Abstract
Teosinte Branched1/Cycloidea/Proliferating cell factors (TCP) genes are key mediators of genetic innovations underlying morphological novelties, stress adaptation, and evolution of immune response in plants. They have a remarkable ability to integrate and translate diverse endogenous, and environmental signals with high fidelity. Compilation of studies, aimed at elucidating the mechanism of TCP functions, shows that it takes an amalgamation and interplay of several different factors, regulatory processes and pathways, instead of individual components, to achieve the incredible functional diversity and specificity, demonstrated by TCP proteins. Through this minireview, we provide a brief description of key structural features and molecular components, known so far, that operate this conglomerate, and highlight the important conceptual challenges and lacunae in TCP research.
Keywords: gene regulation, plant development, plant morphology, stress response, TCP domain, transcription factor
Introduction
TCP (Teosinte Branched1/Cycloidea/Proliferating cell factors) is a plant-specific family of transcription factors (TFs), with the earliest members reported in fresh water charophyte algae (Cubas et al., 1999a; Navaud et al., 2007). TCP proteins are characterized by a non-canonical beta helix-loop-helix (bHLH) domain, known as TCP domain. Although, TCP proteins have little homology with bHLH TFs and bind to DNA elements distinct from those recognized by bHLH TFs; the DNA contacting residues and mechanism of binding seem to be conserved in both the families (Kosugi and Ohashi, 1997). Aggarwal et al. (2010) suggested divergent evolution of TCP domain from the bHLH domain by insertion of a short stretch in the basic region thereby, splitting the long helix into two.
TCP family comprises six genes each in bryophyte species, Selaginella and Physcomitrella (Navaud et al., 2007). Whereas, the size of this family in angiosperms ranges from 12 in the orchid, Orchis italica (De Paolo et al., 2015) to more than 60 in tobacco (Chen et al., 2016) and cotton (Ma et al., 2016).
Multiple sequence alignment revealed two major classes of the TCP family viz., classes I and II. The residue composition in the DNA-binding TCP domain and, supplementary motifs confer specific characteristics to the members of both the classes. Some of the notable differences include a four-amino-acid deletion in the basic region of the class I TCPs and presence of additional motifs, such as glutamic acid-cysteine-glutamic acid (ECE) stretch and/or arginine-rich R-domain in a subset of class II proteins (Cubas et al., 1999a; Navaud et al., 2007). Class II further comprises two distinct subclasses namely, CINCINNATA (CIN) and CYCLOIDEA/TEOSINTE BRANCHED 1 (CYC/TB1). CIN clade is ubiquitous, whereas, CYC/TB1 is restricted to angiosperms and has undergone extensive duplications and diversification giving rise to three different clades: CYC1, CYC2, and CYC3 (Howarth and Donoghue, 2006).
TCP Genes are Key Mediators of Morphological Innovations, Stress Adaptations, and Plant Immunity Evolution
The studies done in experimentally tractable Arabidopsis, and several non-model plant species revealed that TCPs have played key role in generating novel morphologies during plant evolution (Martin-Trillo and Cubas, 2010; Manassero et al., 2013; Li, 2015). Since structural features play important role in determining protein functions, distinctive functions have been associated with members of each class. For example, class I genes (TCP6–9, 11, 14–16, 19–23) mostly act as positive regulators of cell division in diverse biological processes ranging from seed germination, leaf and floral organ development, gametophyte development and senescence (Martin-Trillo and Cubas, 2010; Li, 2015; Nicolas and Cubas, 2016). A recent study involving expression of a dominant repressor form of TCP16 demonstrated the ability of class I genes in modulating meristematic programs and differentiation state of the plant cells (Uberti-Manassero et al., 2016).
Class I TCP genes of rice have been mainly implicated in stress adaptation. PCF2 of rice affects salinity tolerance by positively regulating expression of a Na+/H+ antiporter gene, OsNHX1 (Almeida et al., 2017). Whereas, PCF5 and 6 are involved in drought plus salinity, and cold stress tolerance, respectively (Luo et al., 2012; Wang et al., 2014). OsTCP19, on the other hand, influences both development and abiotic stress tolerance by manipulating abscisic acid (ABA) signaling network (Mukhopadhyay and Tyagi, 2015). Also, mesocotyl elongation in response to darkness in rice has been associated with expression of OsTCP15 (Hu et al., 2014).
Members of the CYC/TB1 clade of class II (TCP1, 12, and 18) are mainly involved in regulating shoot branching, floral transition, organ identity, and development. A mutation in TB1 locus is responsible for the domestication of maize from its wild ancestor, teosinte (Doebley et al., 1995, 1997). Expression of another maize TCP gene BRANCHED ANGLE DEFECTIVE 1 in a grass-specific structure (pulvinus), between main stem and lateral branches of inflorescence, influences lateral branch angle and inflorescence architecture (Bai et al., 2012). The recent studies in non-model systems, cucumber and melon, revealed the role of CYC/TB1 genes in determining tendril identity, as well (Mizuno et al., 2015; Wang C. et al., 2015). A rare single nucleotide polymorphism in a TCP gene TEN is responsible for the tendril-less phenotype in cucumber (Wang S. et al., 2015).
Among the three subgroups of CYC clade, CYC1 genes have retained TB1-like functions across different taxa in regulating branching. Characterization of TB1 orthologs from monocots, such as rice (Fine culm1/OsTB1), barley (INTERMEDIUM-C), Sorghum (SbTB1), and switