Abstract
Denmark is a country with high prevalence of livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) clonal complex (CC) 398 in pigs. Even though pig farming is regarded as the main source of human infection or colonization with MRSA CC398, 10–15% of the human cases appear not to be linked to pigs. Following the recent reports of MRSA CC398 in horses in other European countries and the lack of knowledge on S. aureus carriage in this animal species, we carried out a study to investigate whether horses constitute a reservoir of MRSA CC398 in Denmark, and to gain knowledge on the frequency and genetic diversity of S. aureus in horses, including both methicillin-resistant and -susceptible S. aureus (MSSA). Nasal swabs were collected from 401 horses originating from 74 farms, either at their farms or prior to admission to veterinary clinics. Following culture on selective media, species identification by MALDI-TOF MS and MRSA confirmation by standard PCR-based methods, S. aureus and MRSA were detected in 54 (13%) and 17 (4%) horses originating from 30 (40%) and 7 (9%) farms, respectively. Based on spa typing, MSSA differed genetically from MRSA isolates. The spa type prevalent among MSSA isolates was t127 (CC1), which was detected in 12 horses from 11 farms and represents the most common S. aureus clone isolated from human bacteremia cases in Denmark. Among the 17 MRSA carriers, 10 horses from three farms carried CC398 t011 harboring the immune evasion cluster (IEC), four horses from two farms carried IEC-negative CC398 t034, and three horses from two farms carried a mecC-positive MRSA lineage previously associated with wildlife and domestic ruminants (CC130 t528). Based on whole-genome phylogenetic analysis of the 14 MRSA CC398, t011 isolates belonged to the recently identified horse-adapted clone in Europe and were closely related to human t011 isolates from three Danish equine veterinarians, whereas t034 isolates belonged to pig-adapted clones. Our study confirms that horses carry an equine-specific clone of MRSA CC398 that can be transmitted to veterinary personnel, and reveals that these animals are exposed to MRSA and MSSA clones that are likely to originate from livestock and humans, respectively.
Keywords: horse, MRSA, MSSA, CC398, mecA, mecC, IEC, WGS
Introduction
Over the last decade, livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) belonging to clonal complex (CC) 398 has emerged worldwide in various animal species, especially in pigs as well as in people in contact with MRSA-positive animals (Petinaki and Spiliopoulou, 2012). Zoonotic transmission from pigs to humans has been substantiated by various epidemiological studies (Lewis et al., 2008; Köck et al., 2013). In Denmark the number of human infections caused by MRSA CC398 has increased steeply from 13 cases in 2009 to 240 cases in 2014, and decreased to 208 cases in 2015 (DANMAP, 2014, 2015). Presently, the CC398-associated spa types t034 and t011 account for 19% of all MRSA infections in this country (DANMAP, 2014, 2015), and the majority of cases are found in people with direct or indirect (household members) contact with pigs (Voss et al., 2005). However, a steady proportion of MRSA CC398 cases (10–15% of all cases and 37% of infections) occur in people that are not exposed to livestock (Lekkerkerk et al., 2012; DANMAP, 2014, 2015; Larsen et al., 2015), raising questions about the possible source of infection in these people.
Denmark has a population of ~5.7 million people and produces nearly 31 million pigs per year, according to data from 2015 (Statistics-Denmark, 2015a,b). Approximately 88% of the pig farms are positive for MRSA CC3981. New studies in Denmark in 2015 have found one poultry farm, five veal calf farms and eight mink farms MRSA CC398-positive (2, 5, and 8%, respectively) (DANMAP, 2015). Other potential animal reservoirs in Denmark have not been investigated. However, studies from other countries have shown MRSA CC398 carriage in horses (0.5–11%; Van den Eede et al., 2009, 2012) as well as a high prevalence (8–15%) of this CC amongst equine clinical MRSA isolates (van Duijkeren et al., 2010; Vincze et al., 2014). It appears that a recent shift has occurred in the epidemiology of MRSA in horses, with CC398 gradually replacing CC8 as the most common CC (Loeffler et al., 2009; Van den Eede et al., 2009, 2013; van Duijkeren et al., 2010). In addition to the noticeable spread of MRSA CC398 in horses, recent studies suggest that zoonotic transmission between horses and humans may be enhanced by the presence of the phage ΦSa3 in horse isolates (Cuny et al., 2015; Jung et al., 2017). This phage carries the human immune evasion gene cluster (IEC) that provides S. aureus protection from the immune system of humans and horses (Jung et al., 2017). Data on S. aureus carriage and infection in horses are largely biased in favor of MRSA and very little is not about the prevalence and genetic diversity of methicillin-susceptible (MSSA) strains.
The objectives of this study were (i) to investigate whether horses constitute a reservoir of MRSA CC398 in Denmark and (ii) to gain information on the frequency and genetic diversity of S. aureus in this animal species, including both MRSA and MSSA.
Materials and methods
Study population and sampling
Between April and August 2015, nasal swabs were collected from horses located at their farms of origin or immediately upon arrival at two veterinary clinics involved in the study, prior to entering the clinical environment. Horses were excluded from the study if they showed signs of an upper respiratory tract infection involving nasal discharge that could interfere with S. aureus isolation, if they had visited the horse clinic within 30 days of sampling or if the owner was not present to provide written consent to the sampling. The maximum number of horses included per farm was 20. Sampling was performed by veterinarians by inserting a sterile cotton swab moistened with Stuart's medium (BBL™ CultureSwab™, Becton Dickinson, U.S.A.) into the ventral meatus and the nasal vestibulum of both nostrils (Van den Eede et al., 2013). Samples were sent within 24 h to the Department of Veterinary Disease Biology, University of Copenhagen, and analyzed on the day of arrival.
A questionnaire filled by the horse owners was used to collect information about each horse including age, sex, current health status, and history of antimicrobial therapy in the last 6 months prior to sampling. Information was also retrieved about farm type (small private farm, riding school, or stud farm), presence of other animals on the farm (pigs, dogs, cats, cattle, sheep, or other) and the postal code. In addition to the questionnaire information, data on density of pigs was retrieved for each postal code where the horse farms were located. Number of pigs was retrieved from the Danish Central Husbandry Register (CHR) online database of the Danish Veterinary and Food Administration (Danish Veterinary and Food Administration)2 and postal code areas (sqKm) were provided by the Danish Geo Data Agency (Kortforsyningen)3.
The study was single-blinded meaning that the investigators were not informed about the origin of samples except for the postal code. This study did not require approval by an ethics committee according to the Danish Animal Experimentation Act §1.2.
