Abstract
Differentiation alters molecular properties of stem and progenitor cells, leading to changing shape and movement characteristics. We present a deep neural network that prospectively predicts lineage choice in differentiating primary hematopoietic progenitors, using image patches from brightfield microscopy and cellular movement. Surprisingly, lineage choice can be detected up to three generations before conventional molecular markers are observable. Our approach allows identifying cells with differentially expressed lineage-specifying genes without molecular labeling.
Long-term high throughput time-lapse microscopy is a powerful tool to study the differentiation processes of single cells in unprecedented temporal resolution1. A high frequency of brightfield imaging (typically on the scale of a few minutes) ensures that moving single cells and cell divisions can be accurately tracked and used for the construction of cellular genealogies. Additionally, fluorescent imaging (due to cell phototoxicity typically only possible in intervals of thirty minutes or more2) allows the quantification of molecular lineage markers3,4. However, molecular lineage markers are only available for specific, often already differentiated cell types5,6, hindering an early identification of differentiating cells.
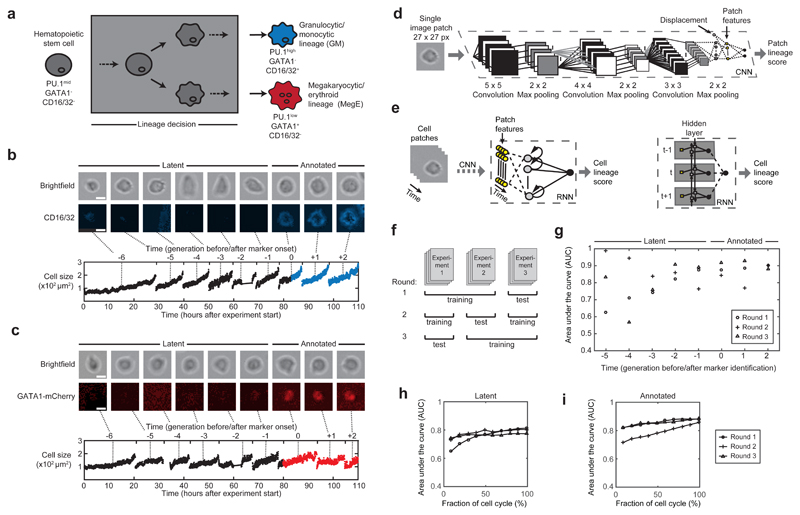
We thus set out to exploit the information in the abundant brightfield images of time-lapse experiments for a prospective detection of lineage commitment. We developed our method on time-lapse experiments of primary murine hematopoietic stem and progenitor cells (HSPCs) differentiating into either the granulocytic/monocytic (GM) or the megakaryocytic/erythroid (MegE) lineage (Fig. 1a, Supplementary Fig. 1). Branches (i.e. a cell and all its predecessors) were generated by automatically linking an image patch of 27x27 pixel covering the mass-centered cell body to every time point of a manual cell track (Fig. 1b,c, Supplementary Note 1). We annotated lineage commitment when the respective lineage marker was detectable in the fluorescent channel (CD16/32 for GM and GATA1-mCherry for MegE lineage) and assigned all tracked cells to one of three categories: i) “annotated” cells with clear marker expression within the cell lifetime, ii) “latent” cells with no immediate marker expression but an expression in a subsequent generation, and iii) “unknown” cells with no marker expression in current or subsequent generations (Supplementary Fig. 2a-c). Our dataset comprised 150 genealogies from 3 independent experiments with a total of 5,922 single cells (Supplementary Fig. 2d-f). Each cell was imaged ~400 times, resulting in more than 2,400,000 image patches.
Figure 1. Prediction of hematopoietic lineage choice up to three generations before molecular marker annotation using deep neural networks.
(a) Hematopoietic stem cells (gray) can differentiate and are annotated as committed towards the granulocytic/monocytic (GM, blue) lineage via detection of CD16/32, or towards the megakaryocytic/erythroid lineage (MegE, red) via GATA1-mCherry expression. These conventional markers necessarily appear after the lineage decision (gray box). (b,c) Exemplary image patches of a branch of single cells committing to either GM (b, upper row) or MegE (c, upper row) lineage (scale bars: 10µm). Cells with no marker expression are called “latent”, cells with marker expression “annotated” (b,c, middle row). Our automatic image processing pipeline allows robust cell identification and thus quantification of movement and morphology (demonstrated with cell size in lower rows of b and c) (d) A single image patch and the according cell’s displacement (white node) with respect to the previous time point are fed into a convolutional neural network (CNN) consisting of convolutional and fully connected layers (see Methods for more details on the network architecture). The last fully connected hidden layer (yellow) can be interpreted as patch-specific features. (e) To account for temporal dependencies we feed the CNN-derived patch features of a cell (yellow) in a recurrent neural network (RNN). The nodes in the hidden layer are connected to output nodes as well as all other hidden nodes across time (left); this temporal dependency is further illustrated in an unrolled representation of the RNN (right), where yellow squares represent the patch feature vectors at a specific time point and forward/backward arrows reflect the bidirectional architecture of the RNN. Every patch is assigned a lineage score between 0 and 1 (0=MegE, 1=GM, 0.5=unsure). (f) Two experiments are used for training, while one experiment is left out to assess generalization quality of the learned model. We repeat this procedure three times in a round-robin fashion. (g) Area under the receiver operating characteristics curve (AUC; 1.0=perfect classification, 0.5=random guessing) determines the performance of the trained models. Annotated cells (generations 0,+1,+2) and latent cells up to three generations before marker onset (generations -3,-2,-1) show AUCs higher than 0.77 (n=3 rounds, 4204 single cells in total). (h,i) AUCs when only (contiguous) subsets of image patches are used to compute the cell lineage score. AUCs over 0.75 were reached when using the first ~25% of timepoints in the cell cycle from latent (h) and annotated cells (i), respectively.
We used these millions of image patches to build a classifier that predicts the lineage choice of a stem cell’s progeny towards the GM or the MegE lineage. To efficiently leverage the information in our dataset we built on recent advances in deep neural networks for image classification. We combine a convolutional neural network (CNN) with a recurrent neural network (RNN) architecture to automatically extract local image features and exploit the temporal information of the single-cell tracks (Fig. 1d,e). Specifically, three connected convolutional layers extract image features, resulting in increasingly global representations of the image patches. As a CNN allows no direct inclusion of features other than pixel information, we introduced a concatenation layer combining the highest-level spatial features with cell displacement, followed by a fully connected layer that can be interpreted as patch features. To train the CNN, this layer is connected to output nodes resulting in a single patch lineage score between 0 and 1. Lineage scores of 0 or 1 indicate a strong similarity to cell patches from the MegE or GM lineage, respectively. Next, in order to classify individual cells as committed to either lineage, we used the patch features as input for the RNN. To model long-range temporal dependencies in the data without suffering from the vanishing-gradient problem7, we used a bidirectional long short term memory (LSTM) architecture8,9 (Fig. 1e).
After filtering out all unknown cells (containing both uncommitted cells and committed cells for which the markers had not yet switched on at the end of the experiments), the dataset to train and evaluate our method consisted of 4,402 single cells (~1,700,000 image patches) with annotated or latent marker onset (34% MegE and 66% GM, Supplementary Fig. 2e,f). To assess the generalization power of our model to reliably predict a cell’s putative lineage choice in independent experiments, we trained our CNN-RNN on 2 experiments and tested the resulting model on the third experiment; we repeated this procedure 3 times in a round-robin fashion (Fig. 1f) and evaluated the performance of the trained model by the area-under-the-curve (AUC) of the receiver-operating characteristic and F1 score (Supplementary Fig. 3). Our method achieved high AUCs of 0.87±0.01 (mean±sd, n=3 rounds) on annotated cells, indicating that morphology and displacement suffice to detect the lineage choice of HSPCs. Interestingly, the reported AUCs for latent cells were also high (0.79±0.02, mean±sd, n=3 rounds), suggesting that latent cells are morphologically different before an identifiable marker expression. We further investigated this finding by analyzing AUCs for every generation separately (Fig. 1g). AUCs stayed at comparable and robust levels from one to three generations before an annotated marker onset (0.84±0.07, 0.86±0.04 and 0.78±0.05 respectively, mean±sd, n=3 rounds). At four and five generations before the marker onset the decline and variance of AUCs (0.74±0.19 and 0.82±0.19 respectively, mean±sd, n=3) suggested that the difference in morphology and displacement did not longer suffice to differentiate GM- and MegE-committed cells. We achieved similar performance when using our classifier on data from genetically non-modified mice (see Methods and Supplementary Fig. 4). To assess the performance of our CNN-RNN on cells where tracking is stopped before cell cycle completion (e.g. in an “online-prediction” scenario where the lineage score is calculated while the experiment runs), we computed AUCs based only on a subset of brightfield patches. While single patches are insufficient for correct prediction, AUCs over 0.75 were reached when using the first ~25% of timepoints in the cell cycle from annotated (Fig. 1h) and latent (Fig. 1i) cells (3, 2 and 1 generation before the identified marker onset).
