Abstract
BACKGROUND/OBJECTIVES
The study was conducted to evaluate the effects of dietary leucine supplementation on mitochondrial biogenesis and energy metabolism in the liver of normal birth weight (NBW) and intrauterine growth-retarded (IUGR) weanling piglets.
MATERIALS/METHODS
A total of sixteen pairs of NBW and IUGR piglets from sixteen sows were selected according to their birth weight. At postnatal day 14, all piglets were weaned and fed either a control diet or a leucine-supplemented diet for 21 d. Thereafter, a 2 × 2 factorial experimental design was used. Each treatment consisted of eight replications with one piglet per replication.
RESULTS
Compared with NBW piglets, IUGR piglets had a decreased (P < 0.05) hepatic adenosine triphosphate (ATP) content. Also, IUGR piglets exhibited reductions (P < 0.05) in the activities of hepatic mitochondrial pyruvate dehydrogenase (PDH), citrate synthase (CS), α-ketoglutarate dehydrogenase (α-KGDH), malate dehydrogenase (MDH), and complexes I and V, along with decreases (P < 0.05) in the concentration of mitochondrial DNA (mtDNA) and the protein expression of hepatic peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α). Dietary leucine supplementation increased (P < 0.05) the content of ATP, and the activities of CS, α-KGDH, MDH, and complex V in the liver of piglets. Furthermore, compared to those fed a control diet, piglets given a leucine-supplemented diet exhibited increases (P < 0.05) in the mtDNA content and in the mRNA expressions of sirtuin 1, PGC-1α, nuclear respiratory factor 1, mitochondrial transcription factor A, and ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide in liver.
CONCLUSIONS
Dietary leucine supplementation may exert beneficial effects on mitochondrial biogenesis and energy metabolism in NBW and IUGR weanling piglets.
Keywords: Leucine, intrauterine growth retardation, weanling piglets, mitochondrial biogenesis, energy metabolism
INTRODUCTION
Intrauterine growth retardation (IUGR) is the impairment of organ development and fetus growth of a mammal during pregnancy, and such retardation increases neonatal morbidity and mortality [1]. Epidemiological evidence shows that IUGR adults are more prone to have hypertension, obesity, type 2 diabetes, and other metabolic diseases [2]. Mitochondria, the main sites of adenosine triphosphate (ATP) production in mammalian cells, have an essential role in energy homeostasis, cellular metabolism, and apoptosis [3,4]. On the basis of several previous observations, we hypothesized that the possible mechanisms responsible for metabolic syndrome in adult IUGR offspring are impairment of mitochondrial biogenesis and energy deficiency [5,6,7]. Indeed, it has been demonstrated that IUGR decreases the concentration of ATP and the transcriptional levels of genes involved in mitochondrial biogenesis, and it impairs activities of mitochondrial enzymes related to oxidative phosphorylation (OXPHOS) in the liver of IUGR animals [7,8,9].
Branched-chain amino acids (BCAA), a set of essential amino acids, have critical roles in protein metabolism and energy production [10,11,12]. Among the BCAA, leucine has been the most extensively investigated due to the varied physiological roles it has in protein synthesis and degradation, stimulation of insulin secretion, and blood glucose homeostasis [13,14,15]. In addition, leucine has been demonstrated to improve hepatic mitochondrial function, as evidenced by increases in citrate synthase (CS) activity and ATP concentration [16]. In vitro experiments have also shown that leucine can increase mitochondrial content and stimulate mitochondrial biogenesis [17,18].
We previously reported that leucine supplementation can alleviate IUGR-induced decreases in final body weight, average daily gain, and feed intake of weanling piglets during the first three weeks post-weaning, along with increasing the relative weight of the liver of piglets at 35 days of age [19]. It is unknown whether these beneficial effects are associated with possible alterations in hepatic mitochondrial biogenesis and energy metabolism following leucine supplementation. Therefore, the current study was conducted to investigate the effects of leucine on hepatic mitochondrial biogenesis and energy metabolism in normal birth weight (NBW) and IUGR weanling piglets. Considering the physiological similarities between humans and pigs [20], this study may provide a new strategy for handling IUGR offspring during the early post-weaning period.
MATERIALS AND METHODS
Ethical statement
The experiments performed in the present study were approved by the Nanjing Agricultural University Institutional Animal Care and Use Committee (NJAU-CAST-2014-065) and followed current animal protection laws [21].