Isolation and characterization of S. aureus
Swabs were enriched 18–24 h at 37°C in 5 ml of Mueller Hinton broth (Sigma-Aldrich, U.S.A.) containing 6.5% NaCl. A 10 μl loopful of broth was plated both onto Brilliance™ MRSA2 agar (Oxoid, UK) and SaSelect™ agar (Bio-Rad, U.S.A) followed by incubation for 24 h at 37°C. One presumptive MRSA colony on Brilliance™ and up to three (if present) presumptive S. aureus colonies on SaSelect™ were sub-cultured onto blood agar. In order to increase the chance of detecting diversity, morphologically distinct colonies were chosen for sub-culturing from the SaSelect™ agar plates when possible. If no presumptive S. aureus was visible on SaSelect™ plates, these were then incubated for another 24 h. Isolates were tested by matrix-assisted laser desorption/ionization time-of-flight mass spectrophotometry (MALDI-TOF MS) (BioMérieux, France) for species confirmation. Confirmed S. aureus isolates were stored at −80°C for further analysis.
A multiplex PCR assay was performed to detect (i) spa encoding the staphylococcal protein A (Kahl et al., 2005), (ii) mecA and mecC encoding methicillin resistance (Oliveira and de Lencastre, 2002; Stegger et al., 2012), (iii) scn, a marker of IEC encoding staphylococcal complement inhibitor protein (scn-F1: 5′TACTTGCGGGAACTTTAGCAA3′ and scn-R1: 5′AATTCATTAGCTAACTTTTCGTTTTGA3′, amplicon size: 130 bp), (iv) the CC398-specific sau1-hsdS1 variant (Stegger et al., 2011) using a newly designed forward primer FP2sau1: 5′GAGAATGATTTTGTTTATAACCCTAG3′ (amplicon size 106 bp), and (v) lukF-PV, a marker of the Panton-Valentine leucocidin (PVL) (Deurenberg et al., 2004). The PCR reactions were carried out in a final volume of 13 μl containing 1 × Qiagen Multiplex PCR Master Mix (Qiagen, Germany), 2 μM of each primer, and 1 μl of bacterial DNA. The following PCR conditions were used: initial denaturation at 94°C for 15 min, followed by 25 cycles consisting of denaturation (94°C for 30 s), annealing (59°C for 60 s), and extension (72°C for 60 s), and a final extension step at 72°C for 10 min. All S. aureus isolates were spa-typed as described previously (Harmsen et al., 2003). BURP cluster analysis of the spa types was performed in the Ridom StaphType software (Ridom GmbH, Germany) using default settings to deduce likely multi-locus sequence types of methicillin-susceptible S. aureus (MSSA) isolates. The staphylococcal cassette chromosome mec (SCCmec) types and subtypes were determined by PCR for all CC398 isolates (Larsen et al., 2016a). The IEC type was determined by the presence of the scn, chp, sak, sea, and sep genes (van Wamel et al., 2006).
Whole genome sequencing (WGS) and phylogenetic analysis of MRSA CC398 isolates
DNA was extracted with the DNeasy Blood & Tissue kit (Qiagen, The Netherlands) and run on an Illumina MiSeq sequencer (Illumina, USA) for 250 bp paired-end sequencing. For comparative purposes, we included in the WGS-based phylogenetic analysis 16 porcine MRSA CC398 isolates from a previous Danish study (Grøntvedt et al., 2016) and 11 human MRSA CC398 isolates from the national MRSA database. The porcine isolates were selected to represent the three major phylogenetic groups in pigs in Denmark (Larsen, unpublished data). The human isolates included all IEC-positive t011 isolates obtained from humans in Denmark in the period from 2004 to 2015 (Table 1). Demographic data and information about risk factors for MRSA acquisition were obtained from written records and through telephone interviews. Use of these data has been approved by the Danish Data Protection Agency (protocol no. 2001-14-0021). Bioinformatic and phylogenetic analyses were performed as described previously (Larsen et al., 2016b). A previously published worldwide collection of 89 S. aureus CC398 isolates were included for comparative purposes (Price et al., 2012). The maximum-likelihood phylogeny was inferred from a total of 3,954 single-nucleotide polymorphisms (SNPs), of which 1,098 were parsimony-informative. We used 100 random bootstraps replicates for calculating branch support with PhyML 3.0 (Guindon et al., 2010). For illustration of the phylogenetic tree, we used R version 3.3.1 with packages ggtree (Yu et al., 2017) and ggplot2 (Wickham, 2009). Genomes were screened for specific genes by mapping sequence reads against reference sequences. References for scn, chp, sak, and sea were obtained from the sequence of S. aureus Newman (GenBank accession no. DQ530361); reference for sep was obtained from S. aureus COL (NC_002951); reference for vWbp Seq1 was obtained from S. aureus DL584 (HM228920); and reference for lukP and lukQ were obtained from S. aureus 3711 (LT671578). Genotypic resistance profiles were obtained with Mykrobe Predictor v0.4.3 build 0-gd6c8714 (Bradley et al., 2015).
Table 1.
Description of human individuals colonized or infected with IEC-positive spa type t011 MRSA CC398 in Denmark.
| Case | Sex | Age, Y | Isolation year | Specimen | Epidemiologic findings |
|---|---|---|---|---|---|
| A | F | 38 | 2010 | Screening (nares) | Wife of pig farmer (family A) |
| B | F | 0 | 2010 | Screening (nares) | Child of pig farmer (family A) |
| C | M | 50 | 2011 | Screening (nares) | Pig farmer |
| D | M | 27 | 2014 | Wound swab | Horse veterinarian (family B) |
| E | M | 61 | 2014 | Screening (nares) | Horse veterinarian (family B) |
| F | M | 24 | 2014 | Abscess | Pig farmer (family C) |
| G | F | 32 | 2014 | Wound swab | Horse contact |
| H | F | 4 | 2014 | Screening (perineum) | Niece of pig farmer |
| I | F | 44 | 2014 | Wound swab | Horse veterinarian |
| J | F | 73 | 2015 | Wound swab | No livestock contact |
| K | M | 58 | 2015 | Screening (nares) | Father of pig farmer (family C) |
Statistical analysis
Occurrence of total MRSA, MRSA CC398, and MRSA CC398 t034 (the only MRSA type of this study associated to pigs) were analyzed at the individual horse level and at the farm level by logistic analysis using the glmer function from the lme4 package (Bates et al., 2015) in R version 3.3.0 (RcoreTeam, 2016). For the horse-level analysis, age, sex, health status, farm type, and antimicrobial treatments were included as fixed effects; and farm was included as a random effect. For the farm-level analysis, farm type, presence of dogs, cats, cows, pigs, other species, postal code, density of pigs, and pig farms in the postal code were included as fixed effects. The 95% confidence interval of the prevalence values were calculated by the modified Wald method using the Graph Pad software Quick Calcs (GraphPad)4. Figure 1 was generated using the mapDK (Barfort, 2015) and ggplot2 (Wickham, 2009) packages in R 3.3.0 (RcoreTeam, 2016).
Figure 1.
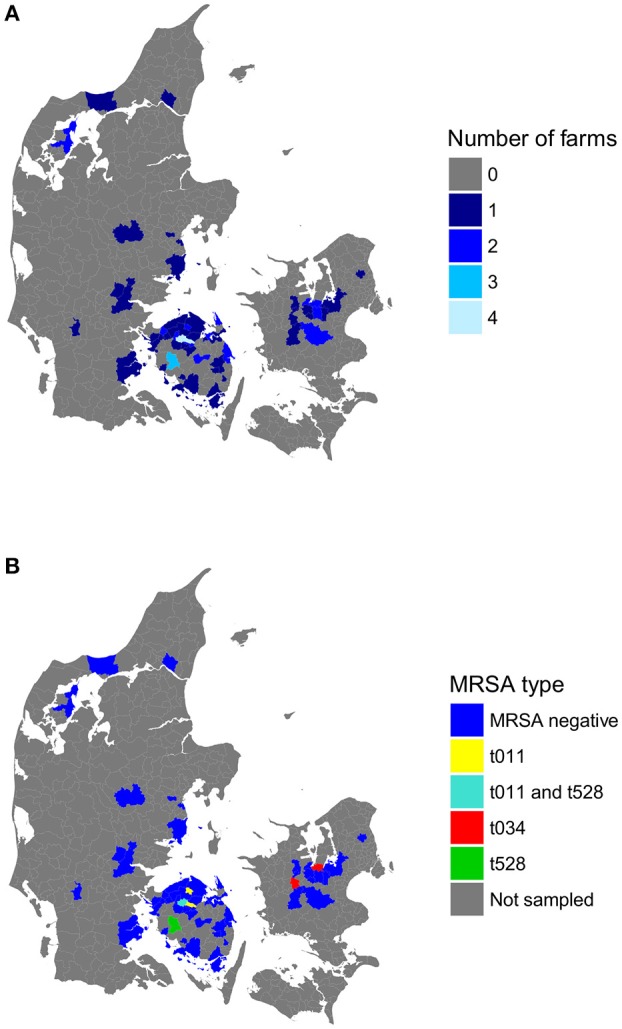
Number of horse farms sampled (A) and MRSA types detected (B) in each of the 53 postal codes included in the study. No farms were sampled for the postal codes colored in gray. Postal codes not associated with MRSA-positive farms are labeled as “MRSA-negative.”
Data access
The whole-genome sequence data from this study have been submitted to the NCBI Sequence Read Archive under BioProject PRJEB19362.
Results
A total of 401 horses originating from 74 farms (41 small private farms, 20 riding schools, and 13 stud farms) were included in the study. The number of horses sampled per farm ranged from 1 to 20 with a median value of four. The sampled population consisted of 211 females and 190 males with a median age of 9 years (range: 3 months to 28 years). Three hundred and ninety-five horses were sampled on the farm where they lived, and six horses were sampled upon arrival to the clinic. Of them, 393 horses did not display any clinical signs at the time of sampling. The remaining eight horses had either laminitis (n = 2), colic/intestine alterations (n = 2), Cushing's syndrome (n = 1), dermatitis (n = 1), infection of the hoof (n = 1), or lung infection (n = 1). Three hundred and fifty-five horses had not been treated with antimicrobial agents in the preceding 6 months. Among the remaining horses, antimicrobial treatment was reported in 26 horses, and the treatment status was unknown for 20 horses (Data Sheet 1). The antimicrobial drugs used included trimethoprim/sulfadiazin (n = 12), cefquinome (n = 6), benzylpenicillinprocain (n = 3), ethacillin (n = 3), ampicillin (n = 1), doxycillin (n = 1), and tobramycin (n = 1). Three of the treated horses had received unknown antimicrobial drugs, and four had been treated with more than one drug. Most farms had cats (n = 52) and/or dogs (n = 51), three had cows, two had pigs, and five farms had other animals including alpaca, deer, and mouflon (n = 1), alpaca only (n = 1), hens (n = 1), rats/mice (n = 1), or fish (n = 1).
A total of 79 S. aureus (MRSA and MSSA) isolates were obtained from 54 horses (13.5%) originating from 30 farms (40.5%). MRSA was isolated from 17 horses (4.2%) from seven farms (9.5%) located in Funen and Zealand (Figure 1B; Table 2). The identified MRSA isolates belonged to three genotypes based on the PCR results: (i) IEC-positive MRSA CC398 t011 (10 horses from three farms), (ii) IEC-negative MRSA CC398 t034 (four horses from two farms), and (iii) IEC-negative mecC-MRSA CC130 t528 (three horses from two farms; Table 3). Each MRSA-positive farm harbored only one of these genotypes. One of the 17 MRSA carriers reported health problems (dermatitis) and unknown antimicrobial history, whereas the remaining 16 carriers were healthy and had received no antimicrobials in the previous 6 months.
Table 2.
Staphylococcus aureus, MRSA and MRSA CC398 prevalence in horses and horse farms.
| Horses (n = 401) | Horse farms (n = 74) | |||
|---|---|---|---|---|
| No. positive | Prevalence% (95% CI) | No. positive | Prevalence% (95% CI) | |
| S. aureus | 54 | 13.5 (10.5–17.2) | 30 | 40.5 (30.1–51.9) |
| MRSA | 17 | 4.2 (2.6–6.7) | 7 | 9.5 (4.4–18.5) |
| MRSA CC398 | 14 | 3.5 (2.1–5.8) | 5 | 6.8 (2.6–15.2) |
Table 3.
