Abstract
Single cell RNA sequencing (scRNA-seq) has become an established and powerful method to investigate transcriptomic cell-to-cell variation, revealing new cell types, and providing insights into developmental processes and transcriptional stochasticity. The array of published scRNA-seq protocols allow one to sequence transcriptomes from minute amounts of starting material. A key question is how these various protocols compare in terms of sensitivity of detection of mRNA molecules, and accuracy of quantification of expression. Here, we present an assessment of sensitivity and accuracy of many published data sets by spike-in standards with uniform data processing, including development of a flexible Unique Molecular Identifier (UMI) counting tool (https://github.com/vals/umis). We computationally compare 15 protocols, and experimentally assess 4 protocols on batch-matched cell populations, as well as investigating the impact of spike-in molecule degradation on two types of spike-ins. Our analysis provides an integrated framework for comparing different scRNA-seq protocols.
Introduction
Recently, there has been an explosion in the development of protocols for RNA-sequencing of individual cells (scRNA-seq)1,2, with different approaches to capture cells, amplify cDNA, minimise biases, and utilise liquid handling platforms. Due to the tiny amount of starting material, considerable amplification is an integral step of all of these protocols. Consequently, it is important to assess the sensitivity and accuracy of the protocols in terms of numbers of RNA molecules detected. An objective strategy to assess the technical variability in these methods is to add exogenous spike-in RNA of known abundances to the individual cell samples. In previous studies, biological features were compared on a limited number of protocols to assess performance3,4. In this study, we assessed the performance of a large number of published scRNA-seq protocols according to their ability to quantify the expression of spike-ins of known concentrations. An ideal protocol is both sensitive and accurate, as well as cost effective, where cost is at least partially reflected in sequencing depth.
We define sensitivity of a method as the minimum number of input RNA molecules required for a spike-in to be detected as expressed, and accuracy as the closeness of estimated relative abundances to ground truth (a priori known abundances of input molecules). High sensitivity permits detection of very lowly expressed genes, while high accuracy implies that detected expression variations reflect true biological differences in mRNA abundance across cells, rather than technical factors.
The standardized ERCC (External RNA Controls Consortium)5 spike-in collections consist of a set of 92 RNA molecule species of varying lengths and GC contents, mixed at known concentrations, and represent 22 abundance levels that are spaced one fold change apart from each other (Supplementary Figure 1). Previously, such spike-ins have been applied to assess standard RNA-sequencing protocol reproducibility6 and performance of differential expression tests in RNA-sequencing data7. In the context of single cell RNA-sequencing protocols, ERCC spike-ins were first published as part of the description of the CEL-seq protocol8.
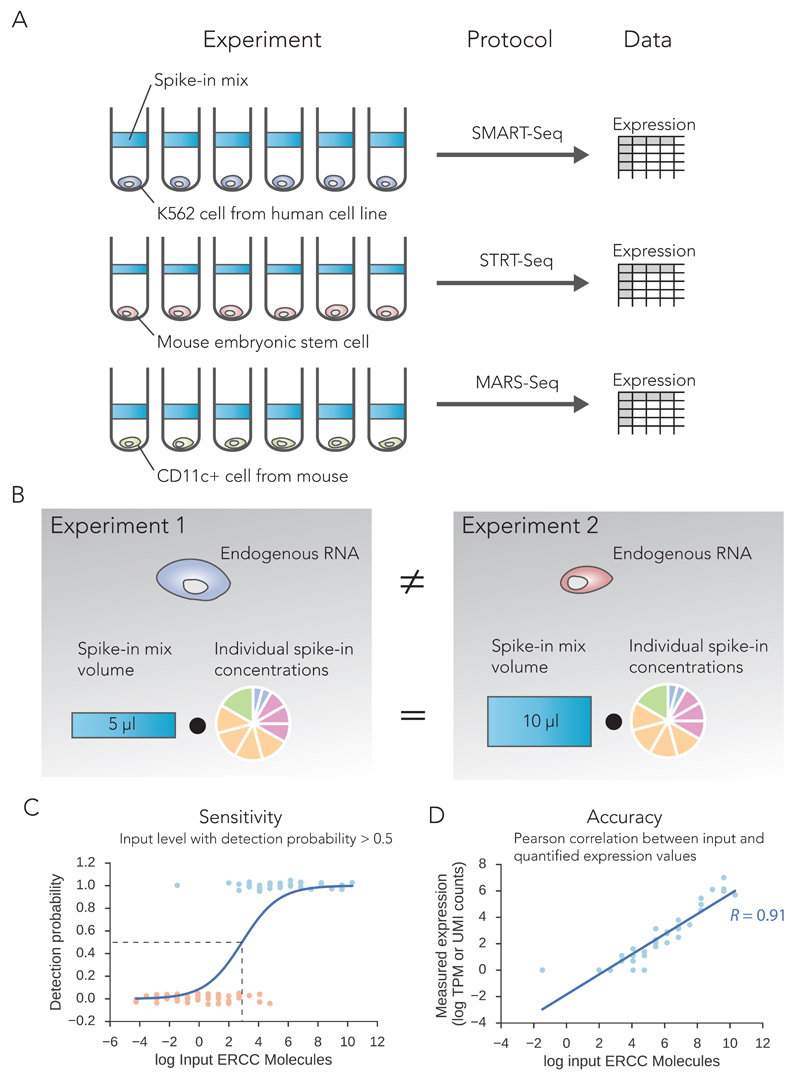
Here, we exploit spike-ins as a means to calculate technical sensitivity and accuracy of different scRNA-seq protocol across various platforms in a comparable manner, independent of the biological cell type investigated (Figure 1A-B). Leveraging the known number of input spike-in molecules allows calculation of the lower molecular detection limit of each sample in each experiment (Figure 1C). By comparing to overall sequencing depth, we ascertain sensitivity of the protocols. The spike-ins also provide a direct way to assess accuracy by comparing a priori input concentration to the measured expression levels by sequencing (Figure 1D). Thus we obtain a unified framework for comparing sensitivities and accuracies of the various protocols at different sequencing depths.
Figure 1. Illustration of protocol comparison strategy.
(A) Schematic illustration highlighting our spike-in comparison strategy of public data sets. (B) The different protocols are compared based on the same standard spike-in molecules and not the endogenous mRNA from the diverse cell types used in these studies. We define two global technical performance metrics based on spike-ins: (C) Sensitivity: the number of input spike-in molecules at the point where the probability of detection reaches 50%. (D) Accuracy: the Pearson product-moment correlation (R) between estimated expression levels and actual input RNA molecule concentration (ground truth).