Experimental animals and design
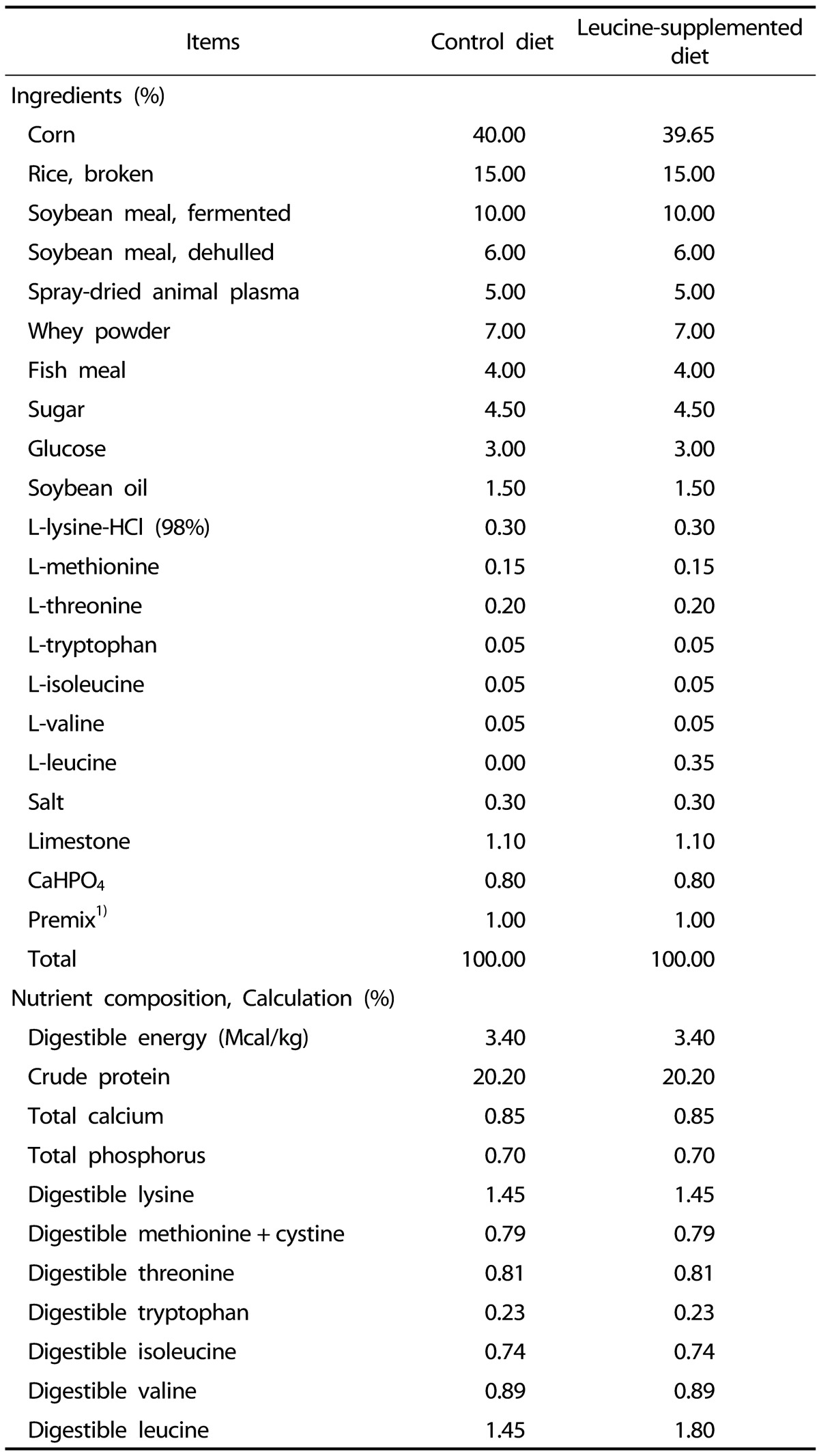
Pregnant sows (Landrace × Yorkshire) with similar parity (2nd or 3rd) and similar expected dates of confinement were selected and fed a commercial diet during pregnancy. At birth, the birth weight (BW) and sex of each newborn piglet (Duroc × (Landrace × Yorkshire)) were recorded. In each litter, piglets with a body weight of 1.52 ± 0.06 kg and 0.87 ± 0.04 kg at birth were defined as NBW and IUGR piglets, as previously reported [7,22]. A total of sixteen pairs of NBW and IUGR piglets from sixteen sows were selected according to their BW. At postnatal day 14, all piglets were weaned and fed either a control diet (CON; 14.5 g leucine per kilogram of feed) or a leucine-supplemented diet (LEU; 18.0 g leucine per kilogram of feed) for 21 d. Thereafter, all piglets were distributed into four groups (NBW-CON, NBW-LEU, IUGR-CON, and IUGR-LEU). Each group consisted of eight replications with one piglet per replication (four males and four females). Dietary leucine supplementation was achieved by adding L-leucine (98%; Sigma-Aldrich, St Louis, MO, USA) into the piglets' feed, replacing the equivalent weight of corn; the concentrations of other nutrients were maintained constant in all experimental diets. The dietary leucine dosage was selected according to an independent study by study colleagues in which the optimum effects of leucine on growth performance in weanling piglets were observed at a dosage of 18.0 g/kg of feed (unpublished). The composition and nutrient levels of the piglets' diets, formulated to meet the nutritional requirements set by the National Research Council (2012) [23], are presented in Table 1. Piglets were housed individually in pens with plastic floors (1 m × 0.6 m) at 28℃ and were given ad libitum access to feed and water.
Table 1. The composition and nutrient levels of diets.
1) In premix, provided per kilogram of diet: Vitamin A, 15,000 IU; Vitamin D3, 3,000 IU; Vitamin E, 150 mg; Vitamin K3, 3 mg; Vitamin B1, 3 mg; Vitamin B2, 6 mg; Vitamin B6, 5 mg; Vitamin B12, 0.03 mg; Niacin, 45 mg; Vitamin C, 250 mg; Calcium pantothenate, 9 mg; Folic acid, 1 mg; Biotin, 0.3 mg; Choline chloride, 500 mg; Fe, 170 mg; Cu, 150 mg; I, 0.90 mg; Se, 0.2 mg; Zn, 150 mg; Mg, 68 mg; Mn, 80 mg; Co, 0.30 mg.
Sample collection
All piglets were sacrificed at postnatal day 35 by intramuscular injection of sodium pentobarbital (50 mg/kg body weight) at 12 h after the last meal. Liver samples were removed immediately after sacrifice, frozen in liquid nitrogen after snipping, and stored at −80℃ prior to analysis.
Measurement of hepatic ATP, adenosine diphosphate (ADP), and adenosine monophosphate (AMP) concentrations
Analyses were carried out by using high-performance liquid chromatography (HPLC) as described in a previous study [24]. Approximately 80 mg of frozen liver samples were homogenized with 1.5 mol/L perchloric acid and centrifuged to obtain supernatants. The collected supernatants were neutralized with 2 mol/L potassium carbonate and centrifuged again to obtain supernatants for analysis by using a Thermo Ultimate 3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) with a C18 chromatographic column (Thermo Fisher Scientific, Waltham, MA, USA). The mobile phase (50 mmol/L K2HPO4-KH2PO4 buffer solution and methanol (HPLC grade); 99:1, v/v; pH = 7.0) was filtered (0.45 µm filter) and degassed before use. All standards and samples were filtered (0.22 µm filter) and injected (20 µL) to the HPLC system. The wavelength for UV detection was 254 nm, the flow rate was 1.0 mL/min, and the column temperature was 40℃. Total adenine nucleotide (TAN) and adenylate energy charges (AEC) were calculated by using the previously reported equations [25]: TAN = ATP + ADP + AMP and AEC = (ATP + 0.5 ADP)/(ATP + ADP + AMP).
Isolation of hepatic mitochondria
Hepatic mitochondria were isolated at 4℃ by using a standard procedure based on differential centrifugation [26]. Approximately 1 g of liver samples were immediately immersed in isolation buffer (0.1 µmol/L EDTA-2Na, 0.01 mol/L Tris-HCl, 0.8% sodium chloride solution, 0.01 mol/L sucrose, pH = 7.4). Tissue samples were then homogenized with an additional 9 volumes (wt/vol) of homogenization media in a homogenizer and centrifuged at 600 g for 10 min. The collected supernatants were centrifuged again at 11,000 g for 15 min. The supernatants were removed and resuspended in isolation buffer and centrifuged again at 11,000 g for 15 min to acquire mitochondria. The concentrations of mitochondrial protein were measured by using the bicinchoninic acid protein assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China).