Characterization of MRSA and MSSA isolates from horses in Denmark.
| spa type | mecA/mecC | IEC | Associated ST/CC (ref) | No. positive horses | No. positive farms | Co-colonization with |
|---|---|---|---|---|---|---|
| t011 | mecA | + | ST398/CC398 (Nemeghaire et al., 2014) | 10 | 3 | MSSA t127b, t865, t15815 |
| t034 | mecA | − | ST398/CC398 (Crombé et al., 2012) | 4 | 2 | |
| − | – | ST398/CC398 (Crombé et al., 2012) | 2 | 1 | MSSA t127 | |
| t127 | − | − | ST1/CC1 (Battisti et al., 2010) | 11 | 10 | MRSA t528; MSSA t034, t549 |
| − | + | ST1/CC1 (Battisti et al., 2010) | 1 | 1 | MRSA t011 | |
| t528 | mecC | − | ST130/CC130 (Espinosa-Gongora et al., 2015) | 3 | 2 | MSSA t127 |
| t549 | − | − | ST1660c (Cuny et al., 2016) | 5 | 3 | MSSA t1508, t3043 |
| t701 | − | + | ST6/CC5 (Yan et al., 2012) | 1 | 1 | |
| t865 | − | − | Unknown | 1 | 1 | MRSA t011 |
| t1166 | − | − | ST133/CC133 (Gharsa et al., 2015) | 2 | 1 | |
| t1294 | − | − | Unknown | 3 | 3 | |
| t1508 | − | − | Unknown | 2 | 2 | MSSA t549, t3043 |
| t1943 | − | + | ST5 or ST130/CC5 or CC130 (Donker et al., 2009) | 1 | 1 | |
| t2420 | − | − | ST133/CC133 (Gómez et al., 2015) | 1 | 1 | |
| t3043 | − | − | ST1660c (Sieber et al., 2011) | 3 | 2 | MSSA t549, t1508 |
| t4735 | − | − | ST133/CC133a | 1 | 1 | |
| t5100 | − | − | ST1/CC1 (Seidl et al., 2015) | 1 | 1 | |
| t15815 | − | − | Unknown | 2 | 1 | MRSA t011 |
| t15816 | − | − | ST1660a,c | 2 | 1 | MSSA t1508 |
Based on clustering by BURP analysis.
IEC-positive.
Does not belong to a clonal complex.
Based on PCR results, 50 of the 79 S. aureus isolates were MSSA belonging to 15 spa types and carried by 41 horses. The population structure of the MSSA isolates differed notably from that of MRSA isolates. None of the MSSA isolates belonged to CC398. The two most prevalent spa types were t127 (12 horses) and t549 (4 horses), which are associated to CC1 and ST1660, respectively (Table 3). Four S. aureus isolates were confirmed by MALDI-TOF but were lost and could not be typed. Two spa types (t15815 and t15816) were identified in this study for the first time. Most S. aureus-positive horses (48 of the 54) carried only one spa type, whereas six horses were found to simultaneously carry two unrelated spa types, either two MSSA or one MSSA and one MRSA (Table 4). In addition to the MRSA CC398 t011 isolates, three MSSA CCs harbored scn: CC1 t127 (n = 2), CC5 t701 (n = 1), and CC5/CC130 t1943 (n = 1). The BURP analysis grouped the spa types into five clusters and provided presumptive ST/CC information of two spa types without previously reported multi-locus sequence type: t4735 (clustering with t2420) presumably belonging to CC133 (Gómez et al., 2015), and t15816 (clustering with t549 and t3043) presumably belonging to ST1660 (Sieber et al., 2011; Cuny et al., 2016; Table 3). None of the isolates carried PVL.
Table 4.
Genotypic features of MRSA CC398 isolates included in this study.
| Isolate ID | Species | spa type | SCCmec type | scn | chp | sak | sea | sep | IEC type | SNP 309-2 | Clade |
|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H42 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H43 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H44 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H46 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H155 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H157 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H160 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H161 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H163 | Horse | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| Case D | Human | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| Case E | Human | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| Case I | Human | t011 | IV(2B)a | + | + | + | − | − | B | + | H |
| H223 | Horse | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| H224 | Horse | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| Case F | Human | t011 | V(5C2&5)c | + | + | + | + | − | A | − | P1 |
| Case H | Human | t011 | V(5C2&5)c | + | + | + | + | − | A | − | P1 |
| Case J | Human | t011 | V(5C2&5)c | + | + | + | - | − | B | − | P1 |
| Case K | Human | t011 | V(5C2&5)c | + | + | + | + | − | A | − | P1 |
| 55-100-009 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| 55-100-067 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| 55-100-079 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| 55-100-124 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| 55-100-133 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P1 |
| 55-100-016 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P2 |
| 55-100-026 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P2 |
| 55-100-056 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P2 |
| 55-100-080 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P2 |
| 55-100-132 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P2 |
| H280 | Horse | t034 | V(5C2)* | − | − | − | − | − | − | − | P3 |
| H285 | Horse | t034 | V(5C2)* | − | − | − | − | − | − | − | P3 |
| Case A | Human | t011 | V(5C2&5)c | + | + | + | − | − | B | − | P3 |
| Case B | Human | t011 | V(5C2&5)c | + | + | + | − | − | B | − | P3 |
| Case C | Human | t011 | V(5C2&5)c | + | + | + | − | − | B | − | P3 |
| Case G | Human | t011 | V(5C2)b | + | + | + | − | − | B | − | P3 |
| 55-100-042 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
| 55-100-073 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
| 55-100-081 | Pig | t011 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
| 55-100-087 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
| 55-100-096 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
| 55-100-137 | Pig | t034 | V(5C2&5)c | − | − | − | − | − | − | − | P3 |
All isolates were negative to vWbp Seq1, lukP, lukQ.
SCCmec subtype non-typeable.
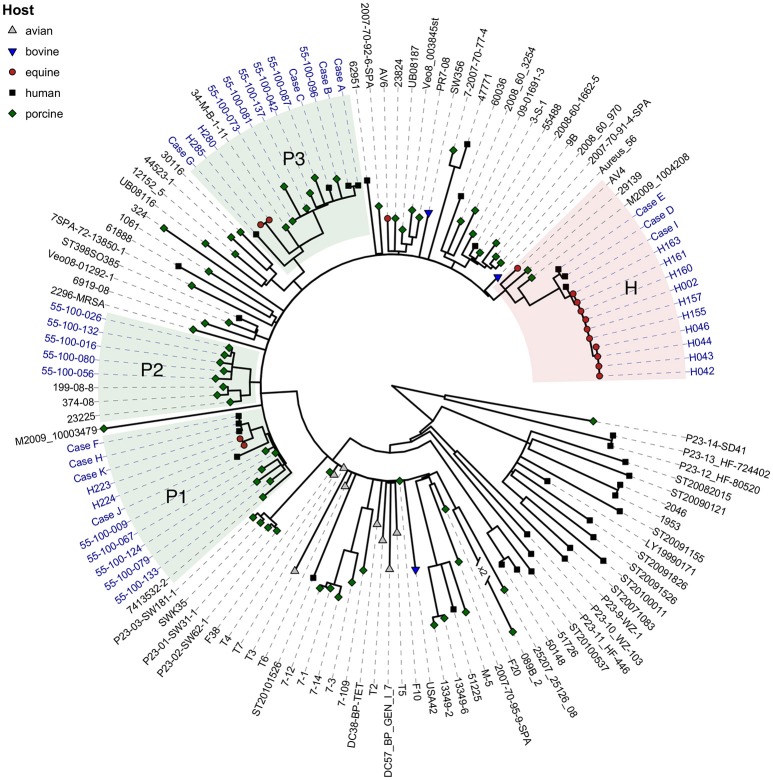
Table 4 lists the genotypic characteristics of the MRSA CC398 isolated in this study. The four equine MRSA CC398 isolates with spa type t034 carried SCCmec V(5C2&5)c, lacked IEC, and clustered with two of the three widespread porcine clones in Denmark (hereafter referred to as pig-associated MRSA CC398). The 10 MRSA t011 from horses carried SCCmec type IV(2B)a and IEC type B, and formed a separate branch in the maximum-likelihood phylogeny (hereafter referred to as horse-adapted MRSA CC398; Figure 2; Table 4). Three of the 11 human MRSA t011 IEC-positive isolates form a monophyletic branch with 100% bootstrap support (Supplementary Image 1) together with all equine isolates with spa type t011. These isolates originated from equine veterinarians, including two members of the same family. The remaining eight human isolates either grouped together with, or were nearest neighbors to the IEC-negative spa type t034 isolates from pigs and horses (Figure 2; Table 4), and originated from two pig farm workers, four family members of farm workers, one person with contact to horses, and one person without any contact to livestock. The 10 horse isolates and the three closely related human isolates carried the unique SNP called 309-2 (Table 4). We also found SNP 309-2 in three IEC-negative MRSA CC398 isolates from the worldwide collection, including two porcine spa type t011 isolates from Hungary and Italy and one equine spa type t1451 isolate from Belgium. These three IEC-negative isolates were located on the same branch as, but basal to, the IEC-positive horse-adapted isolates from Denmark (Figure 2), suggesting that IEC was acquired after diversification of the horse-adapted clone.