Our analysis leveraging spike-in information from published datasets is subject to limitations (explored in depth in the Discussions). We rely on accurate reporting of spike-in volumes and dilution by original authors, and have reconfirmed in few cases (personal communication). In addition, spike-in molecules may not truly reflect endogenous mRNA capture efficiency in scRNA-seq owing to deviation from natural mRNA sequence features such as shorter polyA tails and mRNA binding proteins. Nevertheless, the scale and number of different protocols and platforms with published spike-in data provide a rich resource for mining their comparative performance. In our comprehensive analysis, most protocols are replicated across at least two different cell types and different laboratories (Supplementary Table 1). This reduces potential bias due to a specific cell type or study.
Results
We retrieved and analyzed published scRNA-seq data sets that used ERCC spike-ins for quality control or normalization, and compared both sensitivity and accuracy performance metrics. Our analysis spans 15 distinct experimental protocols encompassing 28 single-cell studies, including 17 studies with traditional whole-transcript coverage based strategy for measuring expression levels and 11 that used strategies based on UMI’s for digital quantification of transcripts (Supplementary Table 1). We additionally performed 4 different scRNA-seq protocols across 3 platforms using dedicated batch-matched mouse embryonic stem cells (mESC) across two replicates, leveraging both ERCC and Spike-in RNA Variant (SIRV) spike-ins. We performed SMARTer and Smart-Seq2 protocols in the first replicate, and performed SMARTer, Smart-Seq2 and STRT-Seq in the second replicate on the Fluidigm C1 platform. We also include a high throughput droplet based 10X Genomics Chromium dataset using ERCC spike-ins and Human Brain Total RNA. All coverage-based datasets have been sequenced using Illumina paired-end sequencing, with read lengths between 75 and 150 base pairs. In total, our analysis consists of 18,123 publicly available samples of 30x109 sequencing reads.
For each data set, the concentration of spike-ins was noted either through the original study or by personal communication with the original authors, including relevant dilution and volumes (Supplementary Table 1). This allowed us to calculate the absolute number of spike-in RNA molecules at different abundance levels across individual cell samples. Thus, permitting comparison of the different experimental data sets on the same scale.
Technical accuracy of quantification of different scRNA-seq protocols
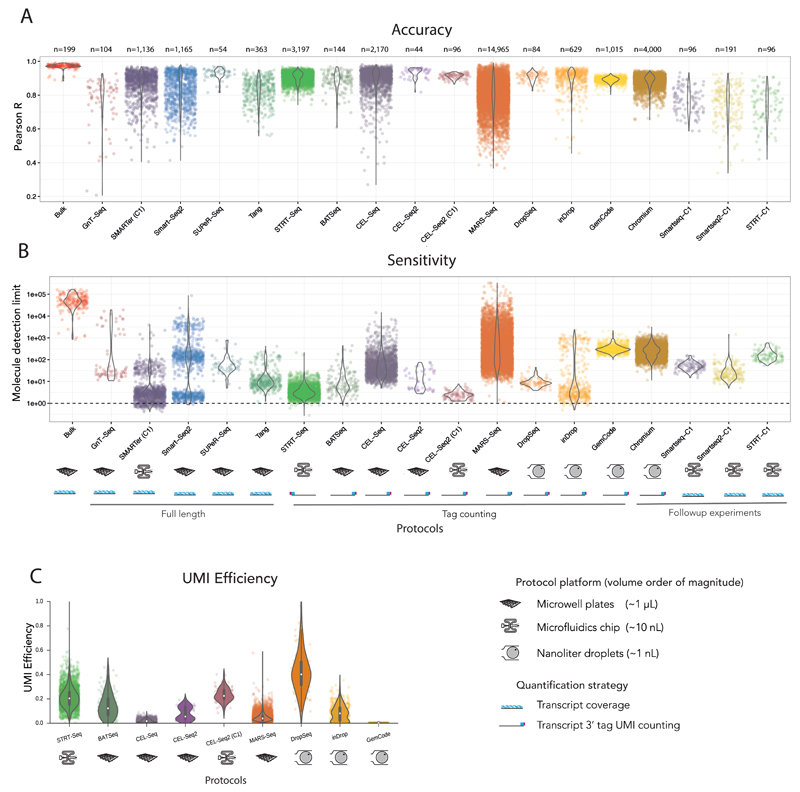
To effectively assess the accuracy of quantification by different protocols, we first used 92 ERCC spike-in RNAs spanning 22 abundance levels with two-fold difference between each level. The Pearson correlation between estimated expression levels and actual input RNA molecule concentration (ground truth) provides a direct measure for quantifying accuracy (Figure 1D). We computed the Pearson product-moment correlation coefficient (R) between log transformed values for estimated expression and input concentration for each individual cell (or sample) and compared these values across all protocols (Figure 2A).
Figure 2. Comparison of performance metrics for different protocols.
(A) Accuracy. Distributions of Pearson correlations (R) of all samples stratified by protocol investigated (without accounting for sequencing depth). (B) Sensitivity. Distributions of molecule detection limits of all samples stratified by protocol (without accounting for sequencing depth). The number of samples (n) is highlighted above the protocols. The implementation platforms and quantification strategies are indicated below the protocols. (C) UMI Efficiency. Distributions of UMI counting efficiencies in UMI-tag counting based samples, stratified by protocols. For the analysed UMI based data, the UMI efficiency recapitulates the logistic regression based molecule detection limit.
The accuracy of conventional bulk RNA-sequencing is higher than scRNA-seq protocols. Remarkably, the accuracy of scRNA-seq protocols is still high, and rarely do individual samples have a Pearson correlation lower than 0.6. The lower accuracy and variable Pearson correlations for individual cells (samples) within protocols (GnT-Seq, CEL-Seq, MARS-Seq), might be indicative of variable success rates of protocols.
Technical sensitivity of different scRNA-seq protocols
To investigate the technical sensitivity of each sample and quantify inter-sample variability, we devise a logistic regression model for detection, with detection of expression as the dependant variable. Our measure of sensitivity is the molecular spike-in input level where probability of detection reaches 50% (Figure 1C). This allows us to measure sensitivity of each sample individually for every protocol investigated, avoiding biases due to uneven batch sizes. Our approach avoids using detected spike-ins ratios at each abundance level which would give poor resolution, because at most seven spike-ins share one abundance level.
Applying logistic regression to all samples, we observe that scRNA-seq protocols are more sensitive than bulk RNA-sequencing and can detect very low numbers of input molecules (Figure 2B). The sensitivity of scRNA-seq protocols varied over four orders of magnitude, and several protocols have the potential to detect as little as single digit input spike-in molecules (SMARTer (C1), CEL-Seq2 (C1), STRT-Seq, and inDrop). We observe high within-protocol variability of sensitivity for protocols. This can be attributed to sequencing depth, which we quantify in a section below to rank the protocols.