Measurement of hepatic mitochondrial pyruvate dehydrogenase (PDH), CS, isocitrate dehydrogenase (ICDH), α-ketoglutarate dehydrogenase (α-KGDH), and malate dehydrogenase (MDH) activities
The activities of hepatic mitochondrial PDH, ICDH, and α-KGDH were determined by using the corresponding diagnostic kits (Suzhou Comin Biotechnology, Suzhou, Jiangsu, China). The activities of hepatic mitochondrial CS and MDH were determined by using corresponding diagnostic kits obtained from the Nanjing Jiancheng Bioengineering Institute (Nanjing, Jiangsu, China). Briefly, the activity of hepatic mitochondrial PDH was measured in the reaction with 2,6-dichlorophenolindophenol (2,6-DCPIP) by using a spectrophotometer and monitoring the reduction of 2,6-DCPIP at 600 nm [27]. One unit of PDH activity was defined as 1 nmol of 2,6-DCPIP reduced per minute at 37℃. The activity of hepatic mitochondrial CS was measured by applying the method of Shepherd et al. [28], which follows the enzyme-catalyzed reaction between oxaloacetic acid and acetyl-CoA. Absorbance was monitored at 412 nm with a spectrophotometer. One unit of CS activity was defined as the amount of enzyme catalyzing 1 nmol citrate per minute at 37℃. The activity of hepatic mitochondrial ICDH was measured by determining the production of NADH at 340 nm [29]. One unit of ICDH activity was defined as the amount of enzyme producing 1 nmol of NADH per minute at 37℃. The activity of hepatic mitochondrial α-KGDH was assayed by measuring the rate of increase of absorbance due to NADH at 340 nm [30]. One unit of α-KGDH activity was defined as the amount of enzyme producing 1 nmol of NADH per minute at 37℃. The activity of hepatic mitochondrial MDH was measured by applying the method of Mehler et al. [31], which follows the decrease in extinction at 340 nm consequent on the oxidation of NADH by oxaloacetate. One unit of MDH activity was defined as the amount of enzyme producing 1 µmol of NAD+ per minute at 37℃.
Measurement of hepatic mitochondrial respiratory chain complex I, II, III, IV, and V activities
Analyses were undertaken by using commercial kits (Suzhou Comin Biotechnology, Suzhou, Jiangsu, China). Briefly, the activity of mitochondrial complex I was measured according to the method described by Ragan [32], which follows the decrease in NADH absorbance (340 nm) that occurs when ubiquinone (CoQ1) is reduced to form ubiquinol. The activity of mitochondrial complex I was expressed as 1 nmol of NADH oxidized per minute per milligram of mitochondrial protein. The activity of mitochondrial complex II was measured according to the method described by Medja et al. [33], which follows the reduction of 2,6-DCPIP at 605 nm at 37℃. The activity of mitochondrial complex II was expressed as 1 nmol of 2,6-DCPIP reduced per minute per milligram of mitochondrial protein. The oxidation of reduced ubiquinone (CoQH2) by mitochondrial complex III was measured by using cytochrome c as an electron acceptor at 550 nm at 37℃ [34]. The activity of mitochondrial complex III was expressed as 1 nmol of CoQH2 oxidized per minute per milligram of mitochondrial protein. The activity of mitochondrial complex IV was measured according to the method described by Wharton et al. [35], which follows the oxidation of cytochrome c by the absorbance (550 nm) at 37℃. The activity of mitochondrial complex IV was expressed as 1 nmol of cytochrome c oxidized per minute per milligram of mitochondrial protein. The activity of mitochondrial complex V was determined by using a microcolorimetric assay that measures the production rate of inorganic phosphorus at 660 nm, which has been utilized for the formation of ATP from ADP [36]. The activity of mitochondrial complex V was expressed as 1 nmol of inorganic phosphorus production per minute per milligram of mitochondrial protein.
Total DNA extraction and analysis of mitochondrial DNA (mtDNA) content
DNAiso reagent (TaKaRa, Otsu, Shiga, Japan) was used to extract total DNA from frozen liver samples. The relative content of mtDNA was determined by co-amplifying mt D-loop and β-actin according to the method in a previous study [37]. Real-time PCR was conducted by using an ABI StepOnePlus™ Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction system was 20 µL in total, which consisted of 1 µL of forward primer, 1 µL of reverse primer, 8 µL of TaqMan Universal Master Mix (TaKaRa, Otsu, Shiga, Japan), 1 µL of probes, 1 µL of enhance solution, 1 µL of DNA template, and 7 µL of double-distilled water. The reaction conditions were as follows: 10 s at 95℃, 50 cycles of 5 s at 95℃, 25 s at 60℃, and 10 s at 95℃. The 2-ΔΔCt method was used to calculate the relative mtDNA content [37]. The primer sequences used to measure the content of mtDNA are shown in Table 2.
Table 2. Primer sequences used in the real-time PCR.
1) SIRT1, sirtuin 1; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α; NRF1, nuclear respiratory factor 1; TFAM, mitochondrial transcription factor A; NDUFA1, NADH dehydrogenase 1α subcomplex, 1; NDUFA13, NADH dehydrogenase 1α subcomplex, 13; NDUFB1, NADH dehydrogenase 1β subcomplex, 1; SDHA, succinate dehydrogenase complex, subunit A; SDHB, succinate dehydrogenase complex, subunit B; COX I, cytochrome c oxidase subunit I; COX IV, cytochrome c oxidase subunit IV; COX V, cytochrome c oxidase subunit V; ATP5G1, ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1; ATP5B, ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide; mt D-loop, mitochondrial displacement loop region.
Total RNA extraction and mRNA quantification
Trizol reagent (TaKaRa, Otsu, Shiga, Japan) was used to extract total RNA from frozen liver samples. After determination of RNA concentration, mRNA was reverse-transcribed into complementary DNA (cDNA) by using a reverse transcription kit (TaKaRa, Otsu, Shiga, Japan). Real-time PCR was conducted by using an ABI StepOnePlus™ Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction system was 20 µL in total, which consisted of 0.4 µL each of forward and reverse primers, 0.4 µL of ROX reference dye, 10 µL of SYBR Premix Ex Taq™ (TaKaRa, Otsu, Shiga, Japan), 6.8 µL of double-distilled water, and 2 µL of cDNA template. Each sample was assayed in duplicate. The reaction conditions were as follows: 30 s at 95℃, 40 cycles of 5 s at 95℃, and 30 s at 60℃. The relative mRNA expression levels were calculated by the 2-ΔΔCt method after normalization to that of β-actin [38]. The values of the NBW-CON group were used as a calibrator. The primer sequences for sirtuin 1 (SIRT1), peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), nuclear respiratory factor 1 (NRF1), mitochondrial transcription factor A (TFAM), NADH dehydrogenase 1α subcomplex, 1 (NDUFA1), NADH dehydrogenase 1α subcomplex, 13 (NDUFA13), NADH dehydrogenase 1β subcomplex, 1 (NDUFB1), succinate dehydrogenase complex, subunit A (SDHA), succinate dehydrogenase complex, subunit B (SDHB), cytochrome c oxidase subunit I (COX I), cytochrome c oxidase subunit IV (COX IV), cytochrome c oxidase subunit V (COX V), ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (ATP5G1), ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide (ATP5B), and β-actin are shown in Table 2.
Western blot
Approximately 40 mg of frozen liver samples were homogenized in RIPA lysis buffer (Beyotime Institute of Biotechnology, Nantong, Jiangsu, China) and phenylmethanesulfonyl fluoride (PMSF, Beyotime Institute of Biotechnology, Nantong, Jiangsu, China) by using a glass homogenizer. The homogenates were centrifuged at 12,000 g, and the supernatants were transferred to new tubes. The protein concentrations were measured by using a commercial kit (Beyotime Institute of Biotechnology, Nantong, Jiangsu, China). Protein (40 µg) from the supernatant of each sample was electrophoresed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were blocked with Tris-buffered saline Tween-20 containing 5% non-fat dry milk for 90 min at room temperature. Then, the membranes were washed and probed with the polyclonal antibodies. The primary antibody was anti-peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α, 1:500; Abcam, Cambridge, MA, USA). The membranes were washed and incubated with a secondary antibody (horseradish peroxidase-conjugated anti-goat IgG, 1:2000; Abcam, Cambridge, MA, USA) for 1 h at room temperature. The blots were developed by using an enhanced chemiluminescence kit (ECL-plus, Beyotime Institute of Biotechnology, Nantong, Jiangsu, China) followed by autoradiography. Images were recorded by a Luminescent Image Analyzer LAS-4000 system (Fujifilm Co. Ltd., Tokyo, Japan) and quantified by using Gel-Pro Analyzer 4.0 software (Media Cybernetics, Silver Spring, MD, USA).
Statistical analysis
Data were analyzed by using SPSS 16.0 software (SPSS, Chicago, IL, USA). The general linear model procedure was used to look for significant differences, comparing the main experimental variables (BW and Diet) and their interaction. If significant differences (P < 0.05) were detected in the interaction of the main effects, Duncan's multiple range test was used to compare the means. P-values less than 0.05 were considered statistically significant, and P-values between 0.05 and 0.10 were considered a trend. Results are expressed as means ± SEM.
RESULTS
Hepatic ATP, ADP, and AMP concentrations
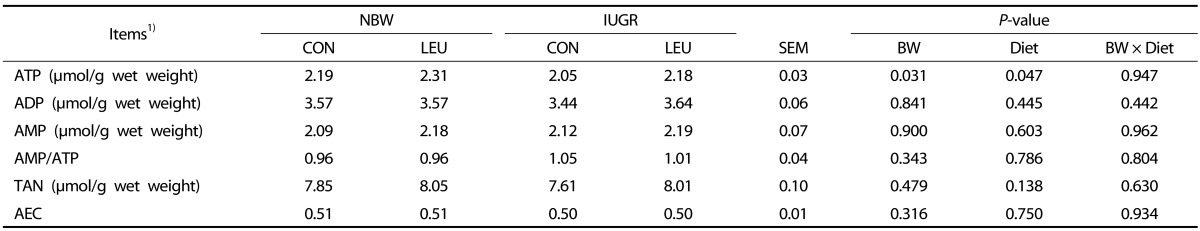
There was a significant decrease (P < 0.05) in hepatic ATP concentration of IUGR piglets compared that in NBW piglets (Table 3). An increased concentration of ATP (P < 0.05) in the liver was observed in piglets given a leucine-supplemented diet compared to piglets given a control diet. There were no differences in other parameters among the groups (P > 0.10).
Table 3. Effects of dietary leucine supplementation on the hepatic metabolite concentrations in normal birth weight and intrauterine growth-retarded weanling piglets.
1) NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight; ATP, adenosine triphosphate; ADP, adenosine diphosphate; AMP, adenosine monophosphate; TAN, total adenine nucleotide, TAN = ATP + ADP + AMP; AEC, adenylate energy charges, AEC = (ATP + 0.5 ADP)/(ATP + ADP + AMP). Data are presented as means ± SEM (n = 8/group). Mean values within a row with unlike superscript letters were significantly different (P < 0.05).
Hepatic mitochondrial PDH, CS, ICDH, α-KGDH, and MDH activities
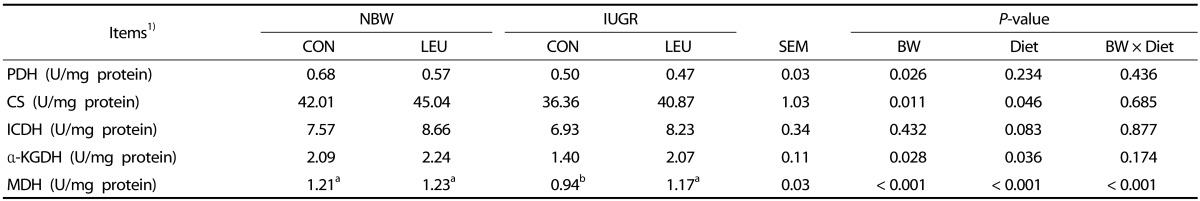
The activities of mitochondrial PDH, CS, α-KGDH, and MDH in the liver of IUGR piglets were significantly lower (P < 0.05) than those in NBW piglets (Table 4). Piglets given a leucine-supplemented diet exhibited increased (P < 0.05) activities of hepatic mitochondrial CS, α-KGDH, and MDH compared with those given a control diet. Dietary leucine supplementation tended to partially recover the decrease (P = 0.083) in the hepatic mitochondrial ICDH activity of weanling piglets. Additionally, BW and Diet had an obvious interaction effect (P < 0.05) on hepatic mitochondrial MDH activity, in which dietary leucine supplementation increased hepatic mitochondrial MDH activity in IUGR piglets rather than NBW piglets.
Table 4. Effects of dietary leucine supplementation on the activities of hepatic mitochondrial enzymes related to energy metabolism in normal birth weight and intrauterine growth-retarded weanling piglets.
1) NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight; PDH, pyruvate dehydrogenase; CS, citrate synthase; ICDH, isocitrate dehydrogenase; α-KGDH, α-ketoglutarate dehydrogenase; MDH, malate dehydrogenase. Data are presented as means ± SEM (n = 8/group). Mean values within a row with unlike superscript letters were significantly different (P < 0.05).