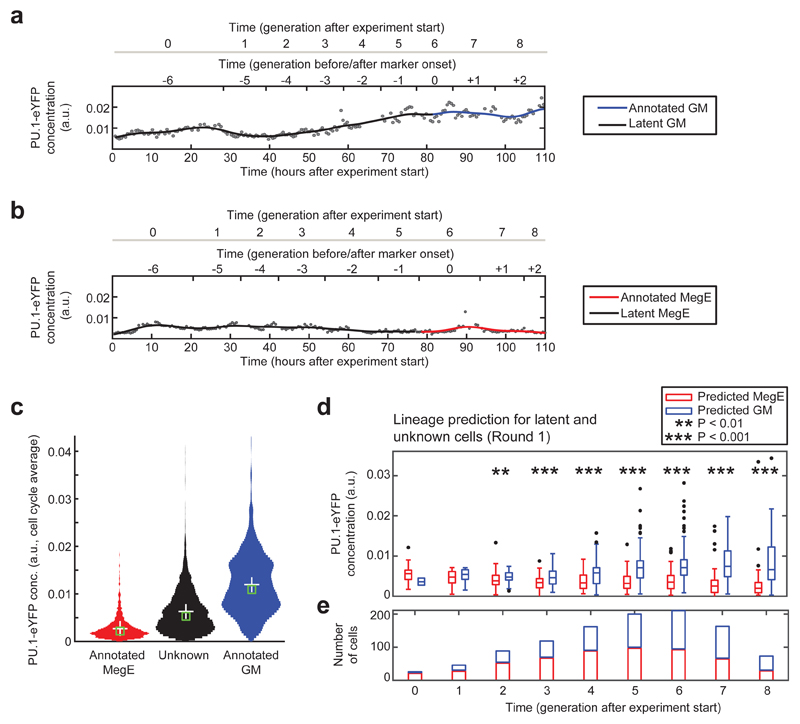
The combination of single-cell transcriptomic profiling10 with our approach would in principle allow to compare expression patterns of early committed cells after stratification in accordance to their lineage score to identify differentially expressed regulators. Since the usefulness of our approach for such an experiment is hard to assess from the AUCs, we evaluated our method with the expression of PU.1, a transcription factor that is tagged with eYFP in our cells (see Methods). As expected6,11, PU.1-eYPF was upregulated in GM-annotated cells (Fig. 2a), downregulated in MegE-annotated cells (Fig. 2b), and showed intermediate expression in cells without annotated marker expression (Fig. 2c). If our proposed method was capable of reliably predicting early lineage choice, we should be able to in silico stratify all cells at every timepoint of a hypothetical experiment into GM- or MegE-committed cells. These groups in turn are expected to differentially express PU.1-eYFP. Thus we classified every latent and unknown cell into two groups by analyzing if its lineage score was above (GM) or below (MegE) a threshold of 0.5 (Fig. 2d, Supplementary Fig. 5a,b). We found the two groups to differentially express PU.1-eYFP from 2 generations after experiment start onwards (P<0.01 in generation 2, P<0.001 in generations 3-8, unpaired Wilcoxon rank-sum test, Fig. 2d) with at least 89 cells per generation (Fig. 2e). As only 2±1% (mean±sd, n=3 rounds) of GM and 15±8% (mean±sd, n=3 rounds) of MegE marker onsets were annotated earlier than four generations after experiment start, our method is clearly superior to lineage identification based on traditional molecular markers in that particular time window (Supplementary Fig. 5c).
Figure 2. Subsets of cells with differential PU.1-eYFP expression can be distinguished two generations after experiment start.
(a) Increase of PU.1-eYFP concentration in a branch with annotated GM marker onset. (b) Decrease of PU.1-eYFP concentration in a branch with annotated MegE marker onset. Concentrations (black dots) are fitted with a B-spline (black/colored line). (c) Cells without marker expression (unknown or latent fate, black) show an intermediate PU.1-eYFP concentration compared to cells annotated for GM (blue) and MegE (red) lineage. (d) Concentration of PU.1-eYFP for unknown and latent cells in generations after experiment start, subdivided into predicted GM (blue) and MegE (red), based on CNN-RNN lineage score for round 1 (see Supplementary Fig. 4 for rounds 2 and 3). PU.1-eYFP concentration is significantly different in generation 2 (P<0.01) and 3-8 (P<0.001, unpaired wilcoxon rank-sum test) after experiment start between the two predicted groups. (e) Significantly different PU.1-eYFP expression in generation 2 after experiment start is detected in 55 MegE (red) vs. 34 GM (blue) predicted cells, respectively.
Different automated methods have been used for single-cell classification12–15. We compared the performance of our CNN-RNN to several other approaches. To this end, we trained a random forest model and a support vector machine (SVM) with a set of 87 morphological features and displacement. In addition, we evaluated the algorithmic information theoretic prediction (AITP)15, a method designed to predict the differentiation fate of retinal progenitor cells using a set of six movement and size features as well as a conditional random field (CRF) approach based on SIFT features13,16. Finally, we quantified the performance of two CNN models, where we averaged patch-wise lineage scores to obtain cell-specific predictions (Supplementary Fig. 6). We evaluated all methods in terms of AUC; in addition, to quantify performance in terms of precision and recall, we further compared the F1 scores of all methods. While our CNN-RNN outperformed the SVM on annotated cells and was on par on latent cells, we found the AUCs for random forest to be slightly higher on both sets (Supplementary Fig. 7a). However, the CNN-RNN achieved considerably higher F1 scores than the random forest, AITP and CRF-based approach (Supplementary Fig. 7b), indicating a poorer calibration of these methods. Using an RNN to model cell dynamics rather than simple averaging in the CNN approach yielded a slight but consistent increase in predictive power (in terms of F1 score). This suggests that the CNN-RNN yields more robust results when applied to new experiments that were not part of the training procedure.
The computation of hand-crafted features can be time-consuming and biased since appropriate features have to be chosen carefully for every new dataset. Instead, our CNN-RNN takes as input only raw brightfield image patches, rendering the explicit computation of features obsolete. Yet, knowing which interpretable features are most important for lineage prediction could support the design of novel experiments to study hematopoietic differentiation2. Since the features implicitly derived within the CNN are difficult to extract and interpret, we evaluated the feature importance reported by the trained random forest model17. We found that multiple features - most importantly displacement and simple morphological features (maximal/mean pixel intensity and cell size) - are required for correct random forest classification (Supplementary Fig. 8). To investigate the relevance of the displacement feature, we re-trained our CNN-RNN model omitting displacement, resulting in a somewhat lower predictive power for latent cells (from 0.79±0.02 to 0.76±0.04), illustrating that displacement is indeed used by the network. Moreover, we found slight differences in displacement and cell diameter for GM vs. MegE predicted cells (Supplementary Fig. 9).
Previously, computational image analysis has been used to predict the fate of rat retinal progenitor cells15 and to identify characteristic features of two populations of progenitor cells in the cerebral cortex18. In the hematopoietic system, long cell cycle times and trailing cellular projections19 as well as reduced proliferation and increased asynchronous divisions20 have been identified as key features of HSPC self-renewal via time-lapse microscopy. Our method allows to prospectively discriminate between two different lineages arising from hematopoietic progenitors and performs robustly on multiple independent time-lapse experiments. For a single cell, this prediction relies on a sequence of brightfield images and the combination of multiple features - single images and single features do not suffice. In future experimental applications, the brightfield-based prediction frees fluorescent channels that are currently used for lineage marker annotation. Moreover, the differential expression of PU.1-eYFP implies that our method can be used to identify important regulators of lineage choice when combined with single-cell profiling. While parameterizing deep neural networks requires large quantities of training data, the application of this approach matches the large amount of labeled image data that emerges from the diligent and careful annotation of time-lapse microscopy movies4,6 and can be used for training the networks. Compared to other machine learning methods, our CNN-RNN method predicts fast and robustly for new experiments not used for training. While it is independent of a cell-type specific, curated feature set and requires no high-level feature calculation, the interpretation of the derived features is challenging and an active field of research. In summary, our approach is versatile and well suited to analyze differentiation processes in biological systems where robustness is pivotal, suitable feature sets are unknown or fast prediction is required.
Online Methods
Generation of knock-in mice
The generation of knock-in mouse lines with reading frames for yellow (enhanced yellow fluorescent protein; eYFP) and red (mCherry) fluorescent proteins knocked into the gene loci for PU.1 and Gata1, respectively has previously been described6. The resulting PU.1eYFP and GATA1mCherry mice were mated to create PU.1eYFPGATA1mCherry mice with no discernible phenotype6.
