Abstract
Hepatocellular Carcinoma (HCC) is one of the most predominant malignancies with high fatality rate. This deadly cancer is rising at an alarming rate because it is quite resistant to radio- and chemotherapy. Different epigenetic mechanisms such as histone modifications, DNA methylation, chromatin remodeling, and expression of noncoding RNAs drive the cell proliferation, invasion, metastasis, initiation, progression, and development of HCC. These epigenetic alterations because of potential reversibility open way towards the development of biomarkers and therapeutics. The contribution of these epigenetic changes to HCC development has not been thoroughly explored yet. Further research on HCC epigenetics is necessary to better understand novel molecular-targeted HCC treatment and prevention. This review highlights latest research progress and current updates regarding epigenetics of HCC, biomarker discovery, and future preventive and therapeutic strategies to combat the increasing risk of HCC.
1. Introduction
Hepatocellular Carcinoma (HCC) is the fifth most common cancer in the world that causes 250,000 to 1 million deaths annually [1]. This alarming incidence is attributed to several different genetic and epigenetic alterations. HBV and HCV infections, smoking, alcohol, dietary exposure to aflatoxins, diabetes, and obesity are the other risk factors of HCC [2]. Epigenetic changes that contribute to HCC metastasis, invasion, and dissemination encompass noncoding RNAs regulation, DNA methylation, and histone modification. All these changes are associated with initiation and progression of HCC [3].
2. Epigenetic Changes in HCC
Epigenetics refers to heritable states of gene expression without alteration to the DNA sequence itself. Epigenetic changes such as DNA hypermethylation or hypomethylation, dysregulation of histone modification patterns, chromatin remodeling, and aberrant expression of micro-RNAs (miRNAs) and long noncoding RNAs (lncRNAs) are associated with HCC [4]. Different epigenetic mechanisms that drive cell proliferation, metastasis, progression, and development of HCC are discussed below.
3. DNA Methylation
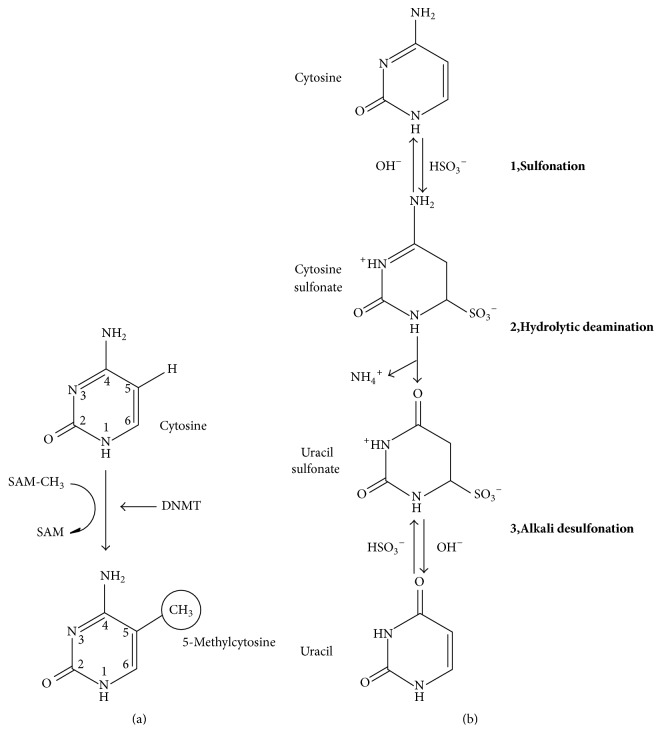
DNA methylation, specifically methylation of cytosine at 5th carbon, is a well characterized epigenetic mechanism of gene regulation that occurs in mammals at promoter-rich region of gene that is, cytosine-phosphate-guanine (CpG) (Figure 1).
Figure 1.
CpG methylation. (a) DNA methylation is catalyzed by three methyl transferase genes (DNMT1, DNMT3a, and DNMT3b) that add methyl group (CH3) at 5th carbon position of pyrimidine ring of cytosine. S-adenosyl methionine (SAM-CH3) acts as a methyl donor. (b) Cytosine to cytosine sulfonate: sulfonation of cytosine causes C to T transition followed by deamination. Cytosine sulfonate to uracil sulfonate: conversion of cytosine sulfonate to uracil sulfonate leads to alkali desulfonation. Uracil sulfonate is converted into uracil. PCR distinguishes methylated CpG from unmethylated CpG because methylated cytosine resists this chemical treatment [124, 125].
CpG dinucleotides occur throughout human genome in nonuniform manner with the frequency of about one per eighty nucleotides [5]. Approximately 1 to 2% of human genome is referred to as CpG islands or CpG-rich regions containing hundred to several thousand base pairs and exists in proximity to different gene promoter regions [6]. Nearly 70% of human genes harbor CpG islands at 5′ region that consist of promoter as well as transcription sites [7].
In HCC and other wide range of tumors, specific promoter hypermethylation and global hypomethylation have been associated with inactivation of tumor-suppressor genes (TSGs) and genomic instability, respectively. Silencing of tumor-related genes and tumor-suppressor genes such as SOCS1, hMLH1, and RASSF1A is achieved by hypermethylation of CpG islands in promoter sequences that downregulates mRNA transcript expression. Epigenetic silenced genes play an important role in molecular pathways of carcinogenesis such as cell adhesion or DNA repair, apoptosis, and cell cycle regulation [8]. Set of proteins known as ten-eleven translocation (Tet1–3) demethylates methylcytosine via hydroxymethylcytosine (hmC) [9]. It has been reported that level of hmC reduces in various types of cancers [10]. However, the mechanism of this downregulation is still to be determined [11].
Different DNA methylome patterns as compared to adjacent normal tissues have been observed in independent genome-wide methylation profiling studies. HCC and non-HCC surrounding liver tissues can be distinguished easily because aberrant DNA hypermethylation is specific to the cancerous tissues. Likewise, a set of hypermethylated gene promoters, for example, FZD7, CDKN2A, RASSFIA, and APC, were able to distinguish nontumor liver tissues from HCC tumors. Another study recruited 27 patients and revealed the significant level of DNA methylation of NFATC1, GSTP1, CDKN2A, and BMP4 genes in HCC tissues [12]. Lambert et al. have recently analyzed the methylation status of set of imprinted genes in HCC and found that 15q11-13 imprinting control region that includes maternally imprinted GABRA5 gene was significantly hypomethylated in tumors compared to their surrounding tissues. The study suggested that imprinted gene methylation acts as a potential marker of environmental exposures [13].
Poor tumor differentiation is attributed to promoter methylation of DNMT1 [14, 15]. S100A8 can be used as prognostic and diagnostic biomarker because of its overexpression observed in MHCC-97H and Huh-7 cell lines [16]. Likewise, HK2 promoter CpG island (HK2-CGI) represents prognostic biomarker of HCC because hypermethylation of HK2-CGI induces HK2-CGI methylation phenotype (HK2-CIMP) [17] (Table 1).
Table 1.
Aberrant DNA methylation markers for HCC.
| Gene and its location | Function | Methylation frequency% in adjacent normal tissue | Methylation frequency% in HCC | Ref. |
|---|---|---|---|---|
| WT1 | Urogenital development | 0 | 54 | [34] |
| 11p13 | ||||
| TIMP3 | Cell adhesion | 0 | 13 | [35] |
| 22q12.3 | ||||
| SOCS-1 | Cytokine inhibitor | 0–7 | 43–65 | [36] |
| 16p13.13 | ||||
| SEMA3B | Apoptosis | — | 83 | [37] |
| 3p21.3 | ||||
| RB | Chromatin structure | 0 | 32 | [34] |
| 13q14.2 | ||||
| RASSF1A | Apoptosis | 0 | 59–75 | [35, 37] |
| 3p21.3 | ||||
| RaR-Beta | Retinoic acid signaling | 7 | 12 | [36] |
| 3p24.2 | ||||
| P73 | Tumor suppressor | 0 | 6 | [36] |
| 1p36.32 | ||||
| P53 | Tumor suppressor | 0 | 14 | [34] |
| 17p13.1 | ||||
| P300 | Growth/cell division | 0 | 65 | [34] |
| 22q13.2 | ||||
| P27 | CDK inhibitor | 0 | 48 | [34] |
| 12p13.1 | ||||
| P21 | CDK inhibitor | 10 | 63 | [34] |
| 6p21.2 | ||||
| P16 INK4a | CDK inhibitor | 0–10 | 16–83 | [35, 36] |
| 9q21.3 | ||||
| P15 | CDK inhibitor | 0 | 42–47 | [36] |
| 18q12.2 | ||||
| P14 | CDK inhibitor | 0 | 6 | [36] |
| 11q13.1 | ||||
| hMLH1 | Mismatch repair | 0 | 0 | [35] |
| 3p21.3 | ||||
| GSTP1 | Glutathione synthesis | 0–7 | 41–76 | [35, 36] |
| 11q13 | ||||
| E-Cadherin | Cell adhesion | 7 | 33–67 | [34–36] |
| 16q22.1 | ||||
| E2F-1 | Transcription factor | 0 | 70 | [34] |
| 20q11.22 | ||||
| DAPK1 | Apoptosis | 0 | 10 | [35] |
| 9q21.33 | ||||
| CPS1 | Urea cycling | 0 | 80 | [38] |
| 2q346 | ||||
| COX2 | Prostaglandin synthesis | 0 | 35–50 | [35] |
| 1q31.1 | ||||
| BLU | Unknown zinc-finger | — | 20 | [37] |
| 3q21.3 | ||||
| APC | Prostaglandin synthesis | 0–14 | 53–81 | [35, 36] |
| 5q22.2 |
Other frequently methylated genes in HCC include RIZ1 (45.2%), CDKN2A (69.7%), SCARA5 (30%), EFEMP1 (50%), TIP30 (47%), WIF1, FBLN1 (50%), DLEC1 (70.6%), FBP1 (80%), ITGA4 (23%), KLK10 (94%), LIFR (47.9%), MTIG (60.4%), HHIP (53.6%), HINT1 (55%), SYK (12%), and TAT (54%) [18, 19].