Mitochondrial respiratory chain complex activities
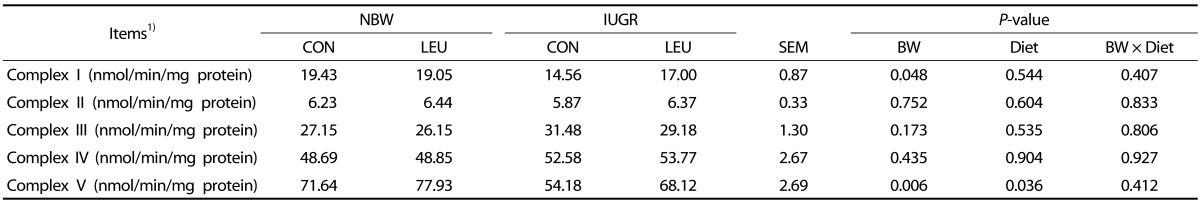
There were obvious decreases (P < 0.05) in the activities of hepatic complexes I and V in IUGR piglets compared to those in NBW piglets (Table 5). In contrast, increased complex V activity (P < 0.05) was observed in piglets given a leucine-supplemented diet compared to piglets given a control diet. There were no significant differences in the activities of complexes II, III, and IV (P > 0.10) among the groups.
Table 5. Effects of dietary leucine supplementation on the hepatic mitochondrial respiratory chain complex activities in normal birth weight and intrauterine growth-retarded weanling piglets.
1) NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight. Data are presented as means ± SEM (n = 8/group). Mean values within a row with unlike superscript letters were significantly different (P < 0.05).
Hepatic mtDNA content
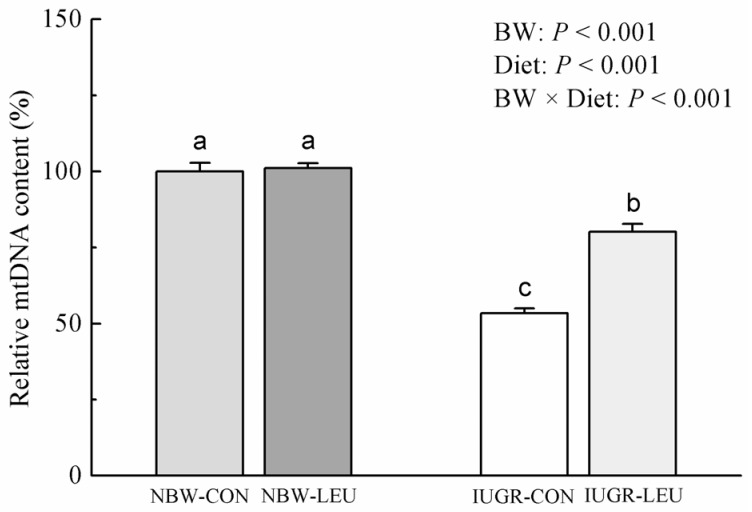
The hepatic mtDNA content was decreased (P < 0.05) in IUGR piglets compared to that in NBW piglets (Fig. 1). Dietary leucine supplementation increased (P < 0.05) the content of hepatic mtDNA in weanling piglets. Additionally, BW and Diet had an obvious interaction effect (P < 0.05) on hepatic mtDNA content, in which dietary leucine supplementation increased hepatic mtDNA content in IUGR piglets rather than NBW piglets.
Fig. 1. Effects of dietary leucine supplementation on the hepatic mitochondrial DNA content in normal birth weight and intrauterine growth-retarded weanling piglets.
NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight; mtDNA, mitochondrial DNA. Data are presented as means ± SEM (n = 8/group). Mean values in columns with unlike superscript letters were significantly different (P < 0.05).
Messenger RNA expressions
Compared with NBW piglets, IUGR piglets had downregulated (P < 0.05) mRNA abundances of SIRT1, PGC-1α, NRF1, TFAM, SDHA, and ATP5B in liver (Fig. 2). A similar trend was observed in hepatic NDUFB1 (P = 0.061) and COX V (P = 0.059) mRNA expressions in IUGR piglets. Dietary leucine supplementation increased (P < 0.05) mRNA expressions of hepatic SIRT1, PGC-1α, NRF1, TFAM, and ATP5B in piglets. In addition, dietary leucine supplementation alleviated (P < 0.05) the IUGR-induced decrease in hepatic PGC-1α and TFAM expressions. There were no significant differences in transcriptional levels of NDUFA1, NDUFA13, SDHB, COX I, COX IV, and ATP5G1 (P > 0.10) among the groups.
Fig. 2. Effects of dietary leucine supplementation on the hepatic mRNA expressions related to mitochondrial biogenesis and energy metabolism in normal birth weight and intrauterine growth-retarded weanling piglets.
(A) Relative mRNA expressions of SIRT1, PGC-1α, NRF1, TFAM, NDUFA1, NDUFA13, and NDUFB1. (B) Relative mRNA expressions of SDHA, SDHB, COX I, COX IV, COX V, ATP5G1, and ATP5B. NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight; SIRT1, sirtuin 1; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α; NRF1, nuclear respiratory factor 1; TFAM, mitochondrial transcription factor A; NDUFA1, NADH dehydrogenase 1α subcomplex, 1; NDUFA13, NADH dehydrogenase 1α subcomplex, 13; NDUFB1, NADH dehydrogenase 1β subcomplex, 1; SDHA, succinate dehydrogenase complex, subunit A; SDHB, succinate dehydrogenase complex, subunit B; COX I, cytochrome c oxidase subunit I; COX IV, cytochrome c oxidase subunit IV; COX V, cytochrome c oxidase subunit V; ATP5G1, ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1; ATP5B, ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide. Data are presented as means ± SEM (n = 8/group). Mean values in columns with unlike superscript letters were significantly different (P < 0.05).