UMI efficiency of tag-counting protocols
The majority of single cell RNA-sequencing protocols utilise a UMI-tag counting strategy to achieve digital quantification of mRNA transcripts. In this strategy, a single tag from a single mRNA molecule is reverse transcribed, and an additional probabilistically unique random identifier sequence is added. These protocols create cDNA libraries with extremely low complexity, which may lead to extreme amplification biases. The UMI on each tag should allow one to remove these biases, as it is added prior to amplification9. The question then remains as to how efficient the entire scRNA-seq process is.
If E is the UMI (counting) efficiency, the underlying assumption is that the number of UMIs of a gene U = E·M, where 0 < E < 1 (Supplementary Figure 2A) and M is the number of RNA molecules of a gene. We fitted this model for every UMI-tag sample and stratified the results across different protocols (Figure 2C). The results recapitulate the logistic regression based measure for sensitivity, as samples with high efficiency have a low molecule detection limit (with the exception of MARS-Seq data, Supplementary Figure 2B).
However, more in depth analysis shows this measure might not be as appropriate as it appears. If we extend the model to be U = E·Mc, the best fit should give values of the molecular exponent c close to 1, if the underlying UMI counting assumption is correct. Instead, we find that the best fit is systematically lower than 1, with a mode of around 0.8 (Supplementary Figure 2C). This implies a saturation of UMI counts as a function of input molecules. This can be explained partially (but not fully) by differences in UMI length between the different protocols (Supplementary Figure 2D). For example, UMIs with length of 4 base pairs can only count up to 256 unique molecules, and have on average a molecular exponent of 0.6. However, even in protocols with longer UMI’s of 10 base pairs (which can count over a million unique molecules), the molecular exponent is still 0.8 per sample on average, and rarely reaches 1.
In conclusion, while UMIs should provide a way of removing amplification biases, the assumed absolute quantification does not seem to hold perfectly.
Endogenous transcripts are more efficiently captured than ERCC spike-ins
It is unclear to what extent the sensitivity and accuracy calculated based on exogenous spike-ins corresponds to the same metrics for endogenous mRNA. On the one hand, ERCC spike-ins have shorter poly(A) tails than typical mRNA from mammalian cells10, making them harder to capture by poly(T) priming. On the other hand, endogenous mRNA have intricate secondary structures and can be bound to proteins, potentially reducing the efficiency of reverse transcription.
To investigate the relationship between scRNA-seq measurements of endogenous mRNA and ERCC spike-ins, we analysed single molecule fluorescent in-situ hybridization (smFISH) data and CEL-seq data from the same mESC line and culture conditions11 (molecule counts from personal communication). Based on smFISH data for 9 endogenous genes, CEL-Seq UMI counts correspond to 5-10% of smFISH counts. This clearly contrasts with the ERCC spike-ins in the same samples, where the average UMI counts correspond to 0.5-1% of input molecule counts (Supplementary Figure 2E).
This data suggests that endogenous RNA is much more efficiently captured and amplified than ERCC spike-in molecules. Overall, this comparison implies that our sensitivity estimates are conservative, and are likely to underestimate rather than overestimate. The accuracy metric is based on relative abundances, and is not affected by this. This difference in efficiency is important to consider if absolute molecule counts are to be inferred based on ERCC spike-ins.
Sensitivity is more dependent on sequencing depth than accuracy
The results of the per-sample accuracy and sensitivity analysis shows a large amount of within-protocol heterogeneity (Figure 2A-B). Seeking to explain performance by technical factors, we find a relation with sequencing depth per sample. This becomes especially relevant from a cost perspective for scRNA-seq experiments, since researchers can control read depth to fit their budgets and needs. We used a linear model that considers a global effect of sequencing depth, including diminishing returns (see Online Methods). The model includes an individual corrected performance parameter for each protocol, which allows protocols to be ranked while accounting for the substantial technical factor of sequencing depth.
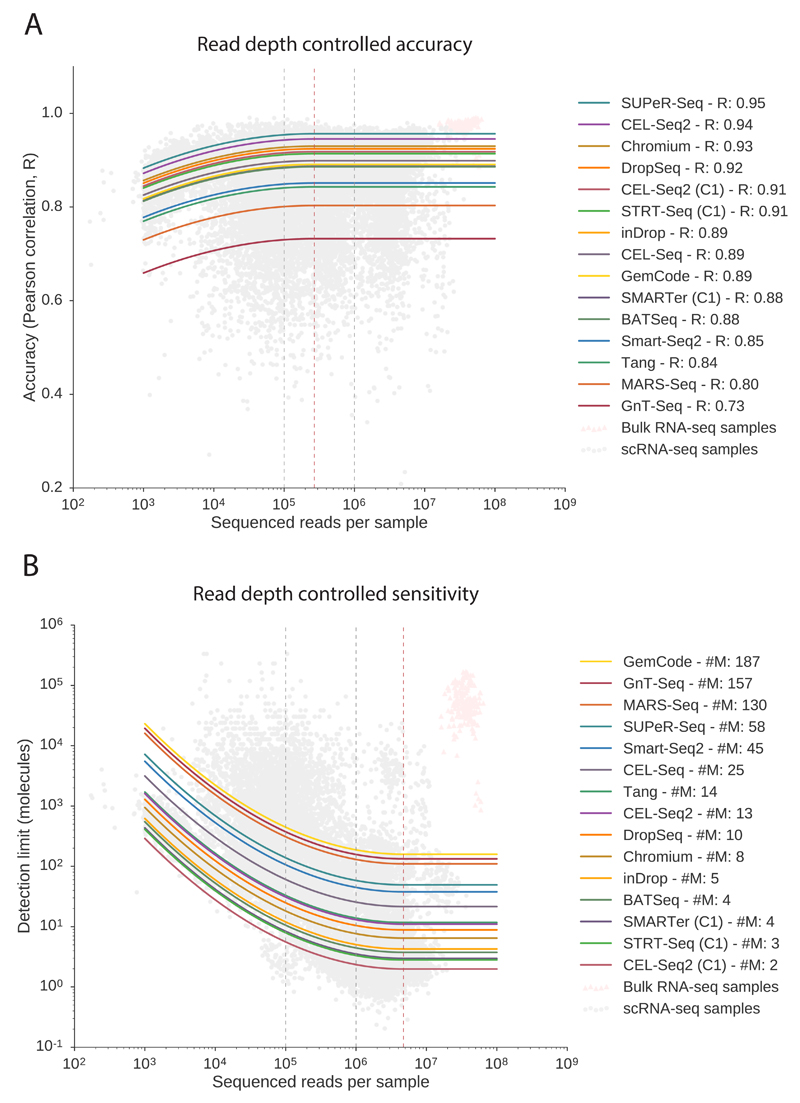
We find that accuracy does not strongly depend on sequencing depth, supporting the theory that heterogeneity of accuracy within a protocol relates to the general reliability of a protocol (Figure 3A). We found that the best performing protocols in terms of accuracy are SUPeR-Seq (R=0.95), the only total-RNA protocol for single cells to date, and CEL-Seq2 (R=0.94), which uses In Vitro Transcription (IVT) rather than PCR to amplify cDNA.