Purification of primary murine hematopoietic stem and progenitor cells
Femurs, tibiae and ilia were removed from 12-14 weeks old mice and bone marrow was extracted. HSPCs were sorted to a technical purity of >95% and an expected functional purity of at least 40%-60% by flow cytometry21,22. Directly after sorting, cells were incubated with CD16/32 Alexa Fluor 647 antibody and seeded on a plastic slide (µ-slide VI coated with Fibronectin, Integrated BioDiagnostics GmbH, Munich, Germany) with physically separated channels in serum-free medium (StemSpan SFEM, StemCell Technologies) supplied with cytokines that only promote differentiation towards myeloid cells. Animal experiments were approved by veterinary office of Canton Basel-Stadt, Switzerland and Regierung von Oberbayern, Germany.
Long-term time-lapse microscopy data
For each experiment, channels of a plastic slide were subdivided into 72-78 overlapping fields of view. Each field of view corresponds to a 1388x1040 pixel image that was saved in 8-bit png format. Images were acquired using Axio Observer Z1 microscopes (Zeiss), equipped with a 0.63x TV-adapter (Zeiss), an AxioCamHRm camera (Zeiss) and a 10x fluar objective (Zeiss). Microscopes were surrounded by an incubator to keep a constant temperature of 37°C, cells were maintained in 5% CO2. Each field of view was imaged in intervals of 60-120 seconds (brightfield channel), 25-40 minutes (PU.1-eYFP and GATA1-mCherry channels) and 120-240 minutes (CD16/32 channel) for up to 8 days (Supplementary Fig. 1). Automatic focusing was achieved using a hardware autofocus (Zeiss), which was set to 18µm below the optimal focal plane to acquire slightly blurred images optimal for cell detection23.
Time-lapse experiments from PU.1eYFPGATA1mCherry mice (3 experiments, 150 genealogies, 5922 cells, 2477784 cell patches, Supplementary Fig. 2) and non-genetically modified C57BL/6J mice (1 experiment, 29 genealogies, 266 cells, 157384 cell patches) were used in this study, comprising a total size of ~1TB of disc space.
Single-cell tracking and annotation of lineage commitment
Single cells and their progeny were manually followed over time using the tTt software24 (Supplementary Fig. 1,2 and Supplementary Video 1). Next, we combine automated segmentation with cell tracks by linking image patches centered on the cell’s center of mass to the nearest track coordinate. To that end, we extended our previously developed automated image processing pipeline that identifies and segments single cells with high accuracy in time-lapse brightfield microscopy25 (Supplementary Video 2). For a detailed description of tracking, identification and segmentation, see Supplementary Notes and Supplementary Fig. 10.
Lineage commitment was initially annotated by experts by visually inspecting the fluorescence signal of CD16/32 (for the GM lineage) and GATA1-mCherry (for the MegE lineage). We amended these annotations by automatically quantifying the concentration of CD16/32 and GATA1-mCherry using a self-written user interface (see Supplementary Fig. 11). Galleries of cells with marker expression are shown in Supplementary Fig. 12. For the genetically non-modified mice, we used CD16/32 (for the GM lineage) and a large morphology (for the MegE lineage due to the absence of GATA1-mCherry).
Deep neural networks
We combined a convolutional neural network (CNN) for automatically extracting shape-based features with a recurrent neural network (RNN) architecture, modeling the dynamics of the cells.
For the CNN, we extended a model from the LeNet family26 by combining three convolutional layers (20 filters with kernel size 5, 60 filters with kernel size 4, and 100 filters with kernel size 3, respectively) with two fully connected layers (500 and 50 nodes, respectively). Each convolutional layer is followed by a non-linear activation function. We chose Rectified Linear Units (ReLU), which have been shown to introduce non-linearities without suffering from the vanishing gradient problem27. In addition, we used max-pooling layers reducing variance and increasing translational invariance by computing the maximum value of a feature over a region28 and dropout layers following the fully connected layer to avoid overfitting. Here, we follow largely Ciresan et al.29, where it is shown that this combination of layers results in fast training times and good performance on a variety of image classification data sets. We further use the recently proposed batch normalization strategy to normalize outputs after each layer30. Finally, in order to be able to account for non-image-based features, we introduce a concatenation layer that combines spatial features with cell displacement (Fig. 1d).
We used a softmax loss function and trained the network using stochastic gradient descend with stratified batches of 128 images. We initialized all weights in the network using the Xavier algorithm, which automatically determines the scale of the initialization based on the number of input- and output nodes31. We used standard values for the base learning rate (0.01), momentum (0.9) and the learning rate policy (stepwise policy decreasing the learning rate every 10,000 iterations32).