Figure 2.
Maximum likelihood phylogeny of MRSA CC398 isolates from Denmark (blue label) and from a worldwide collection (black label; Price et al., 2012). The maximum-likelihood phylogeny was inferred from a total of 3,954 single-nucleotide polymorphisms (SNPs), of which 1,098 were parsimony-informative. Color and shape of the branch endpoints indicate sample origin (see Legend). The three widespread Danish pig clones (clades P1–P3) are highlighted in green and the red-shaded area (clade H) highlights the isolates with the horse-specific marker SNP 309-2. The tree is midpoint rooted and numbers in broken branches indicate reduction factor in individual branch length.
All horse and human isolates within the horse-specific clade carried aacA-aphD, the gentamicin and kanamycin resistance determinant (Data Sheet 2). The gene was otherwise only found in two livestock-associated MRSA isolates from the worldwide collection, including a porcine spa type t011 isolate from Italy and a bovine spa type t567 isolate from Belgium, which is the closest neighbor to the horse-adapted clone. None of the isolates within the horse-specific clade carried the genes encoding LukPQ.
Occurrence of total MRSA, MRSA CC398, or pig-adapted MRSA CC398 t034 showed no significant associations at the individual-level analysis with age, health status, and antimicrobial use in the preceding 6 months, or at the farm-level analysis with farm type, presence of other animals on the farm or density of pigs or pig farms in the same geographical location based on postal code (Supplementary Image 2).
Discussion
We detected MRSA in 4.2% of the horses in the study population, which mainly included healthy horses without recent admission to clinics or antimicrobial treatment. This prevalence is higher than those reported by previous studies in healthy horses from Canada, Belgium, Ireland, and the UK (0–1.7%; Burton et al., 2008; Abbott et al., 2010; Maddox et al., 2012; Van den Eede et al., 2012). A previous Danish study conducted in 2005 failed to detect MRSA amongst 100 horses, including both healthy and diseased horses (Bagcigil et al., 2007). Although, the study populations are not fully comparable, these data suggest an increase in the prevalence of MRSA in the horse population in Denmark over the last decade. The recent spread of MRSA in horses potentially poses a health risk to equine veterinary practitioners, horse owners, and other people in contact with positive animals. It also poses risks to animal health as evidenced by the increasing prevalence of MRSA infections in horses worldwide (Cuny and Witte, 2016).
Our study shows that horses in Denmark are a reservoir for a variety of MRSA genotypes, including both horse-adapted and pig-adapted MRSA CC398 as well as MRSA CC130 harboring mecC. IEC-positive MRSA CC398 t011 harboring SNP 309-2, which is a marker of a previously identified horse-adapted clone (Abdelbary et al., 2014), constituted 59% of the equine MRSA isolates. This clone was recently shown to be the most common MRSA type in clinical samples from horses in central Europe (Abdelbary et al., 2014). The IEC-positive MRSA CC398 t011 isolates shared other characteristics with horse isolates from Germany and the Netherlands in that they carried SCCmec type IV(2B)a and the gentamicin and kanamycin resistance determinant, aacA-aphD (Cuny et al., 2008; van Duijkeren et al., 2010). The latter genotypic feature may reflect the widespread usage of gentamicin in horses.
Whole-genome phylogenetic analysis showed that three of the human isolates (one from healthy carriage and two from infections) were highly similar to those isolated in this study from horses (Figure 2; Table 4). Interestingly, all these isolates originated from equine veterinarians, indicating an occupational risk for MRSA CC398 carriage and infection in veterinary staff. The horse-adapted clone accounted for 0.2% (2/834) of all MRSA CC398 infections during 2004–2015, according to the national MRSA database. In a previous study by Abdelbary et al. (2014), the horse-adapted clone was detected in Austrian veterinary personnel but not in other humans. Danish horse veterinarians have previously been associated to a higher risk of MRSA carriage compared to people not exposed to horses (Moodley et al., 2008). The current MRSA control policy in Denmark includes screening and decolonization of people at risk of MRSA carriage upon hospital admission, e.g., pig farmers. Whether or not horse veterinarians should be included as another risk group requires further epidemiological evidence through studies that assess the prevalence of MRSA carriage and infection in people in contact with horses, or investigate the exposure to horses in MRSA-positive patients.
The equine IEC-negative MRSA CC398 t034 isolates were closely related to two of the three clones that are widespread among pigs in Denmark (Figure 2), suggesting a recent transmission between these two animal hosts. The origin of pig-adapted MRSA CC398 in the horses is unknown, since pigs were not present in any of the positive horse farms. The lack of contact with pigs suggests that horses could be a reservoir for the pig-adapted MRSA CC398 t034 in addition to pigs. Moreover, the presence of SNP 309-2 in the two porcine t011 isolates from Hungary and Italy suggests a possible jump of MRSA CC398 from pigs to horses, where it subsequently acquired IEC through acquisition of the ΦSa3 phage.