Figure 3. Performance metrics after accounting for sequencing depth.
Both accuracy and sensitivity are modeled with a global dependency on sequencing depth considering diminishing returns, with a distinct corrected performance parameter for each protocol. Both models (one for accuracy and one for sensitivity) has 26 parameters, and are fitted to n=20,717 observations (the number of samples). (Bulk data in pink triangles are only displayed for context). Solid curves show the predicted dependence on sequencing depth. (A) Accuracy is only marginally dependent on sequencing depth. Saturation occurs at 270,000 reads per cell in the model (dashed red line). Protocol names are ordered by performance based on predicted correlation R at 1 million reads. (B) Sensitivity is critically dependent on sequencing depth and accounting for read depth is key for fair comparison of protocols. Saturation identified by the model occurs at 4.6 million reads per cell (dashed red line). The gain from 1 to 4 million reads per sample is marginal, while we note that moving from 100,000 reads to 1 millions reads corresponds to an order of magnitude gain in sensitivity (dashed black lines). Protocols are ordered by performance based on predicted detection limit (#M, number of molecules at 1 million reads).
Since the model considers diminishing returns on the sequencing depth, we can identify when the performance metrics saturate from the model parameters. We find that accuracy becomes saturated at a depth as low as 250,000 reads, illustrating that it is not strongly dependent on sequencing depth.This also suggests that for detected mRNAs in scRNA-seq, the expression levels are generally accurate and quantitatively meaningful.
We find that the technical sensitivity is critically dependent on sequencing depth, and sensitivity comparisons without accounting for differences in depth would be misleading (Figure 3B). The sensitivity parameter of the model accounts for sequencing depth to allow for fair comparison, and we used this to rank protocols. The three protocols implemented in a C1 microfluidics system (CEL-Seq2 (C1), STRT-Seq (C1), and SMARTer (C1); number of molecules at a million reads,#M = 2,3,and4respectively) were the top performing protocols in terms of molecule detection. The matched microwell plate implementation of CEL-Seq2 has poorer sensitivity than the C1 implementation (#M = 13).
Based on the model, we find that sensitivity saturates at about 4.5 million reads per sample. The increase in read depth from 1 million reads to 4.5 million reads per sample results in marginally increased sensitivity; less than a fold change. However the increase from 100,000 reads to 1 million reads per sample results in increased sensitivity of an order of magnitude. Thus, we recommend considering 1 million reads per sample as a good target for saturated gene detection.
It should be noted that not all studies need to saturate detection, especially in cases where the genes of interest are highly expressed. It is equally important to note that sequencing depth is a technical feature, and the number of genes detected depend on the depth. Therefore, sequencing depth must be taken into account when performing and computationally analyzing scRNA-seq, even for compositional expression units such as TPM (Transcripts Per Million).
Degradation of spike-ins does not explain performance variation between experiments
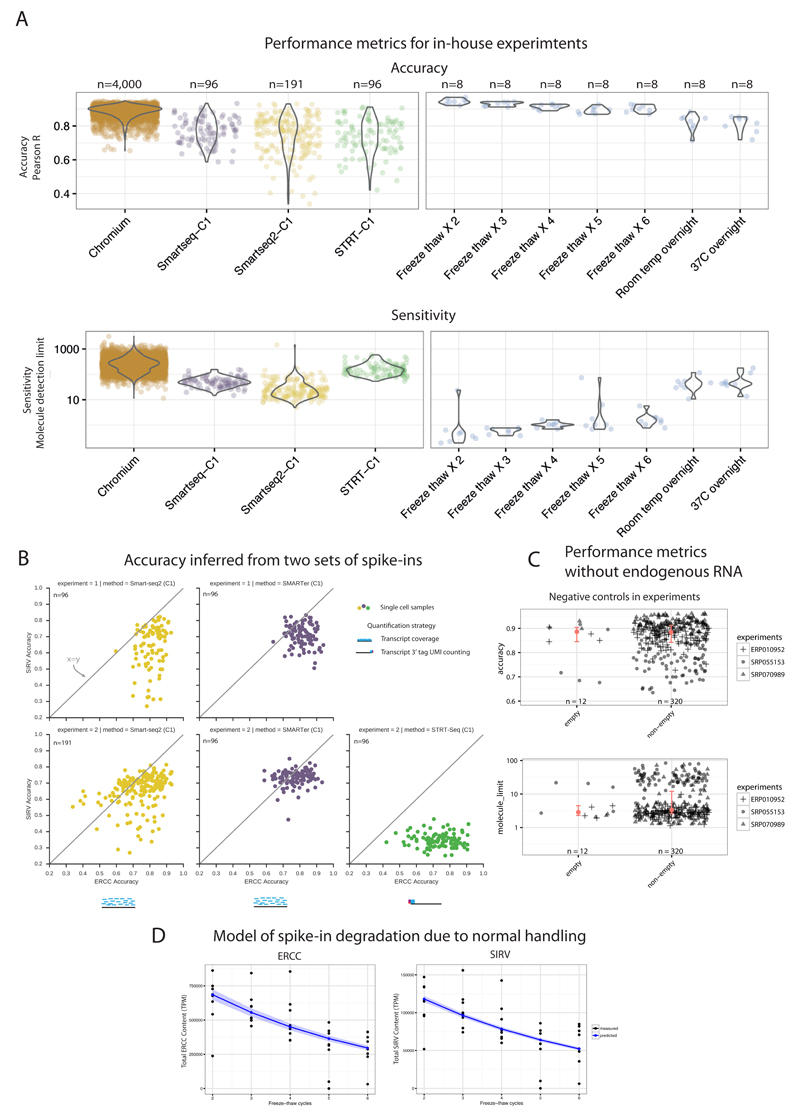
The analyzed performance metrics of the scRNA-seq protocols inherently assumes the gold standard annotation of the spike-ins to be correct. However, due to the labile nature of RNA, it can get degraded during the course of normal reagent handling. To quantify the impact of degradation, we subjected spike-in molecules (both ERCCs and SIRVs) to repeated freeze-thaw cycles (Online Methods). Additionally, as a measure of complete/full degradation, we left the spike-ins either at room temperature or at 37°C overnight. The freeze-thaw cycles emulate normal handling and comparing these to our in-house protocols, we observed that accuracy metric had an overall small effect and was similar between protocols (Figure 4A).