Bead array analysis of 1505 CpG sites in thirty HBV- or HCV-associated HCC infected patients revealed the correlation of specific methylation signatures with tumor progression stage in HCC tumor patients. Hypermethylation of SYK (spleen tyrosine kinase) or CHFR (checkpoint with fork-head associated and ring finger) occurs specifically in advanced stages of HCC, whereas abnormal DNA methylation of p15, GAAD45a, SFRP1, DOK1, CHRNA3, GSTP1, CRABP1, p16, and RASSF1A occurs at all stages of HCC [20–23]. HBV-encoded protein (HBx) affects the methylation and expression by directly interacting with DNMT (DNA-methyltransferase). The prevalence of TSGs methylation, for example, p15, APC, STAT1, GADD45b, and SOCS-1, has been observed to be higher in HCV-positive HCC compared to HCV-negative HCC [24, 25]. Hernandez-Vargas et al. revealed that panel of hypermethylated genes, for example, DCC, CSPG2, and NAT2, was specific to HBV-related HCC [26, 27] and methylation of constitutive androstane receptor (CAR) suppresses CYP2C19 in HBV-associated HCC patients [28]. According to Shih et al. methylation of PAX6 frequently occurs in HCV-associated HCC tissues (61.3%) compared to HBV positive (22.1%) and double negative HCC tissues (33.3%) [29].
A genome-wide methylation study including 69% HBV-associated HCC patients demonstrated that PAX4, WFDC6, SCGB1D1, ATK3, and CCL20 were top five hypomethylated genes, whereas CDKN2A, SPDY1, ZFP41, BMP4, and DAB2IP were found to be top five hypermethylated genes [30]. Nishida et al. demonstrated that DNA methylation is an important mechanism in silencing the set of 8 TSGs that predict HCV progression to HCC [31]. NEFH and SMPD3 have been proved as potent tumor-suppressor genes in HCC [32]. Based on the above findings it can be concluded that early diagnosis and prognosis of HCC can be achieved by methylation profiling because of close association of aberrant gene methylation with clinical outcome and HCC disease stage. However, specific gene methylation signatures must be validated [26]. Recent study has suggested that DNA methylation of miRNA genes may provide a promising strategy for alternative adjuvant therapy in HCC [33].
4. Histone Modification
Histone modifications also known as “histone code” have direct impact on gene expression and chromatin structure. These epigenetic changes are of paramount importance in gene silencing during tumorigenesis [107]. Modifications such as ubiquitination, phosphorylation, methylation, and acetylation that occur at N-terminal tails of nucleosomal histones work together with other epigenetic mechanisms to regulate gene activities and cellular processes. Orderly and coordinated activities of diverse histone modifications regulate cellular processes, for example, DNA repair, DNA replication, and gene transcription. It has been reported that control of chromatin-based processes (responsible for cancer development and oncogenic transformation) is deregulated by functional changes in protein complexes and histone-modifying molecules [108].
DNA methylation is closely associated with histone modification because inhibitors reverse the histone modification changes on H3-K4 and H3-K9 codes [109, 110]. Another distinct histone modification involved in Polycomb-based silencing and X-chromosome inactivation in women is H3K27 trimethylation that is one of the candidates for a silencing mechanism for tumor-suppressor genes [111]. Overexpression of enhancer of zeste homolog 2 (EZH2) is associated with different types of cancer and it has been studied that EZH2 catalyzes histone H3-K27 triMe [112]. Histone methyltransferases (HMT), SUV39H1 and G9a, mediate the histone H3-K9 trimethylation and dimethylation (H3-K9 diMe), respectively [113]. H3K9 methylation is associated with silencing of several tumor-suppressor genes [114]. Significant decrease in histone H2A ubiquitination was noticed in HCC [115].
Researchers have reported several histone modifications that are associated with HCC and alter normal cellular processes; for example, Magerl et al. found negligible expression of dimethylation of histone H3 at lysine 4 (H3K4diMe) in HCC [116] whereas, according to Shon et al., Patt1 (a GNAT family acetyltransferase) is downregulated in HCC and it is overexpressed in healthy liver [117]. Somatic mutation that induced inactivation of MLL1–5 enzymes responsible for H3KYMe has been reported in 1–6% of HCC [118]. Integration of HBV into MLL2 and MLL4 gene loci has also been identified in many cases of HCC [119]. In contrast to this, elevated H3K4me3 is associated with mutations in SMYD3 methyltransferase resulting in poor prognosis specifically during the initial stages of HCC [120]. Likewise, upregulation of SETDB1, another histone methyltransferase for H3K9, was observed in HCC. Overexpression of SETDB1 that is closely associated with metastasis and cancer progression actually is triggered by downregulation of miR-29 and gain of chromosome 1q21 [121, 122]. About 2.6% of cancer patients experience mutations in SETD2 gene [118, 123].
Histone deacetylases (HDACs) are the enzymes that play an important role in regulation of gene expression by removing acetyl group from histones that make the DNA more compact leading to gene silencing. About 18 HDACs are known with activity not limited to just histones because it has been reported that HDACs remove acetyl-lysine on diverse nonhistone proteins like NFkB, transcription factors p53, and many others [128].
Accumulating evidence suggests the correlation of individual HDACs overexpression with poor prognosis in different types of cancer including HCC [129]. Overexpression of HDAC3 was correlated with early recurrence of HCC after surgery and advance tumor stage. Tumor-suppressor role of HDACs has also been noticed as overt HCC that occurred as a result of liver-specific knockdown of HDAC3 [130]. Specifically, persistent inactivation of SMRT, NCOR, or HDAC3 may lead to cancer development and DNA damage by increasing the histone acetylation during S phase [131]. HDACs may act as tumor suppressors and therapeutic targets in developing tumors and advanced cancer, respectively [132]. Inhibition of HDAC may disturb a drug design due to disruption of diverse pathways.
Some histone modifications also act as signature for risk factor exposures; for example, protein arginine methyltransferase 1 (PRMT1) catalyzes histone H4 methylation on arginine 3 and dephosphorylates damage-induced phosphorylation of H2AX (g-H2AX) to repair DNA. In case of HCV infection, Protein Phosphatase 2A (PP2Ac) is overly expressed and inhibits the activity of PRMT1 by binding to it. Overexpression of this phosphatase is considered an important event of viral hepatitis associated hepatocarcinogenesis. Compromised histone H2AX phosphorylation and histone H4 acetylation/methylation occur because of PP2Ac overexpression in HCV-associated HCCs leading to significant changes in gene expression for hepatocarcinogenesis and inhibition of DNA damage repair. Overexpression of this phosphatase is considered a critical early event in hepatocarcinogenesis in the context of chronic viral hepatitis [24, 133].
The direct interaction of HBx with histone acetyltransferase complex CBP/P300 supports the transcription transactivation property of an oncogenic transcription factor HBV-encoded HBx protein that affects the expression of different genes involved in apoptosis or cell cycle control. Recruitment of CBP/P300 complex that is mediated by HBx promotes transactivation and leads to acetylated (active) state of the target cellular genes [117, 134, 135]. Downregulation of CYP2E1 expression in response to deregulated histone modification which resulted in decline in apoptotic potential has been observed in alcohol-associated HCCs [136, 137]. Recent studies suggest that HCC progression can be repressed by inhibition of O-GlcNAcylation [138]. Epigenetic silencing of JMJD5 (jumonji C domain-containing protein 5), another tumor-suppressor gene in HCC pathogenesis, downregulates CDKN1A transcription to promote HCC cell proliferation [139].