We then passed the output of the first fully connected layer together with the displacement feature (which we interpret as patch features extracted by the CNN) to a bidirectional long-short-term memory (LSTM) recurrent neural network (Fig. 1e). Specifically, we trained a LSTM RNN with 20 hidden nodes using the rprop algorithm33 with cross-entropy loss function, taking the mean across all timepoints. We trained the RNN for up to 15 epochs with standard positive update parameter of 1.2 and negative update parameter of 0.5.
In order to avoid over-fitting we divided the training data into a training and validation set and optimized the weights, both of the CNN and the LSTM RNN, until the performance on the validation set started to degrade (early stopping). All images were normalized to mean zero and unit variance, to normalize for possible batch effects.
Quantification of morphodynamics and fluorescence signals
On every extracted image patch (27x27 px around the cell’s center of mass), 87 features (14 basic measurements as provided by MATLAB regionprops method, 27 Zernike moments34, 3 Ray features35, 13 Haralick texture features36, 2 Gabor wavelet features37, 5 Tamura features38, Histograms of oriented Gradients39 with 27 bins) were computed. If a fluorescence image was available, the fluorescence concentration was quantified by summing up all pixel within the segmented cell in the background corrected fluorescence image and dividing by cell size. Quantification errors (clumped cells, dirt, falsely identified as cell, cells lost due to border contact, over-segmented cell fragments) were detected by fitting a B-spline to the cell size over time. We then discarded those time points with residue differences beyond the 98th or below the 2nd percentile of all time points. We computed cell displacement si,t for cell i at time point t as the root mean squared displacement between the frame t and t-1 divided by the time difference between the frames,
where x and y are the spatial coordinates and Tt is the absolute time after experiment start for frame t. We computed s(x,y) for all pairs of adjacent frames for every cell, using the track coordinates for the full cell trajectory.
Feature-based classification
A random forest classifier was trained with 200 trees (default parameters) and evaluated by out-of-bag prediction. We chose BudgetedSVMs40 with the Pegasos algorithm and a radial basis function kernel as a support vector machine (SVM) framework that was able to deal with the millions of single image patches in our dataset. We used a grid search with 5-fold cross-validation for every train-test combination to determine optimal hyperparameters. The best-performing model was then used for predicting the testset. Note that the hyperparameters for SVM had to be determined for every train-test run individually. We trained both methods with a set of 87 morphological features and cell displacement (see above). We applied the same train-test procedure for model evaluation as for the CNN.
AITP was trained as described15, using a set of 6 features for each cell (movement, net movement, movement direction, area and eccentricity of fitted convex hull). As AITP was not able to process the full dataset in a single run, we generated three subsets (n=400 cells per subset) for every train-test round, which we evaluated separately. We used the averaged evaluation results for comparison. As the used version of AITP reported class labels and no prediction scores, we used the macro-averaged F1 score for performance evaluation. It is worth noting that in contrast to all other methods, AITP inherently uses the full cell trajectory for training and prediction.
Recently, conditional random field (CRF) based models have been proposed for sequence labeling in the context of mitosis sequence detection13,14. We adapted the approach of Liu et al.13 and trained a CRF using SIFT features for our cells. Feature extraction was performed based on VLFeat, an open implementation of SIFT features41. The CRF was implemented using the pystruct library42.
Evaluation of model performance
Performance of the trained classification models was determined by receiver-operator characteristics and macro-averaged F1 score.
The receiver-operator characteristic is a function that evaluates the change in true positive (TP) rate with respect to the false positive (FP) rate of a predicted class label in accordance to all possible thresholds of a classification score that can be interpreted as probabilities (as is the case for random forest, SVM and CNN). The area under the curve (AUC) falls in the interval of [0,1] (1 = perfect classification, 0.5 = random guessing) and gives an impression of the general performance of the classifier.
The F1 score combines precision and recall in a single score
where FN is the false negative rate. A perfect classifier would reach a score of 1 and a random classifier would reach a score of 0.5. To account for the classification performance of both classes, the F1 score can be calculated for each class and then averaged resulting in the macro-averaged F1 score
for all class labels C. To determine class membership (i.e. commitment of a cell to GM or MegE lineage) a threshold of 0.5 for the cell lineage score was used for all models.
Implementation
Single-cell identification and quantification was implemented using MATLAB (R2014a). Code from43 was used to compute histograms of oriented gradients. All quantifications were parallelized on single-cell level and processed on a computation cluster (sun grid engine version 6.2u5). The average node architecture was equal to an Intel Xeon 2GHz, 4GB RAM running a 64bit linux-based operating system. Random forest classification was conducted with the python-based scikit-learn package (v0.15). The support vector machine was trained using the code provided with the original publication40. AITP was trained using the latest version (April 1st, 2014) from the website of the authors after slight adaptation of input/output functionality to fit our data. To implement the convolutional neural network, we used the caffe framework32 and trained it on a standard PC equipped with an Intel Core i7-4770 CPU, 32GB working memory and a 6GB Geforce GTX Titan Black graphics card. The recurrent neural network was implemented in Theano44 and trained on that same machine. SIFT features were calculated using VLFeat41, and the CRF was implemented using the pystruct library42.
Supplementary Material
Acknowledgements
We thank Sebastian Pölsterl for comments on the manuscript. This work was supported by the German Federal Ministry of Education and Research (BMBF), the European Research Council starting grant (Latent Causes grant 259294, FJT), the BioSysNet (Bavarian Research Network for Molecular Biosystems, FJT), the German Research Foundation (DFG) within the SPPs 1395 and 1356 to FJT, the UK Medical Research Council (Career Development Fellowship MR/M01536X/1 to FB), and the Swiss National Science Foundation grant 31003A_156431 and SystemsX IPhD grant 2014/244 to TS.
Footnotes
Data and Code Availability
Data and code for cell detection and neural network training and cell fate prediction is available via github.com/QSCD/HematoFatePrediction.
Author Contributions
FBU developed the image processing and machine learning pipeline, tracked cells and analyzed the data. FBT developed the deep neural network approach with MK. MST and MSC contributed to image processing and data analysis. PSH developed and conducted all experiments and tracked cells with ME, TS developed and supervised data generation. DL, KDK, and OH contributed to data generation and analysis. FJT and TS initiated the study. CM supervised the study with FJT, and wrote the manuscript with FBU and FBT. All authors commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Skylaki S, Hilsenbeck O, Schroeder T. Challenges in long-term imaging and quantification of single cell dynamics. Nature Biotechnology. 2016;34:1137–1144. doi: 10.1038/nbt.3713. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder T. Long-term single-cell imaging of mammalian stem cells. Nat Methods. 2011;8:S30–S35. doi: 10.1038/nmeth.1577. [DOI] [PubMed] [Google Scholar]
- 3.Rieger MA, Schroeder T. Exploring hematopoiesis at single cell resolution. Cells Tissues Organs. 2008;188:139–149. doi: 10.1159/000114540. [DOI] [PubMed] [Google Scholar]
- 4.Filipczyk A, et al. Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat Cell Biol. 2015;17:1235–1246. doi: 10.1038/ncb3237. [DOI] [PubMed] [Google Scholar]
- 5.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- 6.Hoppe PS, et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535:299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- 7.Bengio Y, Simard P, Frasconi P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw. 1994;5:157–166. doi: 10.1109/72.279181. [DOI] [PubMed] [Google Scholar]
- 8.Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005;18:602–610. doi: 10.1016/j.neunet.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 9.Graves A, Jaitly N, Mohamed Ar. Hybrid speech recognition with Deep Bidirectional LSTM. Automatic Speech Recognition and Understanding (ASRU), 2013 IEEE Workshop on; 2013. pp. 273–278. [Google Scholar]
- 10.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 11.Hoppe PS, Coutu DL, Schroeder T. Single-cell technologies sharpen up mammalian stem cell research. Nat Cell Biol. 2014;16:919–927. doi: 10.1038/ncb3042. [DOI] [PubMed] [Google Scholar]
- 12.Veta M, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–248. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Liu A-A, Li K, Kanade T. A semi-Markov model for mitosis segmentation in time-lapse phase contrast microscopy image sequences of stem cell populations. IEEE Trans Med Imaging. 2012;31:359–369. doi: 10.1109/TMI.2011.2169495. [DOI] [PubMed] [Google Scholar]
- 14.Huh S, Ker DFE, Bise R, Chen M, Kanade T. Automated Mitosis Detection of Stem Cell Populations in Phase-Contrast Microscopy Images. IEEE Trans Med Imaging. 30:586–596. doi: 10.1109/TMI.2010.2089384. [DOI] [PubMed] [Google Scholar]
- 15.Cohen AR, Gomes FLAF, Roysam B, Cayouette M. Computational prediction of neural progenitor cell fates. Nat Methods. 2010;7:213–218. doi: 10.1038/nmeth.1424. [DOI] [PubMed] [Google Scholar]
- 16.Liu A-A, Li K, Kanade T. Mitosis sequence detection using hidden conditional random fields. 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro; IEEE; pp. 580–583. [Google Scholar]
- 17.Breiman L. Random Forests. Mach Learn. 2001:5–32. [Google Scholar]