Figure 4. Investigation of factors with potential impact on performance metrics.
(A) Batch effects and RNA degradation. Performance distributions for three protocols implemented as a single batch, on the Fluidigm C1 platform (left) and on the 10x Chromium (far-left; different batch). Performance distributions of spike-ins measured after freeze-thaw cycles, normal (2-3 cycles) to critical degradation (6 cycles, left overnight at room temperature). (B) Accuracy estimates across both ERCC and SIRV spike-ins are similar. Accuracy (Pearson correlation R) of both spike-ins (ERCC’s and SIRV’s) inferred across two replicates using multiple protocols. (C) Endogenous mRNA amount does not affect performance metrics. Comparison of performance metrics between empty (lacking endogenous mRNA) and non-empty samples from 3 published datasets shows similar performance and no bias due to presence of endogenous mRNA. Red dot shows median, and red bar shows 95% confidence interval of median, estimated with bootstraps. Empty accuracy CI is 100% contained in non-empty CI, and empty sensitivity CI is 84% contained in non-empty CI. (D) Model of relative spike-in abundance degradation during normal handling. Posterior predictions from Bayesian exponential decay model, for both ERCC’s and SIRV’s. The decay parameter was 19% and 18.5%, respectively. Confidence bands correspond to 95% confidence interval from posterior parameter distribution.
The degradation of spike-ins directly affected the spike-in dilution in a sample, which is a central factor for calculating the technical sensitivity. We observed that normal handling accounts for molecule limit differences within an order of magnitude, even when spike-ins are subjected to as many as six freeze-thaw cycles. The sensitivity metric for samples subjected to conditions as extreme as overnight degradation (room temperature or 37°C) had two orders of magnitude difference compared to other samples, similar to the difference between protocols (Figure 4A).
SIRV spike-ins recapitulate accuracy results based on ERCC spike-ins
All the studies mentioned above are based on the ERCC spike-ins, which have bacterial sequence composition. To ensure that our conclusions are generally applicable to spike-ins, we also analysed an alternative spike-in set: the SIRV spike-in mix consisting of 69 artificial transcripts that are produced to mimic the splicing patterns of 7 human genes and allow RNA isoform assessment. The SIRV mix E2 contains these isoforms across four abundance levels. To assess the accuracy between ERCC and SIRV spike-ins, we performed two matched scRNA-seq comparisons (Smart-seq2, SMARTer and STRT-seq on C1 system) using mESCs with both ERCC and SIRV spike-ins.
We focused on a comparison of accuracies using both sets of spike-ins, as SIRVs only span four abundance levels and are not compatible with sensitivity analysis (Figure 4B). We observe that accuracy is systematically lower when using SIRVs. This is expected, since the ambiguous read assignment to the isoforms introduces a noise element. Overall, we observed a similar pattern of relative accuracy based on SIRV’s and ERCC’s between our SMARTer and Smart-Seq2 experiments. The STRT-Seq samples had very poor accuracy, as expected since the 5’ transcript tags alone cannot distinguish between different mRNA isoforms.
This experiment provides quantitative evidence that mRNA splice form variation can be inferred at the single cell level using the appropriate protocol. Comparing the protocols, accuracy calculated based on SIRV’s recapitulates accuracy based on ERCC’s, indicating that spike-in batch variability does not in general explain differences between protocols.
Endogenous mRNA amount does not affect performance metrics based on spike-ins
Typically, when generating a cDNA library for sequencing, material is created from both endogenous mRNA and spike-in RNA. If the amount of mRNA is higher, we are less likely to sample cDNA fragments from spike-ins during sequencing. To verify that discrepancy in endogenous mRNA levels (due to e.g. cell type differences) does not affect the performance metrics, we investigated published data where information on empty (spike-in RNA alone) and non-empty (mRNA and spike-ins present) samples, was provided for the same batch of cells. We compared accuracy and sensitivity between empty and non-empty samples from three different studies and found equivalent results, confirming that endogenous mRNA content does not affect the performance metrics (Figure 4C). We quantified the equivalence using 95% Confidence Interval (CI) based equivalence analysis12 (Online Methods). We found that empty median CI is 100% contained within the non-empty median CI for accuracy, and 84% for sensitivity.
Impact of freeze-thaw cycles on spike-in abundance
Repeated freeze-thaw cycles degrade RNA, and we set out to quantify the RNA degradation rates from our freeze-thaw experiment. In our investigation of spike-in degradation after repeated freeze-thaw cycles, we added single mESCs to individual wells and performed the Smart-seq2 scRNA-seq protocol. We compared the spike-in content to the endogenous mRNA content within each well, and related this to the number of freeze-thaw cycles.
We made a predictive Bayesian model of mRNA degradation (see Online Methods) with a degradation rate parameter p. Sampling from the posterior distribution of p when applying the model to ERCC spike-ins, we found a degradation rate of 19±0.7% per freeze-thaw cycle (95% CI, Supplementary Figure 3). We also applied the mRNA degradation model to SIRV’s, and found a degradation rate of 18.5±0.1%, which recapitulates ERCC degradation rates. However the SIRVs measurements were more noisy, likely due to mapping uncertainty (Also mentioned in Discussion). The posterior predictions for both ERCC and SIRV degradation are shown in Figure 4D. Overall, our data approximates a 20% degradation rate of spike-ins in each freeze-thaw cycle during normal sample handling.
While we did not observe a large variation in molecular detection limit or accuracy due to normal handling, the relative abundance of spike-ins in a sampling is strongly affected by freeze-thaw cycles. This means inference of total mRNA in cells based on spike-ins might prove challenging. As we also found that the degradation rate was conserved between ERCC and SIRV spike-ins, the approximately 20% degradation rate per freeze-thaw cycle may hold for RNA in general.
Discussion
A previous study showed13 that read alignment to ERCC spike-ins varies widely between libraries and platforms, with some spike-ins having reproducibly poor behavior. This raises the question of suitability of spike-ins for calibration of absolute expression values. The ERCC spike-ins have short poly-A tails ranging from 20 to 26 bases long (the majority are 24 bases) in comparison to eukaryotic mRNAs of 250 base long poly-A tails10. This suggests that poly-T priming of ERCC spike-ins might be less efficient than for endogenous mRNA, possibly making all protocols seem less sensitive than they actually are for endogenous mRNAs. Furthermore, the ERCC spike-ins are not capped at the 5’ end, which may further lead to reduced template switching efficiency (used in several protocols) as compared to endogenous mRNAs14. Lastly, unlike endogenous mRNAs, spike-in RNA are not naturally bound by mRNA-binding proteins or have secondary structures.