5. Chromatin Remodeling
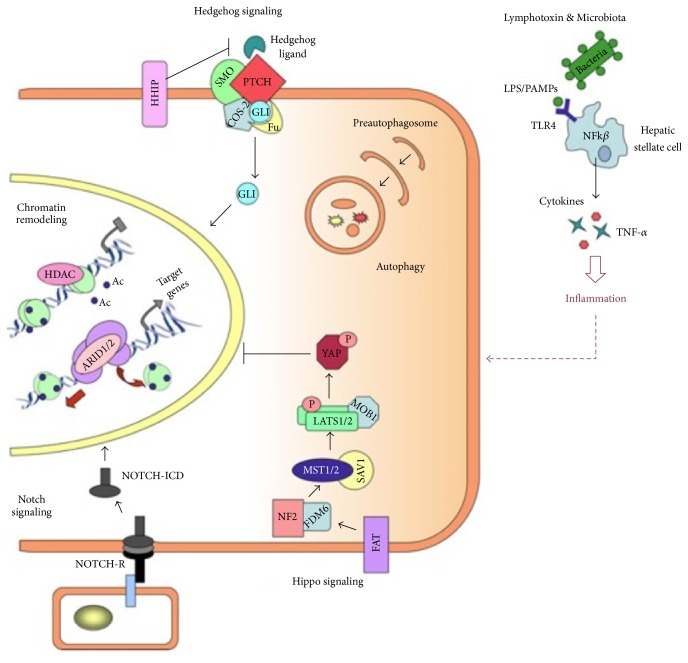
Another important epigenetic mechanism that plays an important role in control of gene expression, differentiation, DNA repair, and proliferation is chromatin remodeling (Figure 2).
Figure 2.
Emerging signaling pathways in HCC: chromatin remodeling: restricting transcriptional and DNA condensation occurs as a result of histone deacetylation catalyzed by HDACs in nucleosome. In contrast, transcriptional activation also occurs using chromatin remodeling complexes by allowing access to transcription machinery via nucleosome restructuring. Notch signaling: NOTCH receptor is cleaved photolytically when protein ligand binds to its extracellular domain. This binding releases its intracellular domain (NOTCH-ICD) that enters into nucleus to modify target gene expression (such as SOX9, HEY, and HES). Hedgehog (Hh) signaling: nuclear translocation of the transcription factor (TF) GLI occurs as a result of PTCH inhibitory effect on SMO and this event takes place in the presence of Hh signaling. Hippo signaling: kinase complexes Lats1/2-Mob1 and MST1/2-SVA1 are activated with phosphorylation of the transcription factor YAP resulting in prevention of its nuclear translocation. This event involves the use of upstream regulators of hippo pathway (i.e., FDM6, NF2, and FAT). Microbiota and lymphotoxins: NF-kβ signaling activates and produces proinflammatory molecules such as TNF-α and cytokines due to recognition of microbial ligands (LPS/PAMPs) by TLKRs (e.g., TLK4) on the hepatic stellate cells [126].
Nucleosomal restructuring by ATP-dependent chromatin remodeling complexes and enzymatic covalent histone modifications are the principal mechanisms involved in chromatin remodeling. Accumulating evidence in recent years has demonstrated that these complexes perform tumor-suppressor roles because of association of different malignancies with inactivated mutations [140]. Recent study based on SWI/SNF chromatin remodeling complex revealed that expression of brahma (BRM) was markedly decreased in HCC samples [141]. ARID1A, ARID1B, and ARID2 components that belong to SWI/SNF-related chromatin remodeling complexes are mutated at frequency of 16.8%, 6.7%, and 5.6%, respectively [142]. ARID1A, ARID1B, and ARID2 mutations are significantly observed in alcohol-associated HCC and HCV-related HCC, respectively [143]. Mutations in other components of SWI/SNF chromatin remodeling complex such as SMARCC2, SMARCC1, SMARCB1, SMARCA4, and SMARCA2 have also been commonly reported in HCCs. SMARCA2 mutations occur at frequency of 2.6% in alcohol-associated HCC [118]. Chromatin remodeler CHD1L that promotes HCC metastasis and progression may act as therapeutic target to control HCC [144]. The analysis of whole exome sequencing of 24 HCCs revealed that chromatin regulators are the third most frequently mutated genes [142].
6. Noncoding RNAs
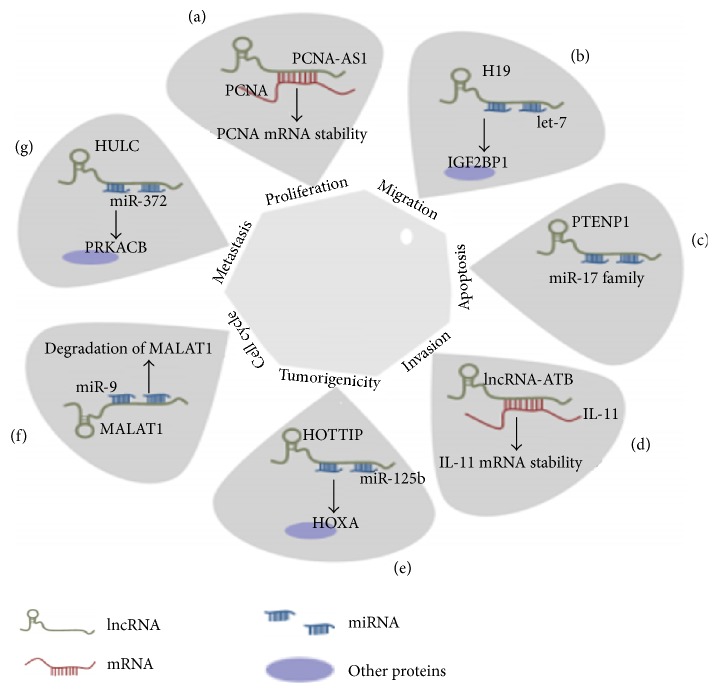
Different human cancers are associated with noncoding RNAs based epigenetic mechanism of regulation of gene expression (Figure 3).
Figure 3.
Role of miRNA, mRNA, and lncRNA in regulation of apoptosis, migration, metastasis, tumorigenicity, cell cycle, invasion, and cell proliferation. PCNA (Proliferating Cell Nuclear Antigen) regulation is related to PCNA-AS1 effects and this event involves the formation of RNA hybridization that increases PCNA mRNA stability. (b) H19 affects let-7 mediated genes involved in promotion of metastasis specifically IGF2BP1 (insulin-like growth factor 2 mRNA-binding protein). (c) Autophagy genes, for example, p62, ATG7 (autophagy-related gene 7), and ULK1 (unccoordinated-51- (unc-51-) like kinase 1), are regulated by PTENP1 that is targeted by miR-17 family. (d) The stability of IL-11 (interleukin-11) mRNA is increased by lncRNA-ATB. (e) miR-125b negatively targeted by HOTTIP. (f) The regulation of MALAT1 is regulated in the nucleus after being binded with miR-9 following AGO2-dependent path. (g) The activity and expression of miR-372 are repressed after getting binded with HULC. Target mRNA of miR-372, that is, Prkacb (cAMP-dependent protein kinase catalytic subunit beta), level is increased in response to mi-372 reduction [127].
Noncoding RNAs are further categorized into two main types based on length: small/short noncoding RNAs that are <200 nucleotides including endogenous siRNAs, snoRNAs, piRNAs, and miRNAs and long noncoding RNAs with length >200 nucleotides [145, 146]. Different functions of miRNA are described in Table 2.
Table 2.
Role of different miRNAs in HCC.