Protein expression of hepatic PGC-1α
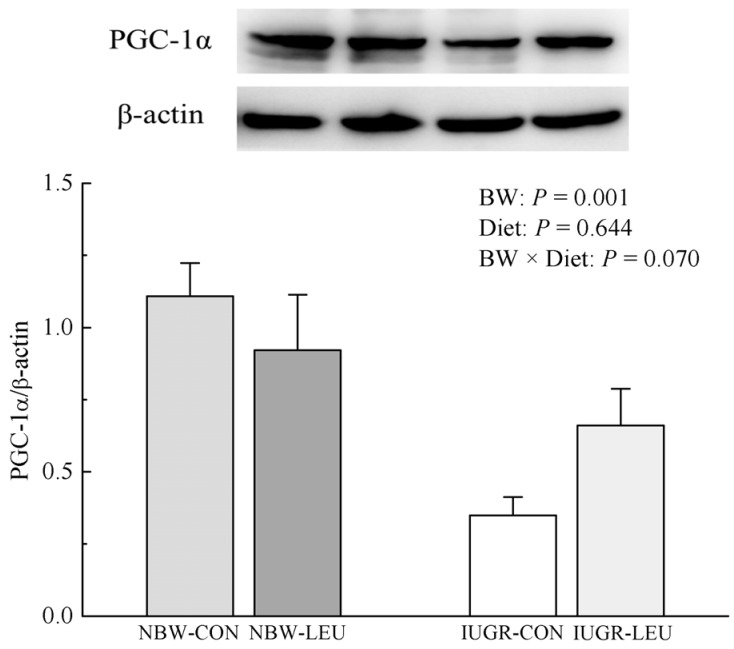
The protein expression of hepatic PGC-1α was decreased (P < 0.05) in IUGR piglets compared to that in NBW piglets (Fig. 3). Dietary leucine supplementation tended to partially recover the decrease (P = 0.070) in protein expression of hepatic PGC-1α induced by IUGR.
Fig. 3. Effects of dietary leucine supplementation on the protein expression of hepatic PGC-1α in normal birth weight and intrauterine growth-retarded weanling piglets.
NBW-CON, normal birth weight group given a control diet; NBW-LEU, normal birth weight group given a leucine-supplemented diet; IUGR-CON, intrauterine growth-retarded group given a control diet; IUGR-LEU, intrauterine growth-retarded group given a leucine-supplemented diet; BW, birth weight; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α. Data are presented as means ± SEM (n = 8/group). Mean values in columns with unlike superscript letters were significantly different (P < 0.05).
DISCUSSION
The results of the present study show that dietary leucine supplementation can increase hepatic ATP production of weanling piglets, which is concomitant with increased mtDNA content, upregulated mRNA abundances of hepatic genes involved in mitochondrial biogenesis, and improved activities of CS, α-KGDH, MDH, and complex V. These changes might further strengthen the beneficial effects that leucine has on mitochondrial biogenesis and energy metabolism in animals [16,39].
It has been demonstrated that IUGR decreases AEC and ATP/ADP in the liver of fetal rats [40], which is similar to the present study's result showing that IUGR offspring had a lower hepatic ATP content than their heavier counterparts. Such a decrease, most likely, resulted from impaired mitochondrial biogenesis and low energy metabolism capacity [41]. Indeed, the present results indicate that IUGR decreases the hepatic mtDNA concentration in piglets, together with a reduction in the influence of multiple transcription factors related to mitochondrial biogenesis. Similarly, Park et al. [5] reported that IUGR decreased hepatic mtDNA levels in rats. The regulation of mitochondrial biogenesis, requiring the interaction of multiple transcriptional factors, is a complex biological process [42]. PGC-1α, a transcriptional coactivator, increases mitochondrial biogenesis and OXPHOS through the replication and transcription of mtDNA [3]. In the present study, IUGR decreased PGC-1α at both the transcription and translation levels. Previous researchers have also shown an obvious decrease in the mRNA expression of PGC-1α in the liver of IUGR offspring during early life [43]. These results indicate that IUGR may cause mitochondrial dysfunction, resulting from an eventual inhibition of hepatic PGC-1α expression.
SIRT1 is a stimulator of mitochondrial biogenesis through PGC-1α deacetylation [44]. In this study, there was a decrease in hepatic SIRT1 mRNA expression in IUGR piglets, suggesting a possible explanation for their low energy metabolism efficiency. Previous researchers have also demonstrated a significantly decreased mRNA expression of SIRT1 in the liver of IUGR offspring [37]. NRF1, a DNA-binding transcription factor, regulates numerous mitochondrial gene expressions [45]. In this study, IUGR downregulated the mRNA expression of hepatic NRF1, which is in agreement with the results of previous research [37]. In addition, a concomitantly reduced transcription of hepatic TFAM was noted in the present study's IUGR piglets. TFAM, a key regulator involved in mtDNA replication and transcription, could be regulated by PGC-1α and NRF1 to activate mitochondrial biogenesis [46,47]. Similarly, Zhang et al. [7] found a decreased transcriptional level of hepatic TFAM in IUGR weanling piglets.
Another manifestation of mitochondrial energy production impairment in the IUGR piglets was the reduction of tricarboxylic acid (TCA) cycle flux. In the present study, the activities of mitochondrial PDH, CS, α-KGDH, and MDH in the liver of IUGR piglets were significantly lower than those in the liver of NBW piglets. Peterside et al. [8] previously reported that IUGR decreased hepatic PDH activity in rats, which is a key entry point for carbon in the TCA cycle. Such a decrease may provide a possible explanation for the observed decreases in CS, α-KGDH, and MDH activities. Zhang et al. [7] obtained similar findings in another study. Previous studies have also suggested that decreased ATP production might result from the low capacity of the electron transport chain in different IUGR animal models [7,40]. In the present study, IUGR induced an obvious reduction in the activities of complexes I and V in the liver of piglets. A similar result has been reported by Simmons et al. [48], who indicated that the activities of complexes I and III were decreased in IUGR rats. Furthermore, control of the OXPHOS process can be regulated by multiple transcriptional factors. The present results indicate that the mRNA expression of SDHA is downregulated in IUGR piglets, which is consistent with previous results [7]. In addition, a lower level of mRNA expression of ATP5B was found in the liver of IUGR piglets. Similarly, Morris et al. [49] revealed that there is functional impairment of the OXPHOS process in IUGR rats, as evidenced by the decreased expression of ATP synthase.
It has been reported that BCAA can improve mtDNA content in both in vivo and in vitro experiments [16,39]. In this study, dietary leucine supplementation prevented the typical drop in mtDNA content induced by IUGR, and the drop in mtDNA may be regulated by multiple transcriptional factors [42]. In fact, leucine treatment alleviated the IUGR-induced decreases in mRNA expression levels of PGC-1α and TFAM, and it upregulated the transcriptional level of NRF1 in the liver of piglets. Similarly, a previous rodent study indicated that leucine can upregulate hepatic PGC-1α, NRF1, and TFAM at the transcriptional level [39]. PGC-1α is a potent coactivator of the central transcription factor NRF1 in regulating transcription of the TFAM gene, which controls the sole mtDNA promoter and replication of mtDNA [3,46]. In addition, an in vitro study confirmed that leucine could stimulate the expressions of PGC-1α and NRF1 via a SIRT1-dependent pathway to induce mitochondrial biogenesis [18]. Thus, the increased mRNA abundance of SIRT1 observed in the liver of piglets fed a leucine-supplemented diet may contribute to the activation of the PGC-1α/NRF1/TFAM pathway.