To assess these trade-offs between the spike-in and endogenous RNA, we compared smFISH values (gold standard for absolute mRNA quantification per cell) to the calculated number of spike-in molecules per cell (Supplementary Figure 2E). This comparison also suggests more efficient capture and amplification of endogenous RNA than spike-ins by about one order of magnitude, and lower sensitivity of protocols than actual. Therefore, it is important to highlight that the “spike-in molecule detection limit” is used only a relative sensitivity measure to rank protocols, and not in absolute terms. For example, the ability to detect 10 spike-in molecules might correspond to a greater sensitivity for endogenous mRNAs. Nevertheless, the global ranking of the protocols remains relevant. The accuracy metric remains unaffected by these issues, as all ERCC spike-ins within a sample are equally affected.
A perfect protocol comparison would be between multiple laboratories implementing each protocol using a single stock of reagents and mRNA dilutions ladders as standards. Having many different scientists carrying out each protocol would allow one to exclude a skill effect on the performance. A control ladder of mRNA would eliminate issues arising from differences between synthetic spike-ins and mRNA. While the majority of the protocols we have investigated here have been reproduced by at least two distinct laboratories (Supplementary Table 1), we cannot completely rule out the impact of technical proficiency on the protocol performance.
We showed that handling and batch variations in ERCC dilutions leads to smaller variations in performance than we see between protocols (Figure 4A). Nevertheless, one needs to be aware that in certain published experiments, spike-ins may have been greatly degraded with an impact on our performance metrics.
These caveats are important when considering the performance metrics of accuracy and sensitivity in our study. Our comparison and assessment is performed on the currently available data across protocols and their performances. A protocol showing poor performance in our comparison does not restrict its suitability and it is indeed possible to get better performance for the same protocol. Our study is the first to provide a detailed comparison across the large number of currently available scRNA-seq protocols.
The different scRNA-seq protocols analyzed provide tremendously powerful and high resolution techniques for unbiased genome-wide dissection of cell populations and their transcriptional regulation. In summary, we show that while these protocols vary widely in their detection sensitivity, with lower limits between 1 and 1,000 molecules per cell, their accuracy in quantification of gene expression is generally high.
While sensitivity critically depends on sequencing depth, this is less critical for accuracy. However, both sensitivity and accuracy are closely dependent on the scRNA-seq protocol used to generate the data. The protocols with high sensitivity are more suitable for analysis of lowly expressed genes, while for other applications, this might be less relevant. The protocols with lower detection limit also allow additional insights into more subtle gene expression differences affecting individual cell states.
Overall, our comparison suggests that miniaturized reaction volumes in scRNA-seq protocols increases sensitivity and provides a good return on investment when sequencing around a million reads per sample. Future improvements of protocols and decreases in the price of sequencing will further boost our ability to answer new questions in biology using single cell transcriptomics.
Accession codes
Primary accessions
ArrayExpress
E-MTAB-5480
E-MTAB-5481
E-MTAB-5482
E-MTAB-5483
E-MTAB-5484
E-MTAB-5485
E-MTAB-5486
Referenced accessions
Gene Expression Omnibus
European Genome-phenome Archive
EGAS0000100120427
European Nucleotide Archive
Sequence Read Archiva
SRP0306173
SRP04173631
SRP03320932
SRP05515333
SRP04542234
SRP04729035
SRP02517136
SRP05049937
SRP07376738
ArrayExpress
Data Availability
All data used in this study have been deposited at ArrayExpress, and summary tables are provided as supplementary files.
Online Methods
Mouse embryonic stem (mES) cells culture
Wildtype E14 mouse ES cells (kindly provided by Pentao Liu, Wellcome Trust Sanger Institute) were cultured on gelatin coated dishes using Knockout DMEM (#10829; Gibco), 15% Fetal Calf Serum (FB-1001/500; batch tested from Labtech), 1x Penicillin-Streptomycin-Glutamine (#10378-016; Gibco), 1x MEM NEAA (11140-035; Gibco), 2-mercaptoethanol (31350-010; Gibco) and 1000U Leukemia Inhibitory Factor (LIF; #ESG1107). Mycoplasma-free tested mESC were passaged every 2-3 days.
SMARTer, Smart-Seq2 and STRT-Seq on C1
E14 mESCs were trypsinized to obtain single cell suspension and passed through 30µm filter (CellTrics; #04-0042-2316). Cells were processed using the C1 Single Cell Auto Prep System (Fluidigm; #100-7000 and #100-6209) following the manufacturers protocol (#100-5950 B1). Briefly, we perform SMARTer, Smart-seq2 and STRT-Seq each across three small C1 Open App IFCs (5-10µm; #100- 5759). The specific sample preparation steps for the three protocols (SMARTer3,28,29,31,32, Smart-seq240 and STRT-Seq9,11,20,23) were downloaded from Fluidigm Script Hub. Dissociated single cells were loaded and captured on C1 Open App IFCs, followed by manual inspection to demarcate empty well, doublets or debris containing wells. Two different spike-in RNA control sets were used for batch-matched comparison of different protocols, 92 ERCC spike-ins (#4456740; Lot# 1411014; Ambion) and 69 SIRV spike-ins (#SKU025.03; E2 Spike-in RNA Variant Control Mixes; Lexogen), were mixed (0.5µl 1:500 diluted ERCCs + 0.6µl 1:500 diluted SIRVs) and added to respective lysis buffer master mixes for SMARTer (20μl), Smart-Seq2 (27μl) and STRT-seq (20μl). 9μl of the respective lysis master mix is added to each Open App C1 IFCs. The subsequent steps (cell lysis, cDNA synthesis by reverse transcription and PCR reaction) are performed as described on Fluidigm Script Hub.
SMARTer and Smart-Seq2 on C1
E14 mESCs were trypsinized to obtain single cell suspension and passed through 30µm filter (CellTrics; #04-0042-2316). Single cell suspension was processed using SMARTer and Smart-seq2 in parallel across two C1 Single Cell Auto Prep System (Fluidigm; #100-7000 and #100-6209) following the manufacturer's protocol (#100-5950 B1). Smart-seq2 protocol was downloaded from Fluidigm Script Hub. The cells were loaded, captured on C1 Open App IFCs, followed by manually inspection. Both ERCC and SIRV spike-ins were mixed (0.5µl 1:500 diluted ERCCs + 0.6µl 1:500 diluted SIRVs) and added to respective Lysis buffer master mixes for SMARTer (20μl) and Smart-Seq2 (27μl). The subsequent steps (cell lysis, cDNA synthesis by reverse transcription and PCR reaction) are performed as described on Fluidigm Script Hub.