- 18.Winter MR, et al. Computational Image Analysis Reveals Intrinsic Multigenerational Differences between Anterior and Posterior Cerebral Cortex Neural Progenitor Cells. Stem Cell Reports. 2015;5:609–620. doi: 10.1016/j.stemcr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dykstra B, et al. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci U S A. 2006;103:8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutolf MP, Doyonnas R, Havenstrite K, Koleckar K, Blau HM. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr Biol. 2009;1:59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osawa M, Hanada K-I, Hamada H, Nakauchi H. Long-Term Lymphohematopoietic Reconstitution by a Single CD34-Low/Negative Hematopoietic Stem Cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 22.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Selinummi J, et al. Bright field microscopy as an alternative to whole cell fluorescence in automated analysis of macrophage images. PLoS One. 2009;4:e7497. doi: 10.1371/journal.pone.0007497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilsenbeck O, et al. Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat Biotechnol. 2016;34:703–706. doi: 10.1038/nbt.3626. [DOI] [PubMed] [Google Scholar]
- 25.Buggenthin F, et al. An automatic method for robust and fast cell detection in bright field images from high-throughput microscopy. BMC Bioinformatics. 2013;14:297. doi: 10.1186/1471-2105-14-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278–2324. [Google Scholar]
- 27.Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. Proceedings of the 27th International Conference on Machine Learning (ICML-10); 2010. pp. 807–814. [Google Scholar]
- 28.Ranzato M, Huang FJ, Boureau Y-L, LeCun Y. Unsupervised Learning of Invariant Feature Hierarchies with Applications to Object Recognition. Computer Vision and Pattern Recognition, 2007. CVPR ’07 IEEE Conference on; 2007. pp. 1–8. [Google Scholar]
- 29.Ciresan DC, Meier U, Masci J, Maria Gambardella L, Schmidhuber J. Flexible, high performance convolutional neural networks for image classification. IJCAI Proceedings-International Joint Conference on Artificial Intelligence 22, 1237; people.idsia.ch; 2011. [Google Scholar]
- 30.Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv [cs.LG] 2015 [Google Scholar]
- 31.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. International conference on artificial intelligence and statistics; machinelearning.wustl.edu; 2010. pp. 249–256. [Google Scholar]
- 32.Jia Y, et al. Caffe: Convolutional Architecture for Fast Feature Embedding. Proceedings of the 22Nd ACM International Conference on Multimedia; ACM; 2014. pp. 675–678. [Google Scholar]
- 33.Braun H, Riedmiller M. RPROP: a fast adaptive learning algorithm. Proceedings of the International Symposium on Computer and Information Science VII; 1992. [Google Scholar]
- 34.Zernike F. Diffraction theory of the cut procedure and its improved form, the phase contrast method. Physica. 1934;1:56. [Google Scholar]
- 35.Smith K, Carleton A, Lepetit V. Fast Ray Features for Learning Irregular Shapes. Proceedings of the International Conference on Computer Vision (ICCV); 2009. [Google Scholar]
- 36.Haralick RM, Dinstein, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3. 1973:610–621. [Google Scholar]
- 37.Gabor D. Theory of communication. Part 1: The analysis of information. Electrical Engineers - Part III: Radio and Communication Engineering. Journal of the Institution. 1946;93:429–441. [Google Scholar]
- 38.Tamura H, Mori S, Yamawaki T. Textural Features Corresponding to Visual Perception. IEEE Trans Syst Man Cybern. 1978;8:460–473. [Google Scholar]
- 39.Dalal N, Triggs W. Histograms of Oriented Gradients for Human Detection. 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition CVPR05; 2004. pp. 886–893. [Google Scholar]
- 40.Djuric N, Lan L, Vucetic S, Wang Z. BudgetedSVM: a toolbox for scalable SVM approximations. J Mach Learn Res. 2013;14:3813–3817. [Google Scholar]
- 41.Vedaldi A, Fulkerson B. VLFeat: An Open and Portable Library of Computer Vision Algorithms. 2008 [Google Scholar]
- 42.Müller AC, Behnke S. PyStruct: learning structured prediction in python. J Mach Learn Res. 2014;15:2055–2060. [Google Scholar]
- 43.Junior OL, Delgado D, Goncalves V, Nunes U, Ludwig O. Trainable classifier-fusion schemes: An application to pedestrian detection. 2009 12th International IEEE Conference on Intelligent Transportation Systems; IEEE; 2009. pp. 1–6. [Google Scholar]
- 44.The Theano Development Team et al. Theano: A Python framework for fast computation of mathematical expressions. arXiv [cs.SC] 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.