the many and fundamental contributions from microbial genetics and in vitro biochemistry. Rather, in this journey, we will emphasize the interplay between microscopy development culminating on super resolution fluorescence microscopy (i.e., nanoscopy) and digital image analysis and its impact on our understanding of DNA duplication. We will interlace the journey with landmark concepts and experiments that have brought the cellular DNA replication field to its present state.
Keywords: DNA, DNA replication, Replication foci, Replicon, Replisome, Super resolution microscopy
The (very) early years
Long after water-filled glass bowls were used to read small letters (Singer 1914), a simple single lens microscope started the microscopic revolution (Bacon 1267) (see Table 1 and Fig. 1). Spurred throughout the ages by accidental inventions (van der Aa 1851), leaps by Galileo (Galilei 1610), and Hooke (1665), it was not until Carl Zeiss started to mass-produce microscopes in 1847 that DNA observation started to take off. Simultaneously, Mendel studied 29,000 pea plants (1866) and Haeckel postulated the containment of hereditary traits in the nucleus (1866) (Dahm 2008; Haeckel 1866), while Miescher put the microscope to good use and purified the nuclei and observed DNA (Miescher 1871). Köhler’s game-changing illumination technique (Köhler 1893) helped to perfect Zeiss UV-microscope together with Siedentopf in 1908. In 1927, shortly after Levene described the nucleic acid structure (1919), Koltsov postulated the semiconservative replication idea (Soyfer 2001).
Table 1.
Chronological list of landmarks in microscopy and DNA replication
| Year | Landmark | Author |
|---|---|---|
| 63 | Water filed glass bowls to read small letters | (Singer 1914) |
| 1267 | The first simple microscope | (Bacon 1267) |
| 1590 | Accidental discovery of the compound microscope with two (or more) lenses by Zacharias Janssen | (van der Aa 1851) |
| 1610 | “Microscope” with ×1000 magnification | (Galilei 1610) |
| 1665 | “Micrographia” | (Hooke 1665; Singer 1914) |
| 1847 | First “mass produced” microscopes in 1847 | |
| 1866 | Hereditary traits in 29,000 pea plants | (Mendel 1866) |
| 1866 | Hereditary traits contained in the nucleus | (Dahm 2008; Haeckel 1866) |
| 1871 | Purified nuclei for the first time and observed DNA | (Miescher 1871) |
| 1893 | Ein neues Beleuchtungsverfahren für mikrophotographische Zwecke | (Köhler 1893) |
| 1907 | On the absorption of antibodies | |
| 1908 | First fluorescence microscopes based on UV-microscopy | |
| 1919 | Identification of the nucleic acid structure | |
| 1927 | “Replicate in a semiconservative fashion using each strand as a template” | (Soyfer 2001) |
| 1932 | Discovery of the electron microscope | (Knoll and Ruska 1932a; Knoll and Ruska 1932b) |
| 1947 | DNA X-ray diffraction images | (Astbury 1947) |
| 1953 | X-ray diffraction “Photo 51” | (Watson and Crick 1953) |
| 1953 | Discovery of the double-helix DNA structure | (Watson and Crick 1953) |
| 1953 | Discovery of phase contrast microscopy | (Zernike 1955) |
| 1958 | Confirmation of the semiconservative DNA replication model | (Meselson and Stahl 1958) |
| 1957 | Discovery of the confocal microscope | (Minsky 1961) |
| 1962 | Extraction, purification, and properties of GFP | (Shimomura et al. 1962) |
| 1963 | DNA unwinding for replication and “replication fork” | (Cairns 1963) |
| 1966 | Autoradiography of chromosomal DNA fibers from Chinese hamster cells. | (Huberman and Riggs 1966) |
| 1966 | On the mechanism of DNA replication in mammalian chromosomes | (Huberman and Riggs 1968) |
| 1967 | First practical application of the “Nipkow disk” in confocal microscopy | (Egger and Petráň 1967; Petráň et al. 1968) |
| 1968 | Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. | (Okazaki et al. 1968) |
| 1968 | Mechanism of DNA chain growth, II. Accumulation of newly synthesized short chains in E. coli infected with ligase-defective T4 phages. | (Sugimoto et al. 1968) |
| 1969 | Duration of the cell cycle | (Van Dilla et al. 1969) |
| 1969 | Mechanism of DNA chain growth, III. Equal annealing of T4 nascent short DNA chains with the separated complementary strands of the phage DNA | (Sugimoto et al. 1969) |
| 1969 | Mechanism of DNA chain growth. IV. Direction of synthesis of T4 short DNA chains as revealed by exonucleolytic degradation. | (Okazaki and Okazaki 1969) |
| 1972 | Bidirectional Replication of Simian Virus 40 DNA | (Danna and Nathans 1972) |
| 1974–1979 | Fork speed, replication speed, and replicon sizes | (Kriegstein and Hogness 1974; Taylor 1977; Taylor and Hozier 1976; Wilson and Wilson 1975; Yurov 1977; Yurov 1978; Yurov 1979; Yurov and Liapunova 1977) |
| 1975 | Continuous cultures of fused cells secreting antibody of predefined specificity. | |
| 1986 | Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus | (Nakamura et al. 1986) |
| 1989 | Three distinctive replication patterns | (Nakayasu and Berezney 1989) |
| 1992 | Dynamic organization of DNA replication in mammalian cell nuclei spatially and temporally defined replication of chromosome | (O’Keefe et al. 1992) |
| 1992 | Progression of DNA synthesis | (Rizzoli et al. 1992) |
| 1993 | Structured Illumination Microscopy (SIM) | (Bailey et al. 1993) |
| 1994 | Green fluorescent protein as a marker for gene expression | (Chalfie et al. 1994) |
| 1994 | 4pi microscope | (Hell 2003; Hell et al. 1994) |
| 1994 | Alignment and sensitive detection of DNA by a moving interface | (Bensimon et al. 1994) |
| 1997 | The replication origin decision point is a mitogen | (Wu and Gilbert 1997) |
| 1997 | Dynamic molecular combing: stretching the whole human genome for high-resolution studies. | (Michalet et al. 1997) |
| 1998 | Replicon clusters are stable units of chromosome structure evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells | (Jackson and Pombo 1998) |
| 1999 | The spatial position and replication timing of chromosomal domains are both established in early G1 phase | (Dimitrova and Gilbert 1999) |
| 1999 | Single molecule analysis of DNA replication. | (Herrick and Bensimon 1999) |
| 2000 | Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci | (Berezney et al. 2000) |
| 2000 | Dynamics of DNA replication factories in living cells | (Leonhardt et al. 2000) |
| 2000 | DNA replication at high resolution | (Keck and Berger 2000) |
| 2000 | Mechanisms of DNA replication | (Davey and O’Donnell 2000) |
| 2001 | Eukaryotic origins | |
| 2001 | Repression of origin assembly in metaphase depends on inhibition of RLF-BCdt1 by geminin | (Tada et al. 2001) |
| 2001 | Visualization of DNA replication on individual Epstein-Barr Virus episomes | (Norio and Schildkraut 2001) |
| 2002 | DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters | (Sporbert et al. 2002) |
| 2002 | DNA replication and chromatin | (Gerbi and Bielinsky 2002) |
| 2002 | Initiation of DNA replication in multicellular eukaryotes | (Gerbi et al. 2002) |
| 2003 | Sequence-independent DNA binding and replication initiation by the human origin recognition complex | (Vashee et al. 2003) |
| 2003 | The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication | (DePamphilis 2003) |
| 2004 | Stable chromosomal units determine the spatial and temporal organization of DNA replication | (Sadoni et al. 2004) |
| 2004 | DNA replication and DNA repair mechanisms most of the replication machinery is also used in DNA repair. | (Sancar and Lindsey-Boltz 2004) |
| 2005 | Preventing rereplication | (Blow and Dutta 2005) |
| 2005 | PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins | (Sporbert et al. 2005) |
| 2005 | Eukaryotic origins of DNA replication: could you please be more specific? | (Cvetic and Walter 2005) |
| 2006 | Origin selection and silent origins | (Patel et al. 2006) |
| 2006 | Regulating the licensing of DNA replication origins in metazoa | (DePamphilis et al. 2006) |
| 2006 | DNA replication: keep moving and don’t mind the gap. | (Langston and O’Donnell 2006) |
| 2007 | Impact of chromatin structure | |
| 2007 | Replisome mechanics: insights into a twin DNA polymerase machine. | (Pomerantz and O’Donnell 2007) |
| 2007 | The many faces of the origin recognition complex | (Sasaki and Gilbert 2007) |
| 2007 | High-throughput mapping of origins of replication in human cells. | (Lucas et al. 2007) |
| 2007 | Characterization of a triple DNA polymerase replisome. | (McInerney et al. 2007) |
| 2007 | Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. | (Hamdan et al. 2007) |
| 2007 | Polymerase switching in DNA replication. | (Lovett 2007) |
| 2008 | 3D–SIM | (Gustafsson et al. 2008) |
| 2008 | Division of labor at the eukaryotic replication fork. | (Nick McElhinny et al. 2008) |
| 2008 | DNA polymerases at the replication fork in eukaryotes | (Stillman 2008) |
| 2008 | Discovery of stimulated emission depletion (STED) | (Schmidt et al. 2008) |
| 2009 | In DNA replication, the early bird catches the worm. | (Boye and Grallert 2009) |
| 2009 | G-quadruplex structures: in vivo evidence and function. | (Lipps and Rhodes 2009) |
| 2009 | Eukaryotic DNA replication control: lock and load, then fire. | (Remus and Diffley 2009) |
| 2010 | Organization of DNA replication | (Chagin et al. 2010) |
| 2010 | Eukaryotic chromosome DNA replication: where, when, and how? | (Masai et al. 2010) |
| 2010 | SCF (Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. | (D’Angiolella et al. 2010) |
| 2010 | Uncoupling of sister replisomes during eukaryotic DNA replication. | (Yardimci et al. 2010) |
| 2010 | DNA replication: making two forks from one prereplication complex. | (Botchan and Berger 2010) |
| 2011 | Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. | (Heller et al. 2011) |
| 2011 | Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. | (Ozeri-Galai et al. 2011) |
| 2011 | Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. | (Fu et al. 2011) |
| 2011 | Genome-wide depletion of replication initiation events in highly transcribed regions. | (Martin et al. 2011) |
| 2011 | Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. | (Tanaka et al. 2011) |
| 2012 | Genome-scale identification of active DNA replication origins. | (Cayrou et al. 2012) |
| 2012 | Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae | (Knott et al. 2012) |
| 2012 | A fragment based click chemistry approach towards hybrid G-quadruplex ligands: design, synthesis and biophysical evaluation | (Ritson and Moses 2012) |
| 2012 | Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. | (Casas-Delucchi et al. 2012) |
| 2012 | OriDB, the DNA replication origin database updated and extended. | (Siow et al. 2012) |
| 2012 | Replication timing: the early bird catches the worm. | (Douglas and Diffley 2012) |
| 2012 | CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy: mechanistic rationale for drug combination therapy. | (Siddiqui-Jain et al. 2012) |
| 2012 | Experimental approaches to identify cellular G-quadruplex structures and functions. | (Di Antonio et al. 2012) |
| 2012 | Activation of the replicative DNA helicase: breaking up is hard to do. | (Boos et al. 2012) |
| 2012 | Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. | (Bianco et al. 2012) |
| 2012 | DeOri: a database of eukaryotic DNA replication origins. | (Gao et al. 2012) |
| 2012 | Replication origins run (ultra) deep. | (Gilbert 2012) |
| 2012 | Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. | (Besnard et al. 2012) |
| 2012 | Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. | (Casas-Delucchi et al. 2012) |
| 2013 | Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. | (Dellino et al. 2013) |
| 2013 | Functional implications of genome topology. | (Cavalli and Misteli 2013) |
| 2013 | Nuclear positioning. | (Gundersen and Worman 2013) |
| 2013 | Chromatin dynamics at the replication fork: there’s more to life than histones. | (Whitehouse and Smith 2013) |
| 2013 | Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. | (McGuffee et al. 2013) |
| 2013 | The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. | (Kubota et al. 2013) |
| 2013 | Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. | (Yamazaki et al. 2013) |
| 2013 | Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins. | (Mesner et al. 2013) |
| 2013 | A personal reflection on the replicon theory: from R1 plasmid to replication timing regulation in human cells. | (Masai 2013) |
| 2013 | From simple bacterial and archaeal replicons to replication N/U-domains. | (Hyrien et al. 2013) |
| 2013 | Genomes and G-quadruplexes: for better or for worse. | (Tarsounas and Tijsterman 2013) |
| 2013 | New insights into replication clamp unloading. | (Ulrich 2013) |
| 2013 | Replication dynamics: biases and robustness of DNA fiber analysis. | (Técher et al. 2013) |
| 2013 | Specification of DNA replication origins and genomic base composition in fission yeasts. | (Mojardín et al. 2013) |
| 2013 | The replication domain model: regulating replicon firing in the context of large-scale chromosome architecture. | (Pope and Gilbert 2013) |
| 2013 | Time to be versatile: regulation of the replication timing program in budding yeast. | (Yoshida et al. 2013) |
| 2013 | Why are there so many diverse replication machineries? | (Forterre 2013) |
| 2014 | Epigenetic control of DNA replication dynamics in mammals | (Casas-Delucchi and Cardoso 2014) |
| 2014 | Lethal effects of short-wavelength visible light on insects. | (Hori et al. 2014) |
| 2014 | Existence and consequences of G-quadruplex structures in DNA. | (Murat and Balasubramanian 2014) |
| 2014 | Histone variants: the tricksters of the chromatin world. | (Volle and Dalal 2014) |
| 2014 | Supercoiling in DNA and chromatin. | (Gilbert and Allan 2014) |
| 2014 | G4 motifs affect origin positioning and efficiency in two vertebrate replicators. | (Valton et al. 2014) |
| 2014 | The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. | (Picard et al. 2014) |
| 2014 | Licensing of DNA replication, cancer, pluripotency and differentiation: an interlinked world? | (Champeris Tsaniras et al. 2014) |
| 2014 | Temporal and spatial regulation of eukaryotic DNA replication: from regulated initiation to genome-scale timing program. | (Renard-Guillet et al. 2014) |
| 2014 | The histone variant H2A. Bbd is enriched at sites of DNA synthesis. | (Sansoni et al. 2014) |
| 2014 | FANCJ promotes DNA synthesis through G-quadruplex structures. | (Castillo Bosch et al. 2014) |
| 2015 | The hunt for origins of DNA replication in multicellular eukaryotes. | (Urban et al. 2015) |
| 2015 | Measuring the effectiveness of scientific gatekeeping. | (Siler et al. 2015) |
| 2015 | Peaks cloaked in the mist: the landscape of mammalian replication origins. | (Hyrien 2015) |
| 2015 | Post-translational modifications of tubulin: pathways to functional diversity of microtubules. | (Song and Brady 2015) |
| 2015 | Regulated eukaryotic DNA replication origin firing with purified proteins. | (Yeeles et al. 2015) |
| 2015 | Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. | (Ticau et al. 2015) |
| 2015 | Single-molecule visualization of MCM2–7 DNA loading: seeing is believing. | (Chistol and Walter 2015) |
| 2015 | High-resolution profiling of Drosophila replication start sites reveals a DNA shape and chromatin signature of metazoan origins. | (Comoglio et al. 2015) |
| 2015 | The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. | (Duzdevich et al. 2015) |
| 2016 | 4D Visualization of replication foci in mammalian cells corresponding to individual replicons | (Chagin et al. 2016) |
| 2016 | 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression | (Löb et al. 2016) |
Fig. 1.
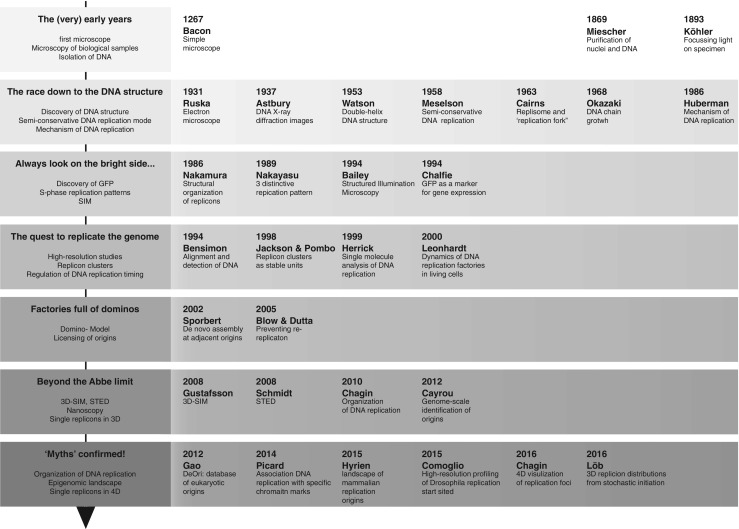
Graphical overview of microscopy developments and their impact on DNA replication studies
The race down to the DNA structure and duplication
Phase contrast microscopy (Zernike 1955) and DNA X-ray diffraction images (Astbury 1947) Franklin, 1952, “Photo 52”) lead to fantastic images, new discoveries, and the description of the double helix DNA structure (Watson and Crick 1953). Meselson and Stahl ingeniously demonstrated the semiconservative mode of DNA replication (Meselson and Stahl 1958). The theoretical description of a confocal microscope and the first practical application of a Nipkow disk in microscopy (Egger and Petráň 1967; Petráň et al. 1968) were clear landmarks of the microscopy revolution.
Radioactive labeling and autoradiography allowed Cairns to observe DNA unwinding and the replication fork (Cairns 1963), and Huberman and Riggs observed similar replication structures in mammalian chromosomes (Huberman and Riggs 1966) and Okazaki described the lagging strand synthesis and “its” fragments (Okazaki et al. 1968; Okazaki and Okazaki 1969; Sugimoto et al. 1969; Sugimoto et al. 1968).
Always look on the bright side
Along came Aequorea victoria green fluorescent protein (Shimomura et al. 1962) and brought light into darkness. Where audioradiography once ruled (Huberman and Riggs 1966; Huberman and Riggs 1968; Taylor et al. 1957), immunofluorescence labeling of fixed cells with monoclonal antibodies to modified nucleotides incorporated into newly synthesized DNA took the stage (e.g., Aten et al. 1992; Cardoso et al. 1993; Jackson and Pombo 1998; Jaunin et al. 1998; Ma et al. 1998; Mazzotti et al. 1990; Nakamura et al. 1986; Raska et al. 1989; Raska et al. 1991) only to be outshined by live cell microscopy of fluorescent fusion proteins (Cardoso et al. 1997; Leonhardt et al. 2000). Cell cycle duration (Van Dilla et al. 1969), fork speed, replication rate, and replicon sizes (Kriegstein and Hogness 1974; Taylor 1977; Taylor and Hozier 1976; Wilson and Wilson 1975; Yurov 1977; Yurov 1978; Yurov 1979; Yurov and Liapunova 1977) were all unearthed from the dark.
In parallel, the first affordable home computers made digital image analysis possible through the help of Wayne S. Rasband who developed the milestone in image analysis ImageJ (then, NIH Image) in 1987 (Schneider et al. 2012).
Extensive microscopic analysis in fixed cells followed and provided a spatiotemporal description of replication sites (replication foci; see Fig. 2) in cells throughout S-phase (Nakamura et al. 1986) along with the three main distinctive early, mid, and late S-phase replication foci patterns (Jackson and Pombo 1998; Mills et al. 1989; Nakayasu and Berezney 1989). Alongside, replication origins (Burhans et al. 1990; Burhans et al. 1991) were also reported.
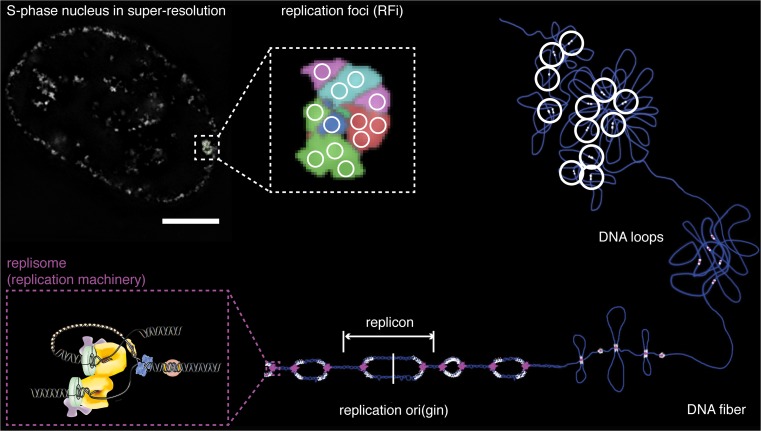
Fig. 2.
Organization of DNA replication from the genome to the individual replisome/replicon. A fluorescently labeled human HeLa Kyoto cell with a typical late S-phase replication pattern is presented in the top left corner (scale bar = 5 μm). Magnified super-resolution replication foci, with white circles representing individual replication sites displayed in the middle of the top row. A scheme of clustered DNA loops with active replication sites (white) is shown on the right. Starting point of DNA replication, the replication origin (ori), and the region replicated from a single origin displayed in the bottom row. Each replicon is replicated by two replication machineries (magenta), composed of various replication proteins, magnified in the bottom left corner. Adapted from (Chagin et al. 2016; Chagin et al. 2010)
The quest to replicate the genome
DNA loops (see Fig. 2) and their “functional” attachments to active transcription units were shown as chromatin organizers during mitosis (Jackson et al. 1992), and replication factories were proposed as clusters of DNA replication sites organized by the nucleoskeleton (Hozák et al. 1993). Molecular combing, refined DNA fiber analysis, and sensitive detection of DNA (Bensimon et al. 1994) opened the door to whole genome stretching and high-resolution studies (Michalet et al. 1997). It allowed analysis of single DNA molecules undergoing replication (see Fig. 2) in a much greater resolution (Herrick and Bensimon 1999) than ever before. Stable replicon clusters were also described as a basis for effective activation and propagation of genome replication during S-phase (Jackson and Pombo 1998) and regulation of replication timing (Dimitrova and Gilbert 1999).
Studies on DNA replication proteins (see Fig. 2) using live-cell fluorescence microscopy produced time lapse movies of replication factories and elucidated basic principles of their dynamic assembly-disassembly behavior (Leonhardt et al. 2000). Different regulatory levels were shown to be necessary to initiate and regulate DNA replication. Not only the chromatin structure, nuclear, and chromosomal locations but also origin recognition complex (ORC) and a whole bunch of other factors were found to define start sites of replication (DePamphilis 2003; Gerbi and Bielinsky 2002; Gerbi et al. 2002; Sasaki and Gilbert 2007).
Factories full of dominos
In addition to the “factory model” (Hozák et al. 1993), more dynamic models ensued (Sadoni et al. 2004; Sporbert et al. 2002) whereby replication at one site induces domino-like activation of neighboring origins, without the need to postulate pre-determined clusters of replicons. The combination with an earlier model postulating that origins of replication would be licensed only during mitosis and this license to replicate would be revoked after one round of replication (Blow and Dutta 2005; Blow and Laskey 1988) elegantly demonstrated how DNA is completely duplicated once, and only once, during each cell cycle. Despite Cvetic wishing for “eukaryotic origins of DNA replication to please be more specific” (Cvetic and Walter 2005), DNA replication origins in higher eukaryotes have been at best elusive. Nonetheless, as a whole, DNA replication is a very robust mechanism and stalled forks can be reactivated or reactivate neighboring origins to close all gaps and provide a perfect copy of billions of nucleotides at every cell division (Langston and O’Donnell 2006; Patel et al. 2006).
The ever elusive origin
The hunt for the elusive consensus motif of DNA replication origins continued with genome-wide high throughput mapping of potential origins and next-generation sequencing methods (Besnard et al. 2012; Cadoret et al. 2008; Cayrou et al. 2012; Dellino et al. 2013; Karnani et al. 2010; Lucas et al. 2007; Martin et al. 2011; Mesner et al. 2013; Mesner et al. 2011; Mukhopadhyay et al. 2014; Picard et al. 2014; Valenzuela et al. 2011) but stalled without a conclusive definition of the mammalian origin of replication. Correlations with specialized DNA structures (e.g., G-quadruplexes) and many others have been suggested but there seems not to be a simple solution and potentially there is no need to have one.
Studies into the epigenomic landscape, epigenetic control of DNA replication, and higher order chromatin organization (Casas-Delucchi and Cardoso 2011; Casas-Delucchi et al. 2012) have provided a link of epigenetic modifications (in particular, histone acetylation level) and temporal control of DNA replication origin firing.
Altogether, even Hyrien’s “Peaks cloaked in the mist,” all out approach was not able to identify possible origins by similarities in thousands of microarrays and/or next-generation sequencing data, suggesting origins form at unspecific DNA sites, but are suppressed by ongoing transcription (Hyrien 2015), which is highly correlated with histone acetylation.
To go where no one has gone before: beyond the Abbe limit
Meanwhile, the microscopy arms race to and beyond the diffraction limit calculated by Abbe continued with the Structured Illumination Microscopy (SIM) (Bailey et al. 1993), the 3D–SIM (Gustafsson et al. 2008) and the stimulated emission depletion (STED) (Schmidt et al. 2008).
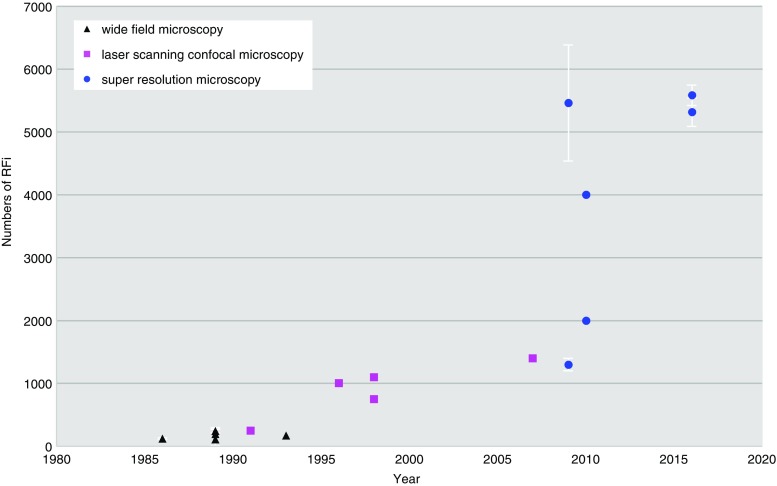
The first attempts to label dating back to 1986 (Nakamura et al. 1986) and quantify replication sites in cells yielded numbers on the low hundreds (see Fig. 3). A decade later with the advent of digital imaging and computational image analysis tools, these numbers grew to around one thousand (Berezney et al. 1996; Fox et al. 1991; Jackson and Pombo 1998; Ma et al. 1998), where they remained for several years (see Fig. 3). Such numbers of replication sites were compatible with a concept of clusters of replicons activated together and, thus, visualized together.
Fig. 3.
Graphical overview of replication foci numbers in correlation with microscopy developments
The dramatic increase in spatial resolution made possible with the new super-resolution microscopy techniques (fluorescence nanoscopy) enabled the visualization, for the first time, of smaller replication structures (Baddeley et al. 2010; Cseresnyes et al. 2009; Ligasová et al. 2009). It was now possible to resolve structures well below the Abbe limit, down to 30 nm and smaller. Nanoscopy (Gustafsson et al. 2008; Hell 2003; Hell et al. 1994) is in full swing and let us go where no one has gone before: beyond the Abbe limit. This, on the other hand, created another level of demand upon image analysis tools.
“Myths” confirmed!
The stage was now set to try and unveil the units of genome replication, i.e., the replicons and their associated machinery, the replisome, in cells.
From the earlier studies using light nanoscopy techniques (Baddeley et al. 2010; Cseresnyes et al. 2009) as well as electron microscopy (Koberna et al. 2005), suitable computational image analysis protocols were developed (Chagin et al. 2015). These combined efforts led to a further increase in the numbers of replication sites measured in cells (see Fig. 3), which was now finally compatible and fitting with the predicted numbers of replicons needed to duplicate the genome in human cells (Chagin et al. 2016; Löb et al. 2016).
The microscopic information age had arrived. Previous efforts by Shaw et al. (2010), together with measurements throughout the years culminating on the visualization and quantification of individual replicons in cells in 4D, all supported by 3D–SIM imaging (Chagin et al. 2016) were all combined in a minimalistic but comprehensive 4D replicon simulation model (Löb et al. 2016) displaying previously published replication polarity gradients, replication timing profiles, N/U domains, topologically associating domains, and timing transition regions (Audit et al. 2013; Baker et al. 2012; Chen et al. 2010; Hyrien et al. 2013; Pope et al. 2014).
Journey into the future
Future work should aim to bridge the ever-increasing genome-wide population data, with single molecule and single-cell microscopic data. Novel ways to combine and relate these very different types of information should be developed to get the highest spatial together with the highest temporal resolution without compromising the data on variability between single cells.
Importantly, the available models should be put to work to predict and test genome replication in different cell types and species and under different stress conditions. This would unleash the value of the existing models and lead us into the in silico DNA replication era.
Acknowledgements
We apologize to the colleagues whose work was not cited due to space constraints. We thank all the past and present members of our laboratory for their many contributions along the years. Last but not least, we thank our collaborators over the years, which have made our work so much more enjoyable.
Our research has been supported by grants of the German Research Foundation (DFG), the Volkswagen Foundation, and the German Ministry for Education and Research (BMBF).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Astbury WT. X-ray studies of nucleic acids. SYM SOC EXP BIOL. 1947;1:66–76. [PubMed] [Google Scholar]
- Aten JA, Bakker PJ, Stap J, Boschman GA, Veenhof CH. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem J. 1992;24:251–259. doi: 10.1007/BF01046839. [DOI] [PubMed] [Google Scholar]
- Audit B, et al. Multiscale analysis of genome-wide replication timing profiles using a wavelet-based signal-processing algorithm. Nat Protoc. 2013;8:98–110. doi: 10.1038/nprot.2012.145. [DOI] [PubMed] [Google Scholar]
- Bacon R (1267) Opus Majus. Vatican Library, Vatican
- Baddeley D, et al. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 2010;38:e8–e8. doi: 10.1093/nar/gkp901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366:44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- Baker A, Chen CL, Julienne H, Audit B, d’Aubenton-Carafa Y, Thermes C, Arneodo A (2012) Linking the DNA strand asymmetry to the spatio-temporal replication program. EUR PHYS J E 35 [DOI] [PubMed]
- Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Berezney R, et al. Connecting nuclear architecture and genomic function. J Cell Biochem. 1996;62:223–226. doi: 10.1002/(SICI)1097-4644(199608)62:2<223::AID-JCB10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Besnard E, et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Bianco JN, et al. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods. 2012;57:149–157. doi: 10.1016/j.ymeth.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. NAT REV MOL CELL BIO. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Boos D, Frigola J, Diffley J. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Mol Cell. 2010;40:860–861. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Grallert B. In DNA replication, the early bird catches the worm. Cell. 2009;136:812–814. doi: 10.1016/j.cell.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Vassilev LT, Caddle MS, Heintz NH, DePamphilis ML. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-O. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Vassilev LT, Wu J, Sogo JM, Nallaseth FS, DePamphilis ML. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret J-C, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. P NATL ACAD SCI USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963;6:208–213. doi: 10.1016/S0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Joseph C, Rahn H-P, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication In Vivo. J Cell Biol. 1997;139:579. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin a and cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Casas-Delucchi CS, Cardoso MC (2011) Epigenetic control of DNA replication dynamics in mammals. Nucleus 2:370–382 [DOI] [PubMed]
- Casas-Delucchi CS, Becker A, Bolius JJ, Cardoso MC. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res. 2012;40:e176–e176. doi: 10.1093/nar/gks784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Delucchi CS et al (2012) Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res 40:159–169 [DOI] [PMC free article] [PubMed]
- Castillo Bosch P, Segura-Bayona S, Koole W, van Heteren JT, Dewar JM, Tijsterman M, Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Grégoire D, Coulombe P, Danis E, Méchali M. Genome-scale identification of active DNA replication origins. Methods. 2012;57:158–164. doi: 10.1016/j.ymeth.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Chagin VO, et al. 4D visualization of replication foci in mammalian cells corresponding to individual replicons. Nat Commun. 2016;7:11231. doi: 10.1038/ncomms11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagin VO, Reinhart M, Cardoso MC. High-resolution analysis of mammalian DNA replication units. METH MOL BIOL. 2015;1300:43–65. doi: 10.1007/978-1-4939-2596-4_3. [DOI] [PubMed] [Google Scholar]
- Chagin VO, Stear JH, Cardoso MC. Organization of DNA replication. Cold Spring Harb Perspect Biol. 2010;2:a000737–a000737. doi: 10.1101/cshperspect.a000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Champeris Tsaniras S, Kanellakis N, Symeonidou IE, Nikolopoulou P, Lygerou Z, Taraviras S. Licensing of DNA replication, cancer, pluripotency and differentiation: an interlinked world? Semin Cell Dev Biol. 2014;30:174–180. doi: 10.1016/j.semcdb.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Chen C-L, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistol G, Walter JC. Single-molecule visualization of MCM2-7 DNA loading: seeing is believing. Cell. 2015;161:429–430. doi: 10.1016/j.cell.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Comoglio F, Schlumpf T, Schmid V, Rohs R, Beisel C, Paro R. High-resolution profiling of drosophila replication start sites reveals a DNA shape and chromatin signature of metazoan origins. Cell Rep. 2015;11:821–834. doi: 10.1016/j.celrep.2015.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cseresnyes Z, Schwarz U, Green CM. Analysis of replication factories in human cells by super-resolution light microscopy. BMC CELL BIOL. 2009;10:88. doi: 10.1186/1471-2121-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- D’Angiolella V, et al. SCF(cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R (2008) Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. vol 122. Human Genetics, vol 6. Springer-Verlag, [DOI] [PubMed]
- Danna KJ, Nathans D. Bidirectional replication of simian virus 40 DNA. P NATL ACAD SCI USA. 1972;69:3097–3100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, O’Donnell M. Mechanisms of DNA replication. Curr Opin Chem Biol. 2000;4:581–586. doi: 10.1016/S1367-5931(00)00134-4. [DOI] [PubMed] [Google Scholar]
- Dellino GI, et al. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23:1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML. The ‘ORC cycle’: a novel pathway for regulating eukaryotic DNA replication. Gene. 2003;310:1–15. doi: 10.1016/S0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Di Antonio M, Rodriguez R, Balasubramanian S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods. 2012;57:84–92. doi: 10.1016/j.ymeth.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/S1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Douglas ME, Diffley J. Replication timing: the early bird catches the worm. Curr Biol. 2012;22:R81–R82. doi: 10.1016/j.cub.2011.12.028. [DOI] [PubMed] [Google Scholar]
- Duzdevich D, Warner MD, Ticau S, Ivica NA, Bell SP, Greene EC. The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Mol Cell. 2015;58:483–494. doi: 10.1016/j.molcel.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger MD, Petráň M. New reflected-light microscope for viewing unstained brain and ganglion cells. Science. 1967;157:305–307. doi: 10.1126/science.157.3786.305. [DOI] [PubMed] [Google Scholar]
- Forterre P. Why are there so many diverse replication machineries? J Mol Biol. 2013;425:4714–4726. doi: 10.1016/j.jmb.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Fox MH, Arndt-Jovin DJ, Jovin TM, Baumann PH, Robert-Nicoud M. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J Cell Sci. 1991;99(Pt 2):247–253. doi: 10.1242/jcs.99.2.247. [DOI] [PubMed] [Google Scholar]
- Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galilei G. Sidereus nuncius. Venedig: Apud Thomam Baglionum; 1610. [Google Scholar]
- Gao F, Luo H, Zhang CT. DeOri: a database of eukaryotic DNA replication origins. Bioinformatics. 2012;28:1551–1552. doi: 10.1093/bioinformatics/bts151. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Bielinsky A-K. DNA replication and chromatin. CURR OPIN GENET DEV. 2002;12:243–248. doi: 10.1016/S0959-437X(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Strezoska Z, Waggener JM. Initiation of DNA replication in multicellular eukaryotes. J Struct Biol. 2002;140:17–30. doi: 10.1016/S1047-8477(02)00538-5. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Replication origins run (ultra) deep. Nat Struct Mol Biol. 2012;19:740–742. doi: 10.1038/nsmb.2352. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Allan J. Supercoiling in DNA and chromatin. CURR OPIN GENET DEV. 2014;25:15–21. doi: 10.1016/j.gde.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E (1866) Generelle Morphologie der Organismen: allgemeine Grundzuge der organischen Formen-Wissenschaft, mechanisch begrundet durch die von Charles Darwin reformirte Descendenz-Theorie., vol 1. Georg Reimer, Berlin
- Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Hell SW. Toward fluorescence nanoscopy. Nat Biotechnol. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- Hell SW, Stelzer EHK, Lindek S, Cremer C. Confocal microscopy with an increased detection aperture: type-B 4Pi confocal microscopy. Opt Lett. 1994;19:222. doi: 10.1364/OL.19.000222. [DOI] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick J, Bensimon A. Single molecule analysis of DNA replication. Biochimie. 1999;81:859–871. doi: 10.1016/S0300-9084(99)00210-2. [DOI] [PubMed] [Google Scholar]
- Hooke R (1665) Micrographia, or, Some physiological descriptions of minute bodies made by magnifying glasses: with observations and inquiries thereupon. J. Martyn and J. Allestry., London
- Hori M, Shibuya K, Sato M, Saito Y. Lethal effects of short-wavelength visible light on insects. Scientific Reports. 2014;4:7383. doi: 10.1038/srep07383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozák P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-I. [DOI] [PubMed] [Google Scholar]
- Huberman JA, Riggs AD. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. P NATL ACAD SCI USA. 1966;55:599–606. doi: 10.1073/pnas.55.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyrien O. Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol. 2015;208:147–160. doi: 10.1083/jcb.201407004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, et al. From simple bacterial and archaeal replicons to replication N/U-domains. J Mol Biol. 2013;425:4673–4689. doi: 10.1016/j.jmb.2013.09.021. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Dolle A, Robertson G, Cook PR. The attachments of chromatin loops to the nucleoskeleton. CELL BIOL INT REP. 1992;16:687–696. doi: 10.1016/S0309-1651(05)80013-X. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaunin F, Visser AE, Cmarko D, Aten JA, Fakan S. A new immunocytochemical technique for ultrastructural analysis of DNA replication in proliferating cells after application of two halogenated deoxyuridines. J Histochem Cytochem. 1998;46:1203–1209. doi: 10.1177/002215549804601014. [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck JL, Berger JM. DNA replication at high resolution. CHEM BIOL. 2000;7:R63–R71. doi: 10.1016/S1074-5521(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Knoll M, Ruska E. Beitrag zur geometrischen Elektronenoptik. I. ANN PHYS-BERLIN. 1932;404:607–640. doi: 10.1002/andp.19324040506. [DOI] [Google Scholar]
- Knoll M, Ruska E. Beitrag zur geometrischen Elektronenoptik. II. ANN PHYS-BERLIN. 1932;404:641–661. doi: 10.1002/andp.19324040602. [DOI] [Google Scholar]
- Knott SRV, et al. Forkhead transcription factors establish origin timing and long-range clustering in S. Cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, et al. Electron microscopy of DNA replication in 3-D: evidence for similar-sized replication foci throughout S-phase. J Cell Biochem. 2005;94:126–138. doi: 10.1002/jcb.20300. [DOI] [PubMed] [Google Scholar]
- Köhler A (1893) Ein neues Beleuchtungsverfahren für mikrophotographische Zwecke. vol 10. vol 4. Zeitschrift fur wissenschaftliche Mikroskopie und fur mikroskopische Technik., Stuttgart
- Kriegstein HJ, Hogness DS. Mechanism of DNA replication in drosophila chromosomes: structure of replication forks and evidence for bidirectionality. P NATL ACAD SCI USA. 1974;71:135. doi: 10.1073/pnas.71.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Langston LD, O’Donnell M. DNA replication: keep moving and don’t mind the gap. Mol Cell. 2006;23:155–160. doi: 10.1016/j.molcel.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligasová A, Raska I, Koberna K. Organization of human replicon: singles or zipping couples? J Struct Biol. 2009;165:204–213. doi: 10.1016/j.jsb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Löb D, Lengert N, Chagin VO, Reinhart M, Casas-Delucchi CS, Cardoso MC, Drossel B. 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat Commun. 2016;7:11207. doi: 10.1038/ncomms11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett ST. Polymerase switching in DNA replication. Mol Cell. 2007;27:523–526. doi: 10.1016/j.molcel.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H. A personal reflection on the replicon theory: from R1 plasmid to replication timing regulation in human cells. J Mol Biol. 2013;425:4663–4672. doi: 10.1016/j.jmb.2013.03.039. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- Mazzotti G, et al. High-resolution detection of newly synthesized DNA by anti-bromodeoxyuridine antibodies identifies specific chromatin domains. J Histochem Cytochem. 1990;38:13–22. doi: 10.1177/38.1.2403578. [DOI] [PubMed] [Google Scholar]
- McGuffee SR, Smith DJ, Whitehouse I. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol Cell. 2013;50:123–135. doi: 10.1016/j.molcel.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Mendel G. Versuche über Pflanzen-Hybriden. Brünn: Verhandlungen des naturforschenden Vereins Brünn; 1866. [Google Scholar]
- Meselson M, Stahl FW. The replication of DNA in Escherichia coli. P NATL ACAD SCI USA. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Cieslik M, Pickin R, Hamlin JL, Bekiranov S. Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins. Genome Res. 2013;23:1774–1788. doi: 10.1101/gr.155218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, Bekiranov S. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 2011;21:377–389. doi: 10.1101/gr.111328.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Miescher JF (1871) Ueber die chemische Zusammensetzung der Eiterzellen. vol 4: 441–60. Medicinsich-chemische Untersuchungen, Felix Hoppe-Seyler, Berlin: Hirschwald
- Mills AD, Blow JJ, White JG, Amos WB, Wilcock D, Laskey RA. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J Cell Sci. 1989;94:471. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- Minsky M (1961) Microscopy Apparatus, US Patent 3,103,467. Washington, DC: U.S. Unite States of America Patent
- Mojardín L, Vázquez E, Antequera F. Specification of DNA replication origins and genomic base composition in fission yeasts. J Mol Biol. 2013;425:4706–4713. doi: 10.1016/j.jmb.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10:e1004319. doi: 10.1371/journal.pgen.1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. CURR OPIN GENET DEV. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus Episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. P NATL ACAD SCI USA. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Okazaki R. Mechanism of DNA chain growth. IV. Direction of synthesis of T4 short DNA chains as revealed by exonucleolytic degradation. P NATL ACAD SCI USA. 1969;64:1242–1248. doi: 10.1073/pnas.64.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17:308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petráň M, Hadravský M, Egger MD, Galambos R. Tandem-scanning reflected-light microscope. J Opt Soc Am. 1968;58:661–664. doi: 10.1364/JOSA.58.000661. [DOI] [Google Scholar]
- Picard F, et al. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Pope BD, Gilbert DM. The replication domain model: regulating replicon firing in the context of large-scale chromosome architecture. J Mol Biol. 2013;425:4690–4695. doi: 10.1016/j.jmb.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Koberna K, Jarník M, Petrasovicová V, Bednár J, Raska K, Bravo R. Ultrastructural immunolocalization of cyclin/PCNA in synchronized 3 T3 cells. Exp Cell Res. 1989;184:81–89. doi: 10.1016/0014-4827(89)90366-2. [DOI] [PubMed] [Google Scholar]
- Raska I, et al. Ultrastructural cryoimmunocytochemistry is a convenient tool for the study of DNA replication in cultured cells. J ELECTRON MICR TECH. 1991;18:91–105. doi: 10.1002/jemt.1060180202. [DOI] [PubMed] [Google Scholar]
- Remus D, Diffley JFX. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Renard-Guillet C, Kanoh Y, Shirahige K, Masai H. Temporal and spatial regulation of eukaryotic DNA replication: from regulated initiation to genome-scale timing program. Semin Cell Dev Biol. 2014;30:110–120. doi: 10.1016/j.semcdb.2014.04.014. [DOI] [PubMed] [Google Scholar]
- Ritson DJ, Moses JE. A fragment based click chemistry approach towards hybrid G-quadruplex ligands: design, synthesis and biophysical evaluation. Tetrahedron. 2012;68:197–203. doi: 10.1016/j.tet.2011.10.066. [DOI] [Google Scholar]
- Rizzoli R, et al. DNA synthesis progression in 3 T3 synchronized fibroblasts: a high resolution approach. Histochemistry. 1992;97:181–187. doi: 10.1007/BF00267309. [DOI] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EHK, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sansoni V, et al. The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 2014;42:6405–6420. doi: 10.1093/nar/gku303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, Hell SW. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A, Olivares-Chauvet P, Maya-Mendoza A, Jackson DA. S-phase progression in mammalian cells: modelling the influence of nuclear organization. Chromosom Res. 2010;18:163–178. doi: 10.1007/s10577-010-9114-2. [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of Aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J CELL COMPAR PHYSL. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A, et al. CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy: mechanistic rationale for drug combination therapy. Mol Cancer Ther. 2012;11:994–1005. doi: 10.1158/1535-7163.MCT-11-0613. [DOI] [PubMed] [Google Scholar]
- Siler K, Lee K, Bero L. Measuring the effectiveness of scientific gatekeeping. P NATL ACAD SCI USA. 2015;112:360–365. doi: 10.1073/pnas.1418218112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer C. Notes on the early history of microscopy. P ROY SOC MED. 1914;7:247–279. [PMC free article] [PubMed] [Google Scholar]
- Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–D686. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Brady ST. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 2015;25:125–136. doi: 10.1016/j.tcb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyfer VN. The consequences of political dictatorship for Russian science. NAT REV GENET. 2001;2:723–729. doi: 10.1038/35088598. [DOI] [PubMed] [Google Scholar]
- Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res. 2005;33:3521–3528. doi: 10.1093/nar/gki665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell. 2002;10:1355–1365. doi: 10.1016/S1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2008;30:259–260. doi: 10.1016/j.molcel.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Okazaki T, Imae Y, Okazaki R. Mechanism of DNA chain growth, III. Equal annealing of T4 nascent short DNA chains with the separated complementary strands of the phage DNA. P NATL ACAD SCI USA. 1969;63:1343–1350. doi: 10.1073/pnas.63.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Okazaki T, Okazaki R. Mechanism of DNA chain growth, II. Accumulation of newly synthesized short chains in E. Coli infected with ligase-defective T4 phages. P NATL ACAD SCI USA. 1968;60:1356–1362. doi: 10.1073/pnas.60.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Méchali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Tijsterman M. Genomes and G-quadruplexes: for better or for worse. J Mol Biol. 2013;425:4782–4789. doi: 10.1016/j.jmb.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Taylor JH. Increase in DNA replication sites in cells held at the beginning of S phase. Chromosoma. 1977;62:291–300. doi: 10.1007/BF00327029. [DOI] [PubMed] [Google Scholar]
- Taylor JH, Hozier JC. Evidence for a four micron replication unit in CHO cells. Chromosoma. 1976;57:341–350. doi: 10.1007/BF00332159. [DOI] [PubMed] [Google Scholar]
- Taylor JH, Woods PS, Hughes WL. The organization and duplication of chromosomes as revealed by autoradiographic studies using tritium-labeled thymidinee. P NATL ACAD SCI USA. 1957;43:122–128. doi: 10.1073/pnas.43.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Técher H, et al. Replication dynamics: biases and robustness of DNA fiber analysis. J Mol Biol. 2013;425:4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161:513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. New insights into replication clamp unloading. J Mol Biol. 2013;425:4727–4732. doi: 10.1016/j.jmb.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Urban JM, Foulk MS, Casella C, Gerbi SA (2015) The hunt for origins of DNA replication in multicellular eukaryotes. F1000Prime Reports 7 [DOI] [PMC free article] [PubMed]
- Valenzuela MS, et al. Preferential localization of human origins of DNA replication at the 5´-ends of expressed genes and at evolutionarily conserved DNA sequences. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton A-L, et al. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aa AJ (1851) Biografisch Woordenboek der Nederlanden. J. J. van Brederode,
- Van Dilla MA, Truiullo TT, Mullaney PF, Coultex JR. Cell microfluorometry: a method for rapid fluorescence measurement. Science. 1969;163:1213. doi: 10.1126/science.163.3872.1213. [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. GENE DEV. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle C, Dalal Y. Histone variants: the tricksters of the chromatin world. CURR OPIN GENET DEV. 2014;25:8–14. doi: 10.1016/j.gde.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Smith DJ. Chromatin dynamics at the replication fork: there’s more to life than histones. CURR OPIN GENET DEV. 2013;23:140–146. doi: 10.1016/j.gde.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B, Wilson BG (1975) DNA replication in the amphibia. Chromosoma 51 [DOI] [PubMed]
- Wu JR, Gilbert DM. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol Cell Biol. 1997;17:4312–4321. doi: 10.1128/MCB.17.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Hayano M, Masai H. Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet. 2013;29:449–460. doi: 10.1016/j.tig.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JTP, Deegan TD, Janska A, Early A, Diffley JFX. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Poveda A, Pasero P. Time to be versatile: regulation of the replication timing program in budding yeast. J Mol Biol. 2013;425:4696–4705. doi: 10.1016/j.jmb.2013.09.020. [DOI] [PubMed] [Google Scholar]
- Yurov YB. DNA replication in human diploid cells of different origin. CELL DIFFER DEV. 1977;6:i–ii. doi: 10.1016/0045-6039(77)90030-6. [DOI] [PubMed] [Google Scholar]
- Yurov YB. Replication of chromosomal DNA in cultured abnormal human cells. Hum Genet. 1978;43:47–52. doi: 10.1007/BF00396477. [DOI] [PubMed] [Google Scholar]
- Yurov YB. The rate of fork movement during DNA replication in mammalian cells. Chromosoma. 1979;74:347–353. doi: 10.1007/BF01190749. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Liapunova NA. The units of DNA replication in the mammalian chromosomes: evidence for a large size of replication units. Chromosoma. 1977;60:253–267. doi: 10.1007/BF00329774. [DOI] [PubMed] [Google Scholar]
- Zernike F. How I discovered phase contrast. Science. 1955;121:345–349. doi: 10.1126/science.121.3141.345. [DOI] [PubMed] [Google Scholar]