Spike-in degradation experiment using Smart-Seq2 on plates
We used new tube of Spike-ins, ERCC (#4456740; Lot# 1412014; Ambion) and SIRV (E2 mix; #SKU025.03; Lot#216651530; Lexogen) for this experiment. Briefly, 1:100 dilutions of ERCCs and SIRVs were mixed together resulting in spike-in master mix (1:200 final dilution; termed ‘x2 Freeze-thaw’). The spike-in master mix was split between three tubes; one left overnight at 37°C (Condition 1), one left overnight at room temperature (Condition 2) and third kept overnight at -80°C. The following day the third tube (from -80°C) was subjected to multiple freeze-thaw cycle wherein the tube was thawed at room temperature for 2-5minutes, an aliquot was taken and re-freezed in dry ice. We repeated this freeze-thaw cycle an additional 5 times (Condition 3 to Condition 7). All the spike-in mixes (Condition 1-7) were subsequently diluted to a final 1:1000,000 dilution. A 96-well plate for Smart-seq2 was prepared by dispensing 2 μl Smart-Seq2 lysis buffer (0.2%Triton, 1:20 RNAse inhibitor, 10mM Oligo-dT30VN, 10mM dNTPs) across each well. 1μl of spike-in mix per condition (Condition 1-7) was added to each well column-wise such that each column represented a single condition with 8 replicate wells. E14 mESCs were filtered through a 30μm filter and FACS sorted (BD Influx; BD Biosciences) into 96-well plate. The first three wells (row-wise) across the 96-well plate received matched bulk 500, 50 and 5 cells, and all other wells received a single cell. The 96-well plate was immediately spun and frozen on dry-ice prior to Smart-seq2 protocol as previously described40.
Library preparation and Sequencing
Representative cDNA from single cells across three C1 runs and Smart-Seq2 (on plates) were assessed using High Sensitivity DNA chips for Bioanalyzer (5067-4626 and 5067-4627; Agilent Technologies). Single cell cDNA from SMARTer3,28,29,31,32 and Smart-Seq2 C1 IFCs and Smart-seq2 (on plates) was tagmented and pooled to make libraries using Illumina Nextera XT DNA sample preparation kit (Illumina; FC-131-1096) with 96 dual barcoded indices (Illumina; FC-131-1002). The library clean-up and sample pooling was performed using AMPure XP beads (Agencourt Biosciences; A63880). All protocols are described in the Fluidigm protocol (100-5950), Fluidigm Script Hub and Smart-seq2 protocol40. The STRT-Seq libraries were made and sequenced at Karolinska Institutet as previously described9,20. The Single cell libraries from SMARTer and Smart-Seq2 C1 IFCs and Smart-seq2 (on plates) was sequenced across 1 lane of HiSeq V4 (Illumina) using 75bp/125bp paired-end sequencing.
10x Genomics Chromium experiment
The Single Cell Gel Bead kit (#120217), Single cell chip kit (#120219) and Single cell library kit (#120218) were used along with 10x GemCode Single Cell Instrument as per manufacturer specifications and manuals (Document # CG00011; Revision B). Equal volumes of control brain RNA (3μl; FirstChoice Human Brain Total RNA; #AM7962) and ERCC spikes (3μl 1:4 dilution; #4456653) were mixed to form a ‘2x Control RNA+ERCC’ master mix. We further diluted this to ‘1x Control RNA+ERCC’ with PCR grade water. We made two single cell master mix preparation using 3μl of ‘2x Control RNA+ERCC’ and ‘1x Control RNA+ERCC’ respectively instead of single cell suspension (adjusted with 34.4 μl Nuclease-Free water). The remaining protocol was followed as per manufacturer's manual (Document # CG00011; Revision B). Each 10x library was sequenced across HiSeq2500 (2x lanes; Rapid Run) as per Wellcome Trust Sanger Institute sequencing guidelines.
Data Sources
Raw read data from published studies was downloaded from either ENA or SRA, as listed with accession numbers in Supplementary Table 1. Information regarding concentration and volume of ERCC mix in each sample was gathered from the original publications (also indicated in Supplementary Table 1) or through direct communication with authors in ambiguous cases.
The expression table for mESC-STRT had non-standard names annotating the ERCC spike-ins, and through personal communication with the authors we were given a table for converting these to the names as provided by Life Technologies. Additionally we were informed by the authors that the final spike-in dilution noted as 1:50000 in Islam et al9 had actually been 1:20000.
The concentrations of the ERCC solution in the Dendritic-MARS table was ambiguous as there were two different values in the GEO table and in the text of the paper. Communication with the authors clarified that these referred to different volumes. The volume and dilution described in the GEO table was used. Thirty samples were excluded as they were annotated as not having had ERCC spike-ins added to them.
For the K562-SMART data it was unclear which data sets had used spike-ins, and personal communication with the authors provided the names of the two batches which had spike-ins added.
A table with notes on individual data sets is provided (Supplementary Table 1).
RNA-Seq data processing
For coverage based data, relative abundances were quantified using Salmon41 0.6.0, with library type parameter -l IU and the optional flag --biasCorrect. The Salmon transcriptome indices were built by adding ERCC sequences to cDNA sequences from Ensembl. For samples with mouse background, this was Ensembl 83 cDNA annotation of GRCm38.p4. For samples with human background, this was cDNA annotation from Ensembl 78 of GRCh38, and for samples with zebrafish background, the Ensembl 77 annotation of Zv9. Finally, for samples with frog background, this was Ensembl 84 annotation of JGI4.2.
In order to process all UMI-based data in a coherent way, we developed a quantification strategy based on pseudo-mapping, and counting up evidence for (transcript, UMI) pairs.
The principle is to transfer information from a (UMI, tag) pair to a (transcript, UMI) pair based on which transcript the tag maps to. Since UMI-based methods only use 3’ or 5’ end tags of cDNA, which can be as short as 25bp, mapping of these tags are commonly ambiguous. Our strategy for this is to weight a (UMI, tag) pair by the number of transcripts the tag maps to. After (UMI, tag) pairs were mapped with either RapMap42 or Kallisto43 in pseudobam mode, only (transcript, UMI) pairs with a user specified minimum amount of evidence are counted (default 1). This can be either on the gene or transcript level. In the 10x Genomics Chromium data we detected 70,000 and 45,000 droplets with respect to the samples. For the sake of computational memory efficiency we uniformly sampled 2000 droplets out of all detected droplets to count the umi tags per droplet.
Code Availability
We implemented the UMI counting strategy in a publicly available command line tool which we call ‘umis’. The tool as available at https://github.com/vals/umis/ as well as in the Python Package Index, and in Bioconda. Version 0.3.0, used for this paper, is submitted as Supplementary Software.
Analysis
An ERCC spike-in was considered detected when the estimated TPM of that ERCC was greater than zero. For UMI-based data, a spike-in is detected when at least one copy of an ERCC molecule is inferred.