| miRNA | Function of miRNA | Gene target involved | Reference |
|---|---|---|---|
| miRNA-221 | Oncogenic | DDIT4, CDKN1C/p57, CDKN1B/p27 | [39] |
| miRNA-125b | Tumor suppressor | LIN28B | [40] |
| miRNA-214 | Tumor suppressor | c-Myc, TCF-1, Cyclin D1 | [41] |
| let-7 family | Tumor suppressor | c-Myc, STAT3 | [42] |
| miRNA-26 | Tumor suppressor | MMP2, cyclin D1, Mcl-1, Bcl-2 | [43] |
| miRNA-122 | Tumor suppressor | ADAM10, igF1R, SRF, Cyclin G1, Wnt1, AKT3, Bcl-w | [44, 45] |
| miRNA-96 | Oncogenic | ephrinA5 | [46] |
| miRNA-101 | Tumor suppressor | Mcl-1 | [47] |
| miRNA-186 | Tumor suppressor | Yes-associated protein 1 | [48] |
| miRNA-29 | Tumor suppressor | Mcl-2, Bcl-2 | [49] |
| miRNA-125b | Tumor suppressor | ILL- 6R, Bcl-w, Mcl-1, Bcl-2 | [50] |
| miRNA-129-2 | Tumor suppressor | High mobility group box 1 | [51] |
| miRNA-193a-3p | Oncogenic | SRSF2 | [52] |
| miRNA-133b | Tumor suppressor | SIRT1 | [53] |
| miRNA-199a-3p | Tumor suppressor | c-Met, mTOR | [54] |
| miRNA-199a-5p | Tumor suppressor | ATG7 | [55] |
| miRNA-155-3p | Oncogenic | FBXW7 | [56] |
| miRNA-222 | Oncogenic | PPP2R2A, p27 | [57] |
| miRNA-21 | Oncogenic | PTEN, kinase 3 | [58] |
| miRNA-449a | Tumor suppressor | ADAM10 | [59] |
| miRNA-139 | Tumor suppressor | ROCK2, Rho Kinase 2 | [60] |
| miRNA-125b | Tumor suppressor | LIN28B, PDZ binding motif, Sirtuin 7 | [40, 50, 60] |
| miRNA-182 | Oncogenic | ephrinA5 | [46] |
| miRNA-125a | Tumor suppressor | VEGF-A, MMP11 | [61] |
| miRNA-1180 | Increase apoptotic resistance to HCC | Through activation of NF-κB pathway | [62] |
| miRNA-200 family | Tumor suppressor | ZEB2, ZEB1 | [63] |
| miRNA-212 | Tumor suppressor | FOXA1 | [64, 65] |
| miRNA-497 | Tumor suppressor | YAP1 | [66] |
| miRNA-519d | Metastasis | PTEN | [67] |
| miRNA-106b | Apoptosis | Bim | [68] |
Significant decrease in miRNA-129-2 with associated inhibition of HMGB1 (high mobility group box 1) has been observed in HCC [51]. miRNA122 that regulates Wnt1, igF1R, SRF, ADAM10, and cyclin G1 to play an active role in cell cycle progression is downregulated in HCC [44]. miRNA-125b and miRNA-26 induce cell-cycle arrest in HCC by targeting oncogenic LIN28B and cyclin D2/cyclin E2, respectively [43, 50]. Overexpression of miRNA-96 is observed in HBV-related HCC. In HCC cell lines, miRNA-101 targets VEGF-C resulting in the suppression of invasion [147]. miRNA-21 has been found upregulated in HCC and its degradation can be used as target in therapeutics [148]. Likewise, recent study has demonstrated the tumor-suppressor activity of miRNA-214 by inhibiting CDK6, CDK3, and E2F2 [149]. In addition to inhibition of cell proliferation of HCC by miRNA-449a, lentivirus mediated overexpression of miRNA-199a, miRNA-133b, and miRNA-185 has also been reported [53, 59, 150, 151]. Oncogenic miRNA-221 and miRNA-1180 target cell cycle inhibitors (CDKN1C/p57, CDKN1B/p27) and repress TNIP2 expression, respectively, resulting in increased proliferation of HCC cells [39, 152]. Likewise, cell proliferation is suppressed in HCC due to let-7a and let-7 g that regulates the oncogenic STAT3 and cMyc, respectively [42]. miRNA-135a targets fork-head box O1 and miRNA-155-3p suppresses FBXW7 leading to an increased invasion and migration in HCC [56, 153]. miRNA-186 targets Yes-associated protein 1 and acts as a potential therapeutic target in treating HCC [48].
Long noncoding RNAs also play an important role in different types of cancers including HCC. Scaffold, signal, guide, and decoy are the molecular mechanisms of lncRNAs [154, 155]. The most widely studied lncRNAs are described in Table 3.
Table 3.
The potential roles of widely studied HCC-related lncRNAs.
| lncRNA | Potential role in HCC | Ref. |
|---|---|---|
| HOTAIR | Overexpression of HOTAIR is associated with progression of HCC via activation of Wnt/β-catenin signaling pathway. HOTAIR upregulates expression of ATG3 and ATG7 that ultimately activate autophagy and promote HCC cell proliferation. | [69, 70] |
| CCAT1 | CCAT1 functions as let-7 sponge and increases HCC progression. | [71] |
| HULC | HULC increases the metastasis and tumorigenesis of HCC through miR-200a-3p/ZEB1 signaling pathway. | [72] |
| H19 | Low expression of H19 decreases HCC progression and metastasis via upregulation of miR-200 family. | [73] |
| HOTTIP | HOTTIP overexpression is associated with metastasis in HCC patients. This lncRNA is negatively regulated by miR-125b. | [74, 75] |
| BANCR | BANCR is considered as an important contributor of progression and initiation of HCC and therefore can be used as biomarker. | [76] |
| MALAT1 | MALAT1 is associated with tumor progression because of its upregulation in HCC cell lines. | [77] |
| HEIH | HEIH is oncogenic in nature and promotes tumor progression. | [78] |
| PTENP1 | PTENP1 represses tumorigenic properties of HCC cells. | [79] |
| SNHG20 | SNHG20 is upregulated in HCC and may serve as prognostic biomarker of HCC. | [80] |
| MEG3 | Tumor suppressor MEG3 is associated with pathogenesis of HCC malignancy. | [81] |
| TUC338 | Upregulation of TUC338 and TUC339 modulates cell growth and increases liver cirrhosis. | [82, 83] |
| LINC-ROR | LINC-ROR acts as mediator of chemotherapeutic response and increases chemosensitivity in HCC because HCC is highly resistant to chemotherapy. It also promotes cell survival during hypoxia. | [84, 85] |
| MVIH | MVIH confirms overall-survival and recurrence-free survival. | [86] |
| lncRNA-ATB | lncRNA-ATB acts as a mediator of TGF-β signaling that increases metastasis in HCC. | [87] |
| TUG1 | Upregulation of TUG1 in HCC and increasing apoptosis and cell growth by epigenetic silencing of KLF2. | [88] |
| URHC | URHC expression is increased in HCC tissues. It regulates apoptosis and cell proliferation via downregulation of ZAK. | [89] |
| FTX | Binds to miR-374a and MCM2 and inhibits metastasis and proliferation in HCC. | [90] |
| PVT1 | High expression level of PVT1 is linked with tumor progression and may act as biomarker of tumor recurrence in HCC patients. | [91] |
| lncRNA-p21 | lncRNA-p21 is downregulated in HCC and its overexpression inhibits tumor invasion by inhibiting Notch signaling and EMT. | [92] |
| UCA1 | Upregulation of UCA1 is associated with progression of HCC via activation of FGFR1-ERK signaling pathway and inhibition of miR-216b. | [93] |
| MT1DP | MT1DP acts as tumor suppressor and inhibits FOXA1 in liver cancer cells because of negative regulation of MT1DP by YAP and RUNx2. | [94] |
| UFC1 | Upregulation of HFC1 promotes cell cycle progression and HCC cell proliferation. | [95] |
| SRHC | Downregulation of SRHC inhibits cancer proliferation; however, the epigenetically silenced SRHC promotes proliferation in HCC. | [96] |
| PCNA-AS1 | PCNA-AS1 can serve as therapeutic target because it promotes tumor growth in HCC. | [97] |
| lncRNA-LET | Downregulation of LET is associated with reduction in HCC metastasis and invasion. | [79] |
| lncRNA-Dreh | lncRNA-Dreh is important in tumor suppression. Downregulation of Dreh inhibits HCC metastasis by targeting vimentin. | [98] |
| UCA1/CUDR | UCA1/CUDR is involved in chemotherapeutic resistance. | [99] |
Potential utilization of noncoding RNAs as novel candidates in treatment, detection, diagnosis, and prognosis of HCC are promising. During the last decade, accumulating evidence suggests the use of noncoding RNAs as potential therapeutic targets for HCC. However, pathological and biological aspects as well as molecular mechanism of noncoding RNAs in HCC are an emerging area of science that needs more research to develop potential therapeutic intervention and treatment against HCC.
7. Epigenetic Biomarkers of HCC
Epigenetic alterations such as DNA hypermethylation, DNA hypomethylation, and noncoding RNAs or histone modifications may serve as diagnostic and prognostic biomarkers of HCC.
Overexpression of histone phosphorylation proteins such as ARK1 and ARK2 and histone-modifying genes, such as histone methyltransferases G9a, EZH2, and SUV39HZ, in HCC tissues predicts tissue invasion and poor prognosis [130, 156].
DNA methylation acts as potential biomarker of HCC because of higher frequency of aberrant methylation found in HCC tissues that can help a clinician or researcher to distinguish healthy liver from cirrhotic liver or liver of HCC patient [157]. Likewise, deregulated expression level of several noncoding RNAs can be used for diagnosis and prognosis of disease. Downregulation of miR-122 is associated with poor prognosis of HCC [158]. Studies showed decreased expression of miR199a/b-3pis and increased expression of miR-21 in HCC tissues [159]. Deregulation of long noncoding RNA has also been reported in HCC tissues. Upregulation of HOTTIP and HOXAIR in HCC tissues has been associated with poor patient survival, tumor progression, and metastasis [74]. Recently, researchers predicted HCC with 100% specificity and 95.6% sensitivity based on DNA methylation level approach in preneoplastic liver tissue [160].
All these findings are encouraging to develop an epigenetic-based biomarker; however, more research regarding specificity, sensitivity, and reproducibility is needed to make the usability of methylated DNA, modified histones, and noncoding RNAs as novel and reliable biomarkers.
8. Nutritional Epigenetics
Occurrence of HCC can be reduced and the development of HCC can be delayed because of epigenetic mechanisms deregulated by nonnutrient dietary bioactive components and several different nutrients. Liver carcinogenesis is under the influence of chemopreventive potential of epigenetic food components such as dietary methyl-group donors, that is, sulforaphane, sodium butyrate, curcumin, resveratrol, and epigallocatechin-3-gallate (ECGC). Food-based deregulation of epigenome contributes towards HCC-related angiogenesis, oxidative stress, apoptosis, inflammation, and cell proliferation.
Several in vivo and in vitro preclinical models of HCC have demonstrated the antihepatocarcinogenic effects of polyphenolic compound curcumin that demethylates DNA and expresses DNMTs to reactivate abnormally silenced cancer related genes [161]. Green tea beverages are enriched with EGCG that have enough potential to inhibit progression and development of HCC via DNA demethylation of abnormally hypermethylated tumor-suppressor genes [162]. Two coffee polyphenols, that is, chlorogenic acid and caffeic acid, inhibited liver carcinogenesis in rat and the inhibition of human DNMT1-mediated enzymatic DNA methylation reaction has also been reported [163, 164].
Abundant HDAC inhibitors are of natural origin and present in different plants such as broccoli (sulforaphane), grapes (resveratrol), blueberries (piceatannol), and garlic (allyl mercaptan) [165]. Bioactive polyphenol resveratrol produced naturally in blueberries, grapes, and strawberries exhibited chemopreventive effects in liver cancer by promoting apoptosis [166]. In addition to this, resveratrol induced anticancer properties such as inflammation and attenuation of oxidative stress in hepatocarcinogenesis [167, 168].
Sodium butyrate produced as a result of metabolic degradation of carbohydrates in human colon and an isothiocyanate sulforaphane found in cruciferous vegetables prevent HCC by inhibiting HDACs [169, 170]. In a nutshell, nutritive epigenetics may serve as potential strategy to prevent HCC progression.
9. Epigenetic-Based Therapeutics for HCC and Future Updates
It is evident that epigenetic alterations play an important role in HCC and therefore can be targeted for treatment. In fact, in recent years the epigenetic drugs are in progress that reverse histone modification and methylation status. Combination of epigenetic drugs may also treat HCC (Table 4).
Table 4.
Drugs that target epigenetic modifications in HCC [3].
| Epigenetic modification | Drugs | Results | Ref. |
|---|---|---|---|
| Histone deacetylation that targets histone deacetylase | Suberoylanilide hydroxamic acid | TNF-related apoptosis-inducing ligand-induced apoptosis. | [100] |
| Belinostat | Tumor stabilization was observed in nonresectable advanced HCC. | [101] | |
| Belinostat | Induction of apoptosis and inhibition of cell growth occurred. | [102] | |
|
| |||
| DNA methylation that targets DNA methyltransferases | 5-Aza-2′-deoxycytidine | Inhibition of telomerase activity and reactivation of c-Myc and p16 were observed. | [103] |
| Zebularine | Induction of apoptosis and inhibition of cell proliferation were observed in HepG2 cell line. | [104] | |
| Zebularine | Tumor growth was inhibited in xenograft models. Genes involved in apoptosis, cell cycle, and tumor suppression were demethylated in KMHC and Huh-7 cell lines. | [105] | |
|
| |||
| Combined epigenetic modifications that target tyrosine kinase inhibitors and histone deacetylase inhibitors | Panobinostat + sorafenib | Combined activity of these two drugs induced apoptosis, increased survival, and decreased tumor density and tumor volume. | [106] |
HHC patients can be treated with HDAC inhibitors such as valproic acid, TSA, panobinostat, ITF2357, resminostat, givinostat, abexinostat, CUDC-101, and pracinostat that have given encouraging results in HCC patients because aberrant expression of HDAC is higher in cancer patients [101, 106, 171–174]. Epigenetic drugs can stimulate the immunity of host by increasing tumor antigen presentation [175].
MiRNAs with tumor-suppressor nature are perfect anticancer agents because of their ability to modulate multiple signaling pathways in cancer growth. Modulation of expression of miRNAs or targeting any tumor-related deregulated ncRNA may offer potential new therapeutic strategies.
Histone-modifying enzymes and DNMTs are the prime candidates for future HCC therapy. Series of clinical studies are currently under research for the development of epigenetic-based therapies to combat life-threatening condition HCC.
10. Conclusion
The above studies provide strong evidence that epigenetic alterations are in close association with disease stage and clinical outcome in HCC. The best-known genetic abnormalities in HCC are dysregulated expression of epigenetic regulatory genes, aberrant expression of noncoding RNAs, promoter methylation, and DNA methylation. Several different epidrugs target these aberrations and control the progression of HCC by reversing the expression of cell cycle and apoptosis related genes. Epigenetic alterations can potentially be considered an alternative option in cancer treatment protocols because epigenetic changes are reversible unlike genetic changes that are irreversible. Epigenetic changes affect cellular transcriptome alterations and result in gene expression and chromatin organization more extensively than genetic changes. Many of drugs with the potential to change the pattern or level of histone modification and DNA methylation have been developed and are now in clinical trials. As prognostic and diagnostic biomarker of HCC, it has been shown that histone modification, DNA methylation, and differential expression of noncoding RNAs help researchers to distinguish between HCC and cirrhotic liver or between tumor and nontumor adjacent tissues. Multikinase inhibitor, Sorafenib, is the first drug that treats HCC. HCC can be suppressed by inhibition of expression or activity of key proteins involved in carcinogenesis; therefore, epigenetic modulation of histones and the expression regulation of miRNAs serve as useful therapeutic strategies against HCC. Detailed mechanisms of HCC-related epigenetic-based therapeutics remain to be explored. Last but not least, targeted and efficient use of epigenetic drugs makes them prime candidates for future HCC therapy.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Herceg Z., Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutation Research/Reviews in Mutation Research. 2011;727(3):55–61. doi: 10.1016/j.mrrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Ma L., Chua M.-S., Andrisani O., So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World Journal of Gastroenterology. 2014;20(2):333–345. doi: 10.3748/wjg.v20.i2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y. Cancer Epigenetics: Risk Assessment, Diagnosis, Treatment, and Prognosis. Springer; 2015. Detection of epigenetic aberrations in the development of hepatocellular carcinoma; pp. 709–731. [DOI] [PubMed] [Google Scholar]
- 5.Hernando-Herraez I., Garcia-Perez R., Sharp A. J., Marques-Bonet T. DNA methylation: insights into human evolution. PLoS Genetics. 2015;11(12) doi: 10.1371/journal.pgen.1005661.e1005661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sceusi E. L., Loose D. S., Wray C. J. Clinical implications of DNA methylation in hepatocellular carcinoma. HPB. 2011;13(6):369–376. doi: 10.1111/j.1477-2574.2011.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber M., Hellmann I., Stadler M. B., et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genetics. 2007;39(4):457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M., Corn P. G., Baylin S. B., Herman J. G. A gene hypermethylation profile of human cancer. Cancer Research. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 9.He Y.-F., Li B.-Z., Li Z., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haffner M. C., Chaux A., Meeker A. K., et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2(8):627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S.-G., Jiang Y., Qiu R., et al. 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Research. 2011;71(24):7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song M.-A., Tiirikainen M., Kwee S., Okimoto G., Yu H., Wong L. L. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0055761.e55761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert M.-P., Ancey P.-B., Esposti D. D., et al. Aberrant DNA methylation of imprinted loci in hepatocellular carcinoma and after in vitro exposure to common risk factors. Clinical Epigenetics. 2015;7(1):p. 1. doi: 10.1186/s13148-015-0053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etoh T., Kanai Y., Ushijima S., et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. The American Journal of Pathology. 2004;164(2):689–699. doi: 10.1016/s0002-9440(10)63156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DNMT1 DNA methyltransferase 1 [Homo sapiens (human)]
- 16.Liu K., Zhang Y., Zhang C., et al. Methylation of S100A8 is a promising diagnosis and prognostic marker in hepatocellular carcinoma. Oncotarget. 2016;7(35):56798–56810. doi: 10.18632/oncotarget.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H. G., Kim H., Son T., et al. Regulation of HK2 expression through alterations in CpG methylation of the HK2 promoter during progression of hepatocellular carcinoma. Oncotarget. 2016;7(27):41798–41810. doi: 10.18632/oncotarget.9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W.-R., Shi Y.-H., Peng Y.-F., Fan J. Epigenetics of hepatocellular carcinoma: a new horizon. Chinese Medical Journal. 2012;125(13):2349–2360. doi: 10.3760/cma.j.issn.0366-6999.2012.13.020. [DOI] [PubMed] [Google Scholar]
- 19.Nishida N., Goe A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Current Genomics. 2011;12(2):130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baylin S. B. DNA methylation and gene silencing in cancer. Nature Clinical Practice Oncology. 2005;2(1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 21.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 22.Sawan C., Vaissière T., Murr R., Herceg Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutation Research—Fundamental and Molecular Mechanisms of Mutagenesis. 2008;642(1-2):1–13. doi: 10.1016/j.mrfmmm.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Lambert M.-P., Paliwal A., Vaissière T., et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. Journal of Hepatology. 2011;54(4):705–715. doi: 10.1016/j.jhep.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Duong F. H. T., Christen V., Lin S., Heim M. H. Hepatitis C virus-induced up-regulation of protein phosphatase 2A inhibits histone modification and DNA damage repair. Hepatology. 2010;51(3):741–751. doi: 10.1002/hep.23388. [DOI] [PubMed] [Google Scholar]