Li et al. [16] reported that dietary inclusion of leucine recovered a high-fat-diet-induced decrease in CS activity. In this study, dietary leucine supplementation increased the mitochondrial activities of CS, α-KGDH, and MDH in the liver of piglets. At the same time, treatment with leucine counteracted the IUGR-induced decreased hepatic mitochondrial MDH activity in the liver of piglets, which has a role in improving the flux of the TCA cycle. Leucine provides carbon skeletons to the TCA cycle at the level of acetyl-CoA, which may enhance cycle flux [50]. Thus, the suppression of CS, α-KGDH, and MDH may be removed via increased acetyl-CoA content from the oxidation of leucine. The present study indicated that dietary leucine supplementation increased the activity of mitochondrial respiratory chain complex V, which was closely related to ATP production and the function of mitochondria in piglets. In addition, dietary leucine supplementation increased the mRNA expression of ATP5B in weanling piglets. It has been reported that ATP5B may exert an effect on energy production and ATP synthesis [51]. The report by D'Antona et al. [39] indicated that BCAA increases the mRNA expression of the β subunit of F1-ATPase. In addition, it has been shown that leucine supplementation increases ATP concentration and prevents mitochondrial dysfunction in high-fat-diet-induced obese mice [16]. Also, an in vitro study on cultured cardiomyocytes revealed that BCAA supplementation increased the production of ATP [39]. In the current study, dietary leucine supplementation increased the content of ATP in the liver of piglets. Taken together, these data indicate that leucine stimulates ATP production by improving mitochondrial biogenesis and energy metabolism in weanling piglets.
In conclusion, the present study demonstrates that leucine may have beneficial effects in hepatic mitochondrial biogenesis and energy metabolism in weanling piglets, which may provide another clue to the mechanism behind the improved growth performance observed previously [19]. The results of this study may be helpful in developing a new feeding strategy for IUGR offspring to attenuate energy deficiency during early life.
ACKNOWLEDGMENTS
The authors thank their laboratory colleagues for their assistance.
Footnotes
This study was supported by the National Basic Research Program of China (973) (no. 2012CB124703) and the National Natural Science Foundation of China (no. 31572418).
CONFLICT OF INTEREST: The authors declare no potential conflicts of interests.
References
- 1.Wu G, Bazer FW, Wallace JM, Spencer TE. Board-invited review: intrauterine growth retardation: implications for the animal sciences. J Anim Sci. 2006;84:2316–2337. doi: 10.2527/jas.2006-156. [DOI] [PubMed] [Google Scholar]
- 2.Valsamakis G, Kanaka-Gantenbein C, Malamitsi-Puchner A, Mastorakos G. Causes of intrauterine growth restriction and the postnatal development of the metabolic syndrome. Ann N Y Acad Sci. 2006;1092:138–147. doi: 10.1196/annals.1365.012. [DOI] [PubMed] [Google Scholar]
- 3.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 4.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Park KS, Kim SK, Kim MS, Cho EY, Lee JH, Lee KU, Pak YK, Lee HK. Fetal and early postnatal protein malnutrition cause long-term changes in rat liver and muscle mitochondria. J Nutr. 2003;133:3085–3090. doi: 10.1093/jn/133.10.3085. [DOI] [PubMed] [Google Scholar]
- 6.Reusens B, Sparre T, Kalbe L, Bouckenooghe T, Theys N, Kruhøffer M, Ørntoft TF, Nerup J, Remacle C. The intrauterine metabolic environment modulates the gene expression pattern in fetal rat islets: prevention by maternal taurine supplementation. Diabetologia. 2008;51:836–845. doi: 10.1007/s00125-008-0956-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Li Y, Hou X, Zhang L, Wang T. Medium-chain TAG improve energy metabolism and mitochondrial biogenesis in the liver of intra-uterine growth-retarded and normal-birth-weight weanling piglets. Br J Nutr. 2016;115:1521–1530. doi: 10.1017/S0007114516000404. [DOI] [PubMed] [Google Scholar]
- 8.Peterside IE, Selak MA, Simmons RA. Impaired oxidative phosphorylation in hepatic mitochondria in growth-retarded rats. Am J Physiol Endocrinol Metab. 2003;285:E1258–E1266. doi: 10.1152/ajpendo.00437.2002. [DOI] [PubMed] [Google Scholar]
- 9.Lane RH, Flozak AS, Ogata ES, Bell GI, Simmons RA. Altered hepatic gene expression of enzymes involved in energy metabolism in the growth-retarded fetal rat. Pediatr Res. 1996;39:390–394. doi: 10.1203/00006450-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Yin Y, Tan B, Kong X, Wu G. Leucine nutrition in animals and humans: mTOR signaling and beyond. Amino Acids. 2011;41:1185–1193. doi: 10.1007/s00726-011-0983-2. [DOI] [PubMed] [Google Scholar]
- 11.Wu G. Dietary requirements of synthesizable amino acids by animals: a paradigm shift in protein nutrition. J Anim Sci Biotechnol. 2014;5:34. doi: 10.1186/2049-1891-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson J, Wilson GJ. Contemporary issues in protein requirements and consumption for resistance trained athletes. J Int Soc Sports Nutr. 2006;3:7–27. doi: 10.1186/1550-2783-3-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garlick PJ. The role of leucine in the regulation of protein metabolism. J Nutr. 2005;135:1553S–1556S. doi: 10.1093/jn/135.6.1553S. [DOI] [PubMed] [Google Scholar]
- 14.Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest. 1998;101:1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layman DK. The role of leucine in weight loss diets and glucose homeostasis. J Nutr. 2003;133:261S–267S. doi: 10.1093/jn/133.1.261S. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Xu M, Lee J, He C, Xie Z. Leucine supplementation increases SIRT1 expression and prevents mitochondrial dysfunction and metabolic disorders in high-fat diet-induced obese mice. Am J Physiol Endocrinol Metab. 2012;303:E1234–E1244. doi: 10.1152/ajpendo.00198.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaughan RA, Garcia-Smith R, Gannon NP, Bisoffi M, Trujillo KA, Conn CA. Leucine treatment enhances oxidative capacity through complete carbohydrate oxidation and increased mitochondrial density in skeletal muscle cells. Amino Acids. 2013;45:901–911. doi: 10.1007/s00726-013-1538-5. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Zemel MB. Leucine modulation of mitochondrial mass and oxygen consumption in skeletal muscle cells and adipocytes. Nutr Metab (Lond) 2009;6:26. doi: 10.1186/1743-7075-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Bai K, He J, Su W, Dong L, Zhang L, Wang T. Leucine improves growth performance of intrauterine growth retardation piglets by modifying gene and protein expression related to protein synthesis. Nutrition. 2016;32:114–121. doi: 10.1016/j.nut.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Institute of Laboratory Animal Resources (US) Guide for the Care and Use of Laboratory Animals. 7th ed. Washington, D.C.: National Academy Press; 1996. [Google Scholar]
- 22.Wang Y, Zhang L, Zhou G, Liao Z, Ahmad H, Liu W, Wang T. Dietary L-arginine supplementation improves the intestinal development through increasing mucosal Akt and mammalian target of rapamycin signals in intra-uterine growth retarded piglets. Br J Nutr. 2012;108:1371–1381. doi: 10.1017/S0007114511006763. [DOI] [PubMed] [Google Scholar]
- 23.National Research Council (US), Committee on Nutrient Requirements of Swine; National Research Council (US), Board on Agriculture and Natural Resources. Nutrient Requirements of Swine. 11th rev. ed. Washington, D.C.: National Academy Press; 2012. [Google Scholar]
- 24.Yi D, Hou Y, Wang L, Ding B, Yang Z, Li J, Long M, Liu Y, Wu G. Dietary N-acetylcysteine supplementation alleviates liver injury in lipopolysaccharide-challenged piglets. Br J Nutr. 2014;111:46–54. doi: 10.1017/S0007114513002171. [DOI] [PubMed] [Google Scholar]
- 25.Atkinson DE. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 26.Weinbach EC. A procedure for isolating stable mitochondria from rat liver and kidney. Anal Biochem. 1961;2:335–343. doi: 10.1016/0003-2697(61)90006-9. [DOI] [PubMed] [Google Scholar]
- 27.Nemeria N, Yan Y, Zhang Z, Brown AM, Arjunan P, Furey W, Guest JR, Jordan F. Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition. J Biol Chem. 2001;276:45969–45978. doi: 10.1074/jbc.M104116200. [DOI] [PubMed] [Google Scholar]
- 28.Shepherd D, Garland PB. The kinetic properties of citrate synthase from rat liver mitochondria. Biochem J. 1969;114:597–610. doi: 10.1042/bj1140597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meixner-Monori B, Kubicek CP, Harrer W, Schreferl G, Rohr M. NADP-specific isocitrate dehydrogenase from the citric acid-accumulating fungus Aspergillus niger. Biochem J. 1986;236:549–557. doi: 10.1042/bj2360549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai JC, Cooper AJ. Brain alpha-ketoglutarate dehydrogenase complex: kinetic properties, regional distribution, and effects of inhibitors. J Neurochem. 1986;47:1376–1386. doi: 10.1111/j.1471-4159.1986.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 31.Mehler AH, Kornberg A, Grisolia S, Ochoa S. The enzymatic mechanism of oxidation-reductions between malate or isocitrate and pyruvate. J Biol Chem. 1948;174:961–977. [PubMed] [Google Scholar]
- 32.Ragan CI. Structure and function of an archetypal respiratory chain complex: NADH-ubiquinone reductase. Biochem Soc Trans. 1990;18:515–516. doi: 10.1042/bst0180515. [DOI] [PubMed] [Google Scholar]
- 33.Medja F, Allouche S, Frachon P, Jardel C, Malgat M, Mousson de Camaret B, Slama A, Lunardi J, Mazat JP, Lombès A. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion. 2009;9:331–339. doi: 10.1016/j.mito.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Krähenbühl S, Talos C, Wiesmann U, Hoppel CL. Development and evaluation of a spectrophotometric assay for complex III in isolated mitochondria, tissues and fibroblasts from rats and humans. Clin Chim Acta. 1994;230:177–187. doi: 10.1016/0009-8981(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 35.Wharton DC, Tzagoloff A. Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 1967;10:245–250. [Google Scholar]
- 36.Taussky HH, Shorr E. A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- 37.Liu J, Yao Y, Yu B, Mao X, Huang Z, Chen D. Effect of folic acid supplementation on hepatic antioxidant function and mitochondrial-related gene expression in weanling intrauterine growth retarded piglets. Livest Sci. 2012;146:123–132. [Google Scholar]
- 38.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F, Caliaro F, Corsetti G, Bottinelli R, Carruba MO, Valerio A, Nisoli E. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metab. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Ogata ES, Swanson SL, Collins JW, Jr, Finley SL. Intrauterine growth retardation: altered hepatic energy and redox states in the fetal rat. Pediatr Res. 1990;27:56–63. doi: 10.1203/00006450-199001000-00017. [DOI] [PubMed] [Google Scholar]
- 41.Lim S, Cho YM, Park KS, Lee HK. Persistent organic pollutants, mitochondrial dysfunction, and metabolic syndrome. Ann N Y Acad Sci. 2010;1201:166–176. doi: 10.1111/j.1749-6632.2010.05622.x. [DOI] [PubMed] [Google Scholar]
- 42.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Yu B, Mao X, He J, Yu J, Zheng P, Huang Z, Chen D. Effects of intrauterine growth retardation and maternal folic acid supplementation on hepatic mitochondrial function and gene expression in piglets. Arch Anim Nutr. 2012;66:357–371. doi: 10.1080/1745039X.2012.710084. [DOI] [PubMed] [Google Scholar]
- 44.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 46.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 47.Maniura-Weber K, Goffart S, Garstka HL, Montoya J, Wiesner RJ. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons RA, Suponitsky-Kroyter I, Selak MA. Progressive accumulation of mitochondrial DNA mutations and decline in mitochondrial function lead to beta-cell failure. J Biol Chem. 2005;280:28785–28791. doi: 10.1074/jbc.M505695200. [DOI] [PubMed] [Google Scholar]
- 49.Morris TJ, Vickers M, Gluckman P, Gilmour S, Affara N. Transcriptional profiling of rats subjected to gestational undernourishment: implications for the developmental variations in metabolic traits. PLoS One. 2009;4:e7271. doi: 10.1371/journal.pone.0007271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatpati LL, Irving BA, Tom A, Bigelow ML, Klaus K, Short KR, Nair KS. The effect of branched chain amino acids on skeletal muscle mitochondrial function in young and elderly adults. J Clin Endocrinol Metab. 2010;95:894–902. doi: 10.1210/jc.2009-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas PJ, Garboczi DN, Pedersen PL. Mutational analysis of the consensus nucleotide binding sequences in the rat liver mitochondrial ATP synthase beta-subunit. J Biol Chem. 1992;267:20331–20338. [PubMed] [Google Scholar]