The mecC-positive MRSA CC130 t528 was detected for the first time in horses in Denmark. mecC-positive MRSA CC130 and CC49 belonging to other spa types have been isolated from horses in France and Germany (Haenni et al., 2015; Cuny et al., 2016). MRSA CC130 is known to have a broad host spectrum, mainly ruminants among domestic animals (Paterson et al., 2012; Loncaric et al., 2013; Monecke et al., 2013; Gomez et al., 2014). In Denmark, mecC-positive MRSA CC130 t528 has been recognized as a cause of human infections (Petersen et al., 2013) and as a commensal in the nasal cavity of cattle (García-Álvarez et al., 2011), small ruminants (Eriksson et al., 2013), and wild animal species at the Copenhagen Zoo (Espinosa-Gongora et al., 2015). Further insight into the phylogenetic relationship between these isolates could shed light on the possible role of horses in the evolution and dissemination of this MRSA clone.
The overall S. aureus carriage rate in the study population was 13.5%, which is slightly higher than the MSSA prevalence reported in healthy horses in Canada (7.9%; Burton et al., 2008). The most common MSSA lineage, CC1 t127, was isolated from 12 horses at 11 farms. This is a ubiquitous spa type that has previously been isolated from horse MRSA infections (Cuny et al., 2008), and in Denmark it is the most common spa type isolated from human cases of S. aureus bacteremia (DANMAP, 2015). The apparent lack of host-specificity of t127 and thereby potential zoonotic relevance requires confirmation by high-resolution genotyping methods such as the WGS approach used in this study for CC398 isolates. The second most common spa type, t549 (ST1660), is considered a horse-adapted S. aureus clone (Cuny et al., 2016).
No significant associations were found between MRSA carriage and the variables covered by the questionnaire (age, health status, recent antimicrobial therapy, farm type, presence of other animals on the farm, and density of pigs or pig farms in the area of the farm). Our data suggest that antimicrobial use is not a major risk factor for MRSA carriage in horses. The factors contributing to the recent spread of MRSA CC398 in the horse population in Europe remain unknown. Further epidemiological studies are needed to assess whether MRSA transmission may be enhanced by movement of horses and horse owners in relation to sport and breeding activities.
To conclude, horses can be healthy carriers of distinct MSSA and MRSA clones that are implicated in human infections. In addition to horse-adapted MRSA CC398, which is a rare cause of human infection, horses in Denmark are exposed to distinct MRSA clones that are likely to originate from other animal species (CC130 t528 and pig-adapted CC398 t034) as well as to a MSSA clone that is prevalent in human cases of S. aureus bacteremia in Denmark (CC1 t127). Further research is needed to determine the stability of MRSA and MSSA carriage in horses, to identify risk factors and elucidate mechanisms of transmission between equine farms, and to quantify the risk of infection in people exposed to horses.
Author contributions
Design of the work: LG, AM, PD, JL; Acquisition, analysis, or interpretation of data: MI, CE, RNS, RM, LH; Drafting the work or revising it critically for important intellectual content: MI, CE, PD, RNS, AM, RS, JL, LG; Final approval of the version to be published (all authors).
Funding
The study was funded by the Danish Pig Levy Fund (Svineafgiftsfonden), and the Knowledge Centre for Agriculture/Danish Pig Research Centre (SEGES). SEGES Horses and the Danish Equestrian Federation funded whole-genome sequencing of MRSA isolates obtained from horses.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Madeleine Nousiainen from the Estonian University of Life Sciences is acknowledged for assistance with sampling and sample processing.
Footnotes
1Danish Veterinary and Food Administration Fødevarestyrelsen. Available online at: https://www.foedevarestyrelsen.dk/Nyheder/Aktuelt/Documents/MRSA%20ekspertgruppe%20-%20resultatene%20forekomst%20af%20husdyr-MRSA%20i%20svin%202016.pdf.
2Danish Veterinary and Food Administration Fødevarestyrelsen, Denmark. Available online at: www.foedevarestyrelsen.dk/.
3Kortforsyningen Danish Geo Data Agency. Available online at: http://kortforsyningen.dk/.
4GraphPad Graph Pad Quick Calcs. Available online at: http://www.graphpad.com/quickcalcs/confInterval1/.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00543/full#supplementary-material
Information about the health and antimicrobial treatments of 49 horses. The remaining 352 horses included in the study were reported as both, healthy and without any antimicrobial treatments in the 6 months prior to the study, and were hence excluded from this table.
Summary table of resistance profiles obtained from Mykrobe Predictor including responsible genes/variants found.
Maximum likelihood phylogeny of relevant clades containing bootstrap values. The phylogeny is drawed as cladogram.
Density of pigs (pigs/sqKm) in each of the 53 postal codes included in the study and the corresponding MRSA type(s) found. Postal codes without identified MRSA-positive horses are labeled as “MRSA negative.”
References
- Abbott Y., Leggett B., Rossney A. S., Leonard F. C., Markey B. K. (2010). Isolation rates of meticillin-resistant Staphylococcus aureus in dogs, cats and horses in Ireland. Vet. Rec. 166, 451–455. 10.1136/vr.b4814 [DOI] [PubMed] [Google Scholar]
- Abdelbary M. M. H., Wittenberg A., Cuny C., Layer F., Kurt K., Wieler L. H., et al. (2014). Phylogenetic analysis of Staphylococcus aureus CC398 reveals a sub-lineage epidemiologically associated with infections in horses. PLoS ONE 9:e88083. 10.1371/journal.pone.0088083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagcigil F. A., Moodley A., Baptiste K. E., Jensen V. F., Guardabassi L. (2007). Occurrence, species distribution, antimicrobial resistance and clonality of methicillin- and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet. Microbiol. 121, 307–315. 10.1016/j.vetmic.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Barfort S. (2015). mapDK: Maps of Denmark. R Package Version 0.3.0.
- Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Battisti A., Franco A., Merialdi G., Hasman H., Iurescia M., Lorenzetti R., et al. (2010). Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 142, 361–366. 10.1016/j.vetmic.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Bradley P., Gordon N. C., Walker T. M., Dunn L., Heys S., Huang B., et al. (2015). Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 6:10063. 10.1038/ncomms10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton S., Reid-Smith R., McClure J. T., Weese J. S. (2008). Staphylococcus aureus colonization in healthy horses in Atlantic Canada. Can. Vet. J. 49, 797–799. [PMC free article] [PubMed] [Google Scholar]
- Crombé F., Willems G., Dispas M., Hallin M., Denis O., Suetens C., et al. (2012). Prevalence and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus among pigs in belgium. Microb. Drug Resist. 18, 125–131. 10.1089/mdr.2011.0138 [DOI] [PubMed] [Google Scholar]
- Cuny C., Abdelbary M. M. H., Köck R., Layer F., Scheidemann W., Werner G., et al. (2016). Methicillin-resistant Staphylococcus aureus from infections in horses in Germany are frequent colonizers of veterinarians but rare among MRSA from infections in humans. One Health 2, 11–17. 10.1016/j.onehlt.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny C., Abdelbary M., Layer F., Werner G., Witte W. (2015). Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 177, 219–223. 10.1016/j.vetmic.2015.02.031 [DOI] [PubMed] [Google Scholar]
- Cuny C., Witte W. (2016). MRSA in equine hospitals and its significance for infections in humans. Vet. Microbiol. 200, 59–64. 10.1016/j.vetmic.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Cuny C., Strommenger B., Witte W., Stanek C. (2008). Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb. Drug Resist. 14, 307–310. 10.1089/mdr.2008.0845 [DOI] [PubMed] [Google Scholar]
- DANMAP (2014). Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. DANMAP. [Google Scholar]
- DANMAP (2015). Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. DANMAP. [Google Scholar]
- Deurenberg R. H., Vink C., Driessen C., Bes M., London N., Etienne J., et al. (2004). Rapid detection of Panton–Valentine leukocidin from clinical isolates of Staphylococcus aureus strains by real-time PCR. FEMS Microbiol. Lett. 240, 1045–1048. 10.1016/j.femsle.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Donker G. A., Deurenberg R. H., Driessen C., Sebastian S., Nys S., Stobberingh E. E. (2009). The population structure of Staphylococcus aureus among general practice patients from The Netherlands. Clin. Microbiol. Infect. 15, 137–143. 10.1111/J.1469-0691.2008.02662.X [DOI] [PubMed] [Google Scholar]
- Eriksson J., Espinosa-Gongora C., Stamphøj I., Larsen A. R., Guardabassi L. (2013). Carriage frequency, diversity and methicillin resistance of Staphylococcus aureus in Danish small ruminants. Vet. Microbiol. 163, 110–115. 10.1016/j.vetmic.2012.12.006 [DOI] [PubMed] [Google Scholar]
- Espinosa-Gongora C., Harrison E. M., Moodley A., Guardabassi L., Holmes M. A. (2015). MRSA carrying mecC in captive mara. J. Antimicrob. Chemother. 70, 1622–1624. 10.1093/jac/dkv024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Álvarez L., Holden M. T., Lindsay H., Webb C. R., Brown D. F., Curran M. D., et al. (2011). Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11, 595–603. 10.1016/S1473-3099(11)70126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharsa H., Ben Slama K., Gómez-Sanz E., Lozano C., Zarazaga M., Messadi L., et al. (2015). Molecular characterization of Staphylococcus aureus from nasal samples of healthy farm animals and pets in Tunisia. Vector Borne Zoonotic Dis. 15, 109–115. 10.1089/vbz.2014.1655 [DOI] [PubMed] [Google Scholar]
- Gomez P., Gonzalez-Barrio D., Benito D., Garcia J. T., Vinuela J., Zarazaga M., et al. (2014). Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J. Antimicrob. Chemother. 69, 2061–2064. 10.1093/jac/dku100 [DOI] [PubMed] [Google Scholar]
- Gómez P., Lozano C., González-Barrio D., Zarazaga M., Ruiz-Fons F., Torres C. (2015). High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in a semi-extensive red deer (Cervus elaphus hispanicus) farm in Southern Spain. Vet. Microbiol. 177, 326–331. 10.1016/j.vetmic.2015.03.029 [DOI] [PubMed] [Google Scholar]
- Grøntvedt C. A., Elstrøm P., Stegger M., Skov R. L., Skytt Andersen P., Larssen K. W., et al. (2016). Methicillin-resistant Staphylococcus aureus CC398 in humans and pigs in norway: a “one health” perspective on introduction and transmission. Clin. Infect. Dis. 63, 1431–1438. 10.1093/cid/ciw552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Haenni M., Châtre P., Dupieux C., Métayer V., Maillard K., Bes M., et al. (2015). mecC-positive MRSA in horses. J. Antimicrob. Chemother. 70:dkv278. 10.1093/jac/dkv278 [DOI] [PubMed] [Google Scholar]
- Harmsen D., Claus H., Witte W., Rothgänger J., Claus H., Turnwald D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P., Abdelbary M. M. H., Kraushaar B., Fetsch A., Geisel J., Herrmann M., et al. (2017). Impact of bacteriophage Saint3 carriage on the immune evasion capacity and hemolytic potential of Staphylococcus aureus CC398. Vet. Microbiol. 200, 46–51. 10.1016/j.vetmic.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Kahl B. C., Mellmann A., Deiwick S., Peters G., Harmsen D. (2005). Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 43, 502–505. 10.1128/JCM.43.1.502-505.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck R., Schaumburg F., Mellmann A., Köksal M., Jurke A., Becker K., et al. (2013). Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS ONE 8:e55040. 10.1371/journal.pone.0055040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J., Clasen J., Hansen J. E., Paulander W., Petersen A., Larsen A. R., et al. (2016a). Copresence of tet(K) and tet(M) in livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 is associated with increased fitness during exposure to sublethal concentrations of tetracycline. Antimicrob. Agents Chemother. 60, 4401–4403. 10.1128/AAC.00426-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J., Petersen A., Sørum M., Stegger M., van Alphen L., Valentiner-Branth P., et al. (2015). Meticillin-resistant Staphylococcus aureus CC398 is an increasing cause of disease in people with no livestock contact in Denmark, 1999 to 2011. Euro Surveill. 20. 10.2807/1560-7917.ES.2015.20.37.30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J., Stegger M., Andersen P. S., Petersen A., Larsen A. R., Westh H., et al. (2016b). Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 63, 1349–1352. 10.1093/cid/ciw532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkerkerk W. S. N., van de Sande-Bruinsma N., van der Sande M. A. B., Tjon-A-Tsien A., Groenheide A., Haenen A., et al. (2012). Emergence of MRSA of unknown origin in the Netherlands. Clin. Microbiol. Infect. 18, 656–661. 10.1111/j.1469-0691.2011.03662.x [DOI] [PubMed] [Google Scholar]