- 25.Higgs M. R., Lerat H., Pawlotsky J.-M. Downregulation of Gadd45β expression by hepatitis C virus leads to defective cell cycle arrest. Cancer Research. 2010;70(12):4901–4911. doi: 10.1158/0008-5472.CAN-09-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez-Vargas H., Lambert M.-P., Le Calvez-Kelm F., et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009749.e9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye C., Tao R., Cao Q., et al. Whole-genome DNA methylation and hydroxymethylation profiling for HBV-related hepatocellular carcinoma. Age (Years) 2016;41(53):p. 40. doi: 10.3892/ijo.2016.3535. [DOI] [PubMed] [Google Scholar]
- 28.Tang X., Ge L., Chen Z., et al. Methylation of the constitutive androstane receptor is involved in the suppression of CYP2C19 in hepatitis B Virus-associated hepatocellular carcinoma. Drug Metabolism and Disposition. 2016;44(10):1643–1652. doi: 10.1124/dmd.116.070243. [DOI] [PubMed] [Google Scholar]
- 29.Shih Y.-L., Kuo C.-C., Yan M.-D., Lin Y.-W., Hsieh C.-B., Hsieh T.-Y. Quantitative methylation analysis reveals distinct association between PAX6 methylation and clinical characteristics with different viral infections in hepatocellular carcinoma. Clinical Epigenetics. 2016;8(1, article 41) doi: 10.1186/s13148-016-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J., Wang S., Zhang Y.-J., et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55(6):1799–1808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida N., Kudo M., Nagasaka T., Ikai I., Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56(3):994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- 32.Revill K., Wang T., Lachenmayer A., et al. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145(6):1424.e25–1435.e25. doi: 10.1053/j.gastro.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anwar S. L., Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World Journal of Gastroenterology. 2014;20(24):7894–7913. doi: 10.3748/wjg.v20.i24.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Dong H., Robertson K., Liu C. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. The American Journal of Pathology. 2011;178(2):652–661. doi: 10.1016/j.ajpath.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tischoff I., Markwarth A., Witzigmann H., et al. Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. International Journal of Cancer. 2005;115(5):684–689. doi: 10.1002/ijc.20944. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C., Guo X., Jiang G., et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. International Journal of Cancer. 2008;123(5):998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 37.Tomasi M. L., Li T. W. H., Li M., Mato J. M., Lu S. C. Inhibition of human methionine adenosyltransferase 1A transcription by coding region methylation. Journal of Cellular Physiology. 2012;227(4):1583–1591. doi: 10.1002/jcp.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warburg O., Wind F., Neglers F. Metabolism of Tumours. London, UK: Constable & Co; 1930. The metabolism of tumours; pp. 254–270. [Google Scholar]
- 39.Rong M., Chen G., Dang Y. Increased MiR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13(1, article 21) doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Fang L., Yu W., Wang Y. MicroRNA-125b suppresses the migration and invasion of hepatocellular carcinoma cells by targeting transcriptional coactivator with PDZ-binding motif. Oncology Letters. 2015;9(4):1971–1975. doi: 10.3892/ol.2015.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L.-L., Guo Y.-J., Zhao C.-N., Gao J.-Y. Effects and mechanism of miR-214 on hepatocellular carcinoma. Asian Pacific Journal of Tropical Medicine. 2015;8(5):392–398. doi: 10.1016/S1995-7645(14)60350-3. [DOI] [PubMed] [Google Scholar]
- 42.Xie K., Liu J., Zhu L., et al. A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma. Cancer Epidemiology. 2013;37(6):998–1002. doi: 10.1016/j.canep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Yang X., Liang L., Zhang X.-F., et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58(1):158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 44.Nassirpour R., Mehta P. P., Yin M.-J. miR-122 regulates tumorigenesis in hepatocellular carcinoma by targeting AKT3. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079655.e79655 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Lin C. J.-F., Gong H.-Y., Tseng H.-C., Wang W.-L., Wu J.-L. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochemical and Biophysical Research Communications. 2008;375(3):315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 46.Wang T.-H., Yeh C.-T., Ho J.-Y., Ng K.-F., Chen T.-C. OncomiR miR-96 and miR-182 promote cell proliferation and invasion through targeting ephrinA5 in hepatocellular carcinoma. Molecular Carcinogenesis. 2016;55(4):366–375. doi: 10.1002/mc.22286. [DOI] [PubMed] [Google Scholar]
- 47.Cao K., Li J., Zhao Y., et al. miR-101 inhibiting cell proliferation, migration and invasion in hepatocellular carcinoma through downregulating girdin. Molecules and Cells. 2016;39(2):96–102. doi: 10.14348/molcells.2016.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan T., He X., Yu J., Hang Z. MicroRNA-186 targets yes-associated protein 1 to inhibit hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncology Letters. 2016;11(4):2941–2945. doi: 10.3892/ol.2016.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parpart S., Roessler S., Dong F., et al. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology. 2014;60(3):872–883. doi: 10.1002/hep.27200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L., Wang W. miR-125b suppresses the proliferation of hepatocellular carcinoma cells by targeting Sirtuin7. International Journal of Clinical and Experimental Medicine. 2015;8(10):18469–18475. [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z., Dou C., Yao B., et al. Methylation-mediated repression of microRNA-129-2 suppresses cell aggressiveness by inhibiting high mobility group box 1 in human hepatocellular carcinoma. Oncotarget. 2016;7(24):36909–36923. doi: 10.18632/oncotarget.9377. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Kelong M., He Y., Zhang H., et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. Journal of Biological Chemistry. 2012;287(8):5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian Z., Jiang H., Liu Y., et al. MicroRNA-133b inhibits hepatocellular carcinoma cell progression by targeting Sirt1. Experimental Cell Research. 2016;343(2):135–147. doi: 10.1016/j.yexcr.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 54.Luo Z., Feng C., Hu P., et al. Serum microRNA-199a/b-3p as a predictive biomarker for treatment response in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. OncoTargets and Therapy. 2016;9:2667–2674. doi: 10.2147/ott.s98408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Q., Cicinnati V. R., Zhang X., et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Molecular Cancer. 2010;9(1, article 227) doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang B., Lei B., Qi G., et al. MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. Journal of Experimental and Clinical Cancer Research. 2016;35(1, article 93) doi: 10.1186/s13046-016-0371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y.-F., Wang F., Xiao J.-J., et al. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. International Journal of Clinical and Experimental Medicine. 2014;7(4):893–902. [PMC free article] [PubMed] [Google Scholar]
- 58.Xu G., Zhang Y., Wei J., et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13(1, article 469) doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S., Liu K., Zhang W., et al. MiR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma. American Journal of Translational Research. 2016;8(6):2609–2619. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Wong C. C.-L., Wong C., Tung E. K., et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating rho-kinase 2. Gastroenterology. 2011;140(1):322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Bi Q., Tang S., Xia L., et al. Ectopic expression of miR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0040169.e40169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan G., Wu L., Tan J., et al. MIR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF-κB signaling pathway. Scientific Reports. 2016;6, article 22328 doi: 10.1038/srep22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L., Yang F., Yuan J.-H., et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34(3):577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 64.Tu H., Wei G., Cai Q., et al. MicroRNA-212 inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting FOXA1. OncoTargets and Therapy. 2015;8:2227–2235. doi: 10.2147/ott.s87976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Dou C., Wang Y., Li C., et al. MicroRNA-212 suppresses tumor growth of human hepatocellular carcinoma by targeting FOXA1. Oncotarget. 2015;6(15):13216–13228. doi: 10.18632/oncotarget.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L., Yu Z., Xian Y., Lin X. microRNA-497 inhibits cell proliferation and induces apoptosis by targeting YAP1 in human hepatocellular carcinoma. FEBS Open Bio. 2016;6(2):155–164. doi: 10.1002/2211-5463.12032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Fornari F., Milazzo M., Chieco P., et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. Journal of Pathology. 2012;227(3):275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 68.Li Y., Tan W., Neo T. W. L., et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Science. 2009;100(7):1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 69.Yang L., Zhang X., Li H., Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Molecular BioSystems. 2016;12(8):2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 70.Gao J.-Z., Li J., Du J.-L., Li X.-L. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncology Letters. 2016;11(3):1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng L., Yang S.-B., Xu F.-F., Zhang J.-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. Journal of Experimental and Clinical Cancer Research. 2015;34(1, article 18) doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S., Xu H., Yu Y., et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J., Qin Y., Li B., He W.-Z., Sun Z.-L. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics. 2008;91(5):443–450. doi: 10.1016/j.ygeno.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 74.Quagliata L., Matter M. S., Piscuoglio S., et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsang F. H. C., Au S. L. K., Wei L., et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver International. 