The amount of input spike-in molecules for each spike, for each sample, in each experiment was calculated from the final concentration of ERCC spike-in mix in the sample.
Calculation of the accuracy of an individual sample was done by the Pearson correlation between input concentration of the spike-ins and the measured expression values. If less than 8 spike-ins were observed, the accuracy was set to infinity, as we consider this to be insufficient evidence to estimate the accuracy.
For the logistic regression model of each sample’s detection limit, the probability of detecting a spike-in at a given input level is modeled by the logistic function:
We used the LogisticRegression class from the linear_model module of the machine learning package scikit-learn44. The fit was performed with the liblinear solver and the optional argument fit_intercept=True. The logistic regression analysis was limited to samples with at least eight spike-ins detected. The detection limit was chosen as the molecular abundance where the logistic regression model passes 50% detection probability:
To investigate the UMI efficiency of UMI based protocols, we used a linear model where the only parameter was the efficiency:
As we mention in the text though, the data fits a model much better where there is a non-one exponent parameter on the number of input molecules:
When we model the relation between read depth and performance metrics for individual protocols, we use a linear model with a quadratic term for read depth to capture diminishing returns on investment. The model considers the read depth effect to be global, and has a categorical performance parameter for each protocol:
Here the performance metric will plateau and saturate when
The linear models were fitted and analysed using the OLS regression function in the statsmodels Python package.
In the spike-in degradation model the degradation rate p and the cellular fraction F were inferred by a Bayesian approach using Stan45 (R package rstan v 2.10.1). The model was specified as the following: p was sampled from a uniform distribution between 0 and 1, Fi for each spike-in i was drawn from a normal distribution with mean 0.5 and standard deviation 1. Fij was estimated by a normal distribution with mean Fi *(1-p)j, where j was the j-th freeze-thaw cycle and standard deviation sigma sampled from a uniform distribution between 0 and 20. The model was run with 5000 iteration steps, 1000 warm up steps and 4 chains.
Confidence intervals with regard to accuracy and sensitivity for non-empty and empty wells were estimated by bootstrapping. Therefore, study SRP055153, ERP010952 and SRP070989 were pooled together separating non-empty and empty wells, respectively. For each group, sample sizes of 20 were randomly picked with replacement and median of the bootstrapped samples was determined. This process was repeated with a number of 1,000 iterations. Having sorted the bootstrapped estimates, we determined the median and the 2.5th and 97.5th percentiles of the distributions for non-empty and empty wells. All data needed for our analysis is provided as Supplementary Table 2.
Supplementary Material
Acknowledgements
We are grateful to O Stegle and J K Kim for helpful discussions and comments on the manuscript. We thank M Lynch for support with the C1 experiments, X Chen for spike-in discussions and M Quail for help with 10x Chromium experiments. We extend our gratitude to S Linnarsson and A Zeisel for invaluable support in implementing STRT-Seq in our lab and help with sequencing the STRT-library. We also thank D Grun for sharing smFISH molecule counts. Finally we thank R Kirchner for many improvements to the umis tool. The study was supported by Cancer Research UK grant number C45041/A14953 to A Cvejic and C Labalette, European Research Council project 677501 - ZF_Blood to A Cvejic and a core support grant from the Wellcome Trust and MRC to the Wellcome Trust – Medical Research Council Cambridge Stem Cell Institute. The ERC grant ThSWITCH to S A Teichmann (grant no. 260507) and a Lister Institute Research Prize to S A Teichmann. K N Natarajan was supported by the Wellcome Trust Strategic Award “Single cell genomics of mouse gastrulation”.
Footnotes
Author Contributions
VS and SAT conceived the study. VS and LL annotated and processed all data. VS conceived and implemented the umis tool. VS conceived and performed the performance modeling of the data. VS, RJM, and KNN. designed the in-house experiments. KNN optimised and implemented the protocols. The degradation experiments were designed by VS, ICM, RJM, and KNN, who performed the experiments. IM and CL performed zebrafish experiments under the supervision of AC. VS and LL designed the degradation model, and LL implemented the model. VS, KNN, and SAT wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 3.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegenhain C, et al. Comparative analysis of single-cell RNA sequencing methods. bioRxiv. 2016:035758. doi: 10.1101/035758. [DOI] [PubMed] [Google Scholar]
- 5.External RNA Controls Consortium. Proposed methods for testing and selecting the ERCC external RNA controls. BMC Genomics. 2005;6:150. doi: 10.1186/1471-2164-6-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munro SA, et al. Assessing technical performance in differential gene expression experiments with external spike-in RNA control ratio mixtures. Nat Commun. 2014;5:5125. doi: 10.1038/ncomms6125. [DOI] [PubMed] [Google Scholar]
- 8.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 10.Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–2433. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 12.Walker E, Nowacki AS. Understanding equivalence and noninferiority testing. J Gen Intern Med. 2011;26:192–196. doi: 10.1007/s11606-010-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapteyn J, He R, McDowell ET, Gang DR. Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genomics. 2010;11:413. doi: 10.1186/1471-2164-11-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira T, et al. Silencing of odorant receptor genes by G protein βγ signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron. 2014;81:847–859. doi: 10.1016/j.neuron.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens NDL, et al. Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell Rep. 2016;14:632–647. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llorens-Bobadilla E, et al. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Fan X, et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang Y, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17:130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 21.Velten L, et al. Single-cell polyadenylation site mapping reveals 3’ isoform choice variability. Mol Syst Biol. 2015;11:812. doi: 10.15252/msb.20156198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul F, et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein AM, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 28.Mahata B, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7:1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015 doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 30.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padovan-Merhar O, et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sansom SN, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24:1918–1931. doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson NK, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streets AM, et al. Microfluidic single-cell whole-transcriptome sequencing. Proc Natl Acad Sci U S A. 2014;111:7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo F, et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. bioRxiv. 2016:065912. doi: 10.1101/065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennecke P, et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol. 2015;16:933–941. doi: 10.1038/ni.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 41.Patro R, Duggal G, Kingsford C. Salmon: Accurate, Versatile and Ultrafast Quantification from RNA-seq Data using Lightweight-Alignment. bioRxiv. 2015:021592. doi: 10.1101/021592. [DOI] [Google Scholar]
- 42.Srivastava A, Sarkar H, Gupta N, Patro R. RapMap: a rapid, sensitive and accurate tool for mapping RNA-seq reads to transcriptomes. Bioinformatics. 2016;32:i192–i200. doi: 10.1093/bioinformatics/btw277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 44.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 45.Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B. Stan: A probabilistic programming language. J Stat Softw. 2016 doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study have been deposited at ArrayExpress, and summary tables are provided as supplementary files.