- Lewis H. C., Mølbak K., Reese C., Aarestrup F. M., Selchau M., Sørum M., et al. (2008). Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg. Infect. Dis. 14, 1383–1389. 10.3201/eid1409.071576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler A., Kearns A. M., Ellington M. J., Smith L. J., Unt V. E., Lindsay J. A., et al. (2009). First isolation of MRSA ST398 from UK animals: a new challenge for infection control teams? J. Hosp. Infect. 72, 269–271. 10.1016/j.jhin.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Loncaric I., Kubber-Heiss A., Posautz A., Stalder G. L., Hoffmann D., Rosengarten R., et al. (2013). Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J. Antimicrob. Chemother. 68, 2222–2225. 10.1093/jac/dkt186 [DOI] [PubMed] [Google Scholar]
- Maddox T. W., Clegg P. D., Diggle P. J., Wedley A. L., Dawson S., Pinchbeck G. L., et al. (2012). Cross-sectional study of antimicrobial-resistant bacteria in horses. Part 1: prevalence of antimicrobial-resistant Escherichia coli and methicillin-resistant Staphylococcus aureus. Equine Vet. J. 44, 289–296. 10.1111/j.2042-3306.2011.00441.x [DOI] [PubMed] [Google Scholar]
- Monecke S., Gavier-Widen D., Mattsson R., Rangstrup-Christensen L., Lazaris A., Coleman D. C., et al. (2013). Detection of mecC-positive Staphylococcus aureus (CC130-MRSA-XI) in diseased european hedgehogs (Erinaceus europaeus) in Sweden. PLoS ONE 8:e66166. 10.1371/journal.pone.0066166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley A., Nightingale E. C., Stegger M., Nielsen S. S., Skov R. L., Guardabassi L. (2008). High risk for nasal carriage of methicillin-resistant Staphylococcus aureus among Danish veterinary practitioners. Scand. J. Work Environ. Health 34, 151–157. 10.5271/sjweh.1219 [DOI] [PubMed] [Google Scholar]
- Nemeghaire S., Argudín M., Haesebrouck F., Butaye P., Tenhagen B., Köster G., et al. (2014). Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet. Res. 10:153. 10.1186/1746-6148-10-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. C., de Lencastre H. (2002). Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 2155–2161. 10.1128/AAC.46.7.2155-2161.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson G. K., Larsen A. R., Robb A., Edwards G. E., Pennycott T. W., Foster G., et al. (2012). The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J. Antimicrob. Chemother. 67, 2809–2813. 10.1093/jac/dks329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A., Stegger M., Heltberg O., Christensen J., Zeuthen A., Knudsen L. K., et al. (2013). Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin. Microbiol. Infect. 19, E16–E22. 10.1111/1469-0691.12036 [DOI] [PubMed] [Google Scholar]
- Petinaki E., Spiliopoulou I. (2012). Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin. Microbiol. Infect. 18, 626–634. 10.1111/j.1469-0691.2012.03881.x [DOI] [PubMed] [Google Scholar]
- Price L. B., Stegger M., Hasman H., Aziz M., Larsen J., Andersen P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3:e00305-11. 10.1128/mbio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RcoreTeam (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Available online at: https://www.r-project.org/ [Google Scholar]
- Seidl K., Leimer N., Palheiros Marques M., Furrer A., Holzmann-Bürgel A., Senn G., et al. (2015). Clonality and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus at the University Hospital Zurich, Switzerland between 2012 and 2014. Ann. Clin. Microbiol. Antimicrob. 14, 14. 10.1186/s12941-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber S., Gerber V., Jandova V., Rossano A., Evison J. M., Perreten V. (2011). Evolution of multidrug-resistant Staphylococcus aureus infections in horses and colonized personnel in an equine clinic between 2005 and 2010. Microb. Drug Resist. 17, 471–478. 10.1089/mdr.2010.0188 [DOI] [PubMed] [Google Scholar]
- Statistics-Denmark (2015a). ANI51: Slaughterings and Production of Pigs by Category and Unit. Statistics-Denmark. [Google Scholar]
- Statistics-Denmark (2015b). FOLK1A: Population at the First Day of the Quarter by Region, Sex, Age and Marital Status. Statistics-Denmark. [Google Scholar]
- Stegger M., Andersen P. S., Kearns A., Pichon B., Holmes M. A., Edwards G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 18, 395–400. 10.1111/j.1469-0691.2011.03715.x [DOI] [PubMed] [Google Scholar]
- Stegger M., Lindsay J. A., Moodley A., Skov R., Broens E. M., Guardabassi L. (2011). Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J. Clin. Microbiol. 49, 732–734. 10.1128/JCM.01970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eede A., Hermans K., Van den Abeele A., Floré K., Dewulf J., Vanderhaeghen W., et al. (2013). The nasal vestibulum is the optimal sampling site for MRSA screening in hospitalised horses. Vet. J. 197, 415–419. 10.1016/j.tvjl.2013.01.031 [DOI] [PubMed] [Google Scholar]
- Van den Eede A., Martens A., Feryn I., Vanderhaeghen W., Lipinska U., Gasthuys F., et al. (2012). Low MRSA prevalence in horses at farm level. BMC Vet. Res. 8:213. 10.1186/1746-6148-8-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eede A., Martens A., Lipinska U., Struelens M., Deplano A., Denis O., et al. (2009). High occurrence of methicillin-resistant Staphylococcus aureus ST398 in equine nasal samples. Vet. Microbiol. 133, 138–144. 10.1016/j.vetmic.2008.06.021 [DOI] [PubMed] [Google Scholar]
- van Duijkeren E., Moleman M., Sloet van Oldruitenborgh-Oosterbaan M. M., Multem J., Troelstra A., Fluit A. C., et al. (2010). Methicillin-resistant Staphylococcus aureus in horses and horse personnel: an investigation of several outbreaks. Vet. Microbiol. 141, 96–102. 10.1016/j.vetmic.2009.08.009 [DOI] [PubMed] [Google Scholar]
- van Wamel W. J. B., Rooijakkers S. H. M., Ruyken M., van Kessel K. P. M., van Strijp J. A. G. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J. Bacteriol. 188, 1310–1315. 10.1128/JB.188.4.1310-1315.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincze S., Stamm I., Kopp P. A., Hermes J., Adlhoch C., Semmler T., et al. (2014). Alarming proportions of methicillin-resistant Staphylococcus aureus (MRSA) in wound samples from companion animals, Germany 2010-2012. PLoS ONE 9:e85656. 10.1371/journal.pone.0085656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966. 10.3201/eid1112.050428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag. [Google Scholar]
- Yan X., Wang B., Tao X., Hu Q., Cui Z., Zhang J., et al. (2012). Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 78, 6637–6642. 10.1128/AEM.01165-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y. (2017). GGTREE : an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information about the health and antimicrobial treatments of 49 horses. The remaining 352 horses included in the study were reported as both, healthy and without any antimicrobial treatments in the 6 months prior to the study, and were hence excluded from this table.
Summary table of resistance profiles obtained from Mykrobe Predictor including responsible genes/variants found.
Maximum likelihood phylogeny of relevant clades containing bootstrap values. The phylogeny is drawed as cladogram.
Density of pigs (pigs/sqKm) in each of the 53 postal codes included in the study and the corresponding MRSA type(s) found. Postal codes without identified MRSA-positive horses are labeled as “MRSA negative.”