2015;35(5):1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 76.Zhou T., Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World Journal of Surgical Oncology. 2016;14(1, article 8) doi: 10.1186/s12957-015-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lai M.-C., Yang Z., Zhou L., et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical Oncology. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 78.Yang F., Zhang L., Huo X.-S., et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54(5):1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 79.Chen C.-L., Tseng Y.-W., Wu J.-C., et al. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials. 2015;44:71–81. doi: 10.1016/j.biomaterials.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 80.Zhang D., Cao C., Liu L., Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. Journal of Cancer. 2016;7(5):608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhuo H., Tang J., Lin Z., et al. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Molecular Carcinogenesis. 2016;55(2):209–219. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- 82.Kogure T., Yan I. K., Lin W.-L., Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes and Cancer. 2013;4(7-8):261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braconi C., Valeri N., Kogure T., et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(2):786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi K., Yan I. K., Haga H., Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. Journal of Cell Science. 2014;127(7):1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takahashi K., Yan I. K., Kogure T., Haga H., Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4(1):458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan S.-X., Yang F., Yang Y., et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56(6):2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 87.Yuan J.-H., Yang F., Wang F., et al. A Long Noncoding RNA Activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 88.Huang M.-D., Chen W.-M., Qi F.-Z., et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Molecular Cancer. 2015;14(1, article 165) doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu W.-H., Zhang J.-B., Dang Z., et al. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. International Journal of Biological Sciences. 2014;10(7):664–676. doi: 10.7150/ijbs.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu F., Yuan J.-H., Huang J.-F., et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35(41):5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 91.Ding C., Yang Z., Lv Z., et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncology Letters. 2015;9(2):955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia M., Jiang L., Wang Y.-D., Huang J.-Z., Yu M., Xue H.-Z. LincRNA‐p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling induced epithelial‐mesenchymal transition. Hepatology Research. 2016;46(11):1137–1144. doi: 10.1111/hepr.12659. [DOI] [PubMed] [Google Scholar]
- 93.Wang F., Ying H.-Q., He B.-S., et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu W., Qiao Y., Tang X., et al. Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cellular Signalling. 2014;26(12):2961–2968. doi: 10.1016/j.cellsig.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 95.Cao C., Sun J., Zhang D., et al. The long intergenic noncoding RNA UFC1, a target of microRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148(2):415–426. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 96.Zheng H., Yang S., Yang Y., et al. Epigenetically silenced long noncoding-SRHC promotes proliferation of hepatocellular carcinoma. Journal of Cancer Research and Clinical Oncology. 2014;141(7):1195–1203. doi: 10.1007/s00432-014-1871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yuan S.-X., Tao Q.-F., Wang J., et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Letters. 2014;349(1):87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 98.Huang J.-F., Guo Y.-J., Zhao C.-X., et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57(5):1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 99.Tsang W. P., Wong T. W. L., Cheung A. H. H., Co C. N. N., Kwok T. T. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;13(6):890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carlisi D., Lauricella M., D'Anneo A., et al. The histone deacetylase inhibitor suberoylanilide hydroxamic acid sensitises human hepatocellular carcinoma cells to TRAIL-induced apoptosis by TRAIL-DISC activation. European Journal of Cancer. 2009;45(13):2425–2438. doi: 10.1016/j.ejca.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 101.Yeo W., Chung H. C., Chan S. L., et al. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the mayo phase II consortium and the cancer therapeutics research group. Journal of Clinical Oncology. 2012;30(27):3361–3367. doi: 10.1200/jco.2011.41.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma B. B. Y., Sung F., Tao Q., et al. The preclinical activity of the histone deacetylase inhibitor PXD101 (belinostat) in hepatocellular carcinoma cell lines. Investigational New Drugs. 2010;28(2):107–114. doi: 10.1007/s10637-009-9219-7. [DOI] [PubMed] [Google Scholar]
- 103.Tao S.-F., Zhang C.-S., Guo X.-L., et al. Anti-tumor effect of 5-aza-2′-deoxycytidine by inhibiting telomerase activity in hepatocellular carcinoma cells. World Journal of Gastroenterology. 2012;18(19):2334–2343. doi: 10.3748/wjg.v18.i19.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakamura K., Aizawa K., Nakabayashi K., et al. DNA methyltransferase inhibitor zebularine inhibits human hepatic carcinoma cells proliferation and induces apoptosis. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0054036.e54036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andersen J. B., Factor V. M., Marquardt J. U., et al. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Science Translational Medicine. 2010;2(54):p. 54ra77. doi: 10.1126/scitranslmed.3001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lachenmayer A., Toffanin S., Cabellos L., et al. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. Journal of Hepatology. 2012;56(6):1343–1350. doi: 10.1016/j.jhep.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones P. A., Baylin S. B. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 108.van Attikum H., Gasser S. M. Crosstalk between histone modifications during the DNA damage response. Trends in Cell Biology. 2009;19(5):207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 109.Nguyen C. T., Weisenberger D. J., Velicescu M., et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Research. 2002;62(22):6456–6461. [PubMed] [Google Scholar]
- 110.Chen H., Zhang T., Sheng Y., et al. Methylation profiling of multiple tumor suppressor genes in hepatocellular carcinoma and the epigenetic mechanism of 3OST2 regulation. Journal of Cancer. 2015;6(8):740–749. doi: 10.7150/jca.11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cao R., Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Current Opinion in Genetics and Development. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 112.Bracken A. P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. EZH2 is downstream of the pRB‐E2F pathway, essential for proliferation and amplified in cancer. The EMBO Journal. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lachner M., O'Sullivan R. J., Jenuwein T. An epigenetic road map for histone lysine methylation. Journal of Cell Science. 2003;116(11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 114.Varambally S., Dhanasekaran S. M., Zhou M., et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 115.Bhattacharya S., Reddy D., Ingle A., Khade B., Gupta S. Brief Communication: Featured Article: Histone H2A mono-ubiquitination and cellular transformation are inversely related in N-nitrosodiethylamine-induced hepatocellular carcinoma. Experimental biology and Medicine (Maywood, N.J.) 2016;241(16):1739–1744. doi: 10.1177/1535370216649262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Magerl C., Ellinger J., Braunschweig T., et al. H3K4 dimethylation in hepatocellular carcinoma is rare compared with other hepatobiliary and gastrointestinal carcinomas and correlates with expression of the methylase Ash2 and the demethylase LSD1. Human Pathology. 2010;41(2):181–189. doi: 10.1016/j.humpath.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 117.Shon J. K., Shon B. H., Park I. Y., et al. Hepatitis B virus-X protein recruits histone deacetylase 1 to repress insulin-like growth factor binding protein 3 transcription. Virus Research. 2009;139(1):14–21. doi: 10.1016/j.virusres.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 118.Schulze K., Imbeaud S., Letouzé E., et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nature Genetics. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sung W.-K., Zheng H., Li S., et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nature Genetics. 2012;44(7):765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 120.He C., Xu J., Zhang J., et al. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Human Pathology. 2012;43(9):1425–1435. doi: 10.1016/j.humpath.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 121.Wong C. M., Wei L., Law C. T., et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 122.Zhang X., Wu J., Choiniere J., et al. Arsenic silences hepatic PDK4 expression through activation of histone H3K9 methylatransferase G9a. Toxicology and Applied Pharmacology. 2016;304:42–47. doi: 10.1016/j.taap.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Longerich T. Dysregulation of the epigenetic regulator SETDB1 in liver carcinogenesis—more than one way to skin a cat. Chinese Clinical Oncology. 2016;5(6):72–72. doi: 10.21037/cco.2016.03.18. [DOI] [PubMed] [Google Scholar]
- 124.Zhu J. DNA methylation and hepatocellular carcinoma. Journal of Hepato-Biliary-Pancreatic Surgery. 2006;13(4):265–273. doi: 10.1007/s00534-005-1054-4. [DOI] [PubMed] [Google Scholar]
- 125.Liao Y.-J., Liu S.-P., Lee C.-M., et al. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: implications of the gender disparity in liver cancer susceptibility. International Journal of Cancer. 2009;124(4):816–826. doi: 10.1002/ijc.23979. [DOI] [PubMed] [Google Scholar]
- 126.Moeini A., Cornellà H., Villanueva A. Emerging signaling pathways in hepatocellular carcinoma. Liver Cancer. 2012;1(2):83–93. doi: 10.1159/000342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang D., Fang J., Luo G. Roles of long noncoding RNAs in Hepatocellular Carcinoma. Open Life Sciences. 2016;11(1):91–97. doi: 10.1515/biol-2016-0012. [DOI] [Google Scholar]
- 128.Falkenberg K. J., Johnstone R. W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature Reviews Drug Discovery. 2014;13(9):673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 129.Rikimaru T., Taketomi A., Yamashita Y.-I., et al. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72(1-2):69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- 130.Liu C., Liu L., Shan J., et al. Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Letters. 2013;339(1):60–69. doi: 10.1016/j.canlet.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 131.Bhaskara S., Knutson S. K., Jiang G., et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18(5):436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ceccacci E., Minucci S. Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia. British Journal of Cancer. 2016;114(6):605–611. doi: 10.1038/bjc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang J. Current progress in epigenetic research for hepatocarcinomagenesis. Science in China, Series C: Life Sciences. 2009;52(1):31–42. doi: 10.1007/s11427-009-0014-7. [DOI] [PubMed] [Google Scholar]
- 134.Cougot D., Wu Y., Cairo S., et al. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. The Journal of Biological Chemistry. 2007;282(7):4277–4287. doi: 10.1074/jbc.m606774200. [DOI] [PubMed] [Google Scholar]
- 135.Zheng D.-L., Zhang L., Cheng N., et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. Journal of Hepatology. 2009;50(2):377–387. doi: 10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 136.Pal-Bhadra M., Bhadra U., Jackson D. E., Mamatha L., Park P.-H., Shukla S. D. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sciences. 2007;81(12):979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang H., Nie Y., Li Y., Wan Y.-J. Y. Histone modification-mediated CYP2E1 gene expression and apoptosis of HepG2 cells. Experimental Biology and Medicine. 2010;235(1):32–39. doi: 10.1258/ebm.2009.009252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhu G., Tao T., Zhang D., et al. O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology. 2016;26(8):820–833. doi: 10.1093/glycob/cww025. [DOI] [PubMed] [Google Scholar]
- 139.Wu B.-H., Chen H., Cai C.-M., et al. Epigenetic silencing of JMJD5 promotes the proliferation of hepatocellular carcinoma cells by down-regulating the transcription of CDKN1A. Oncotarget. 2016;7(6):6847–6863. doi: 10.18632/oncotarget.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wilson B. G., Roberts C. W. M. SWI/SNF nucleosome remodellers and cancer. Nature Reviews Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 141.Endo M., Yasui K., Zen Y., et al. Alterations of the SWI/SNF chromatin remodelling subunit-BRG1 and BRM in hepatocellular carcinoma. Liver International. 2013;33(1):105–117. doi: 10.1111/liv.12005. [DOI] [PubMed] [Google Scholar]
- 142.Guichard C., Amaddeo G., Imbeaud S., et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature Genetics. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li M., Zhao H., Zhang X., et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nature Genetics. 2011;43(9):828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu M., Chen L., Ma N.-F., et al. CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors. Hepatology. 2016;63(5):1544–1559. doi: 10.1002/hep.28437. [DOI] [PubMed] [Google Scholar]
- 145.Wang Z., Li X. The role of noncoding RNA in hepatocellular carcinoma. Gland Surgery. 2013;2(1):p. 25. doi: 10.3978/j.issn.2227-684X.2013.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.George J., Patel T. Seminars in Liver Disease. Stuttgart, Germany: Thieme Medical Publishers; 2015. Noncoding RNA as therapeutic targets for hepatocellular carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu Z., Wang J., Mao Y., Zou B., Fan X. MicroRNA-101 suppresses migration and invasion via targeting vascular endothelial growth factor-C in hepatocellular carcinoma cells. Oncology Letters. 2016;11(1):433–438. doi: 10.3892/ol.2015.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Najafi Z., Sharifi M., Javadi G. Degradation of miR-21 induces apoptosis and inhibits cell proliferation in human hepatocellular carcinoma. Cancer Gene Therapy. 2015;22(11):530–535. doi: 10.1038/cgt.2015.51. [DOI] [PubMed] [Google Scholar]
- 149.Wang P., Chen S., Fang H., et al. miR-214/199a/199a∗ cluster levels predict poor survival in hepatocellular carcinoma through interference with cell-cycle regulators. Oncotarget. 2016;7(1):929–945. doi: 10.18632/oncotarget.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jia X. Q., Cheng H. Q., Qian X., et al. Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochemistry and Biophysics. 2012;62(1):237–244. doi: 10.1007/s12013-011-9263-8. [DOI] [PubMed] [Google Scholar]
- 151.Zhu S., Chen C. M., Jiang Z. Y., et al. MicroRNA-185 inhibits cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. European Review for Medical and Pharmacological Sciences. 2016;20(9):1712–1719. [PubMed] [Google Scholar]
- 152.Zhou X., Zhu H.-Q., Ma C.-Q., et al. MiR-1180 promoted the proliferation of hepatocellular carcinoma cells by repressing TNIP2 expression. Biomedicine & Pharmacotherapy. 2016;79:315–320. doi: 10.1016/j.biopha.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 153.Zeng Y.-B., Liang X.-H., Zhang G.-X., et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell International. 2016;16(1, article 63) doi: 10.1186/s12935-016-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.He Y., Meng X.-M., Huang C., et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Letters. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 155.Gao Y., Chen G., Zeng Y., et al. Invasion and metastasis-related long noncoding RNA expression profiles in hepatocellular carcinoma. Tumor Biology. 2015;36(10):7409–7422. doi: 10.1007/s13277-015-3408-0. [DOI] [PubMed] [Google Scholar]
- 156.Ngo-Yin Fan D., Ho-Ching Tsang F., Hoi-Kam Tam A., et al. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57(2):637–647. doi: 10.1002/hep.26083. [DOI] [PubMed] [Google Scholar]
- 157.Mah W., Lee C. G. DNA methylation: potential biomarker in Hepatocellular Carcinoma. Biomarker Research. 2014;2(1, article 5) doi: 10.1186/2050-7771-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kojima K., Takata A., Vadnais C., et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nature Communications. 2011;2, article 338 doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- 159.Hou J., Lin L., Zhou W., et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 160.Nagashio R., Arai E., Ojima H., Kosuge T., Kondo Y., Kanai Y. Carcinogenetic risk estimation based on quantification of DNA methylation levels in liver tissue at the precancerous stage. International Journal of Cancer. 2011;129(5):1170–1179. doi: 10.1002/ijc.26061. [DOI] [PubMed] [Google Scholar]
- 161.Teiten M.-H., Dicato M., Diederich M. Curcumin as a regulator of epigenetic events. Molecular Nutrition and Food Research. 2013;57(9):1619–1629. doi: 10.1002/mnfr.201300201. [DOI] [PubMed] [Google Scholar]
- 162.Darvesh A. S., Bishayee A. Chemopreventive and therapeutic potential of tea polyphenols in hepatocellular cancer. Nutrition and Cancer. 2013;65(3):329–344. doi: 10.1080/01635581.2013.767367. [DOI] [PubMed] [Google Scholar]
- 163.Lee W. J., Zhu B. T. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006;27(2):269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 164.Furtado K. S., Polletini J., Dias M. C., Rodrigues M. A. M., Barbisan L. F. Prevention of rat liver fibrosis and carcinogenesis by coffee and caffeine. Food and Chemical Toxicology. 2014;64:20–26. doi: 10.1016/j.fct.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 165.Verslype C. Interfering with epigenetics in hepatocellular carcinoma: out of the shelter? Journal of Hepatology. 2016;65(2):243–244. doi: 10.1016/j.jhep.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 166.Bishayee A., Dhir N. Resveratrol-mediated chemoprevention of diethylnitrosamine-initiated hepatocarcinogenesis: inhibition of cell proliferation and induction of apoptosis. Chemico-Biological Interactions. 2009;179(2-3):131–144. doi: 10.1016/j.cbi.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 167.Bishayee A., Waghray A., Barnes K. F., et al. Suppression of the inflammatory cascade is implicated in resveratrol chemoprevention of experimental hepatocarcinogenesis. Pharmaceutical Research. 2010;27(6):1080–1091. doi: 10.1007/s11095-010-0144-4. [DOI] [PubMed] [Google Scholar]
- 168.Bishayee A., Politis T., Darvesh A. S. Resveratrol in the chemoprevention and treatment of hepatocellular carcinoma. Cancer Treatment Reviews. 2010;36(1):43–53. doi: 10.1016/j.ctrv.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 169.Wang H.-G., Huang X.-D., Shen P., Li L.-R., Xue H.-T., Ji G.-Z. Anticancer effects of sodium butyrate on hepatocellular carcinoma cells in vitro. International Journal of Molecular Medicine. 2013;31(4):967–974. doi: 10.3892/ijmm.2013.1285. [DOI] [PubMed] [Google Scholar]
- 170.Clarke J. D., Dashwood R. H., Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Letters. 2008;269(2):291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mottamal M., Zheng S., Huang T. L., Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015;20(3):3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Buurman R., Grlevik E., Schffer V., et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143(3):811–820.e15. doi: 10.1053/j.gastro.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 173.Armeanu S., Pathil A., Venturelli S., et al. Apoptosis on hepatoma cells but not on primary hepatocytes by histone deacetylase inhibitors valproate and ITF2357. Journal of Hepatology. 2005;42(2):210–217. doi: 10.1016/j.jhep.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 174.Pathil A., Armeanu S., Venturelli S., et al. HDAC inhibitor treatment of hepatoma cells induces both TRAIL-independent apoptosis and restoration of sensitivity to TRAIL. Hepatology. 2006;43(3):425–434. doi: 10.1002/hep.21054. [DOI] [PubMed] [Google Scholar]
- 175.Guo Z. S., Hong J. A., Irvine K. R., et al. De novo induction of a cancer/testis antigen by 5-aza-2′- deoxycytidine augments adoptive immunotherapy in a murine tumor model. Cancer Research. 2006;66(2):1105–1113. doi: 10.1158/0008-5472.CAN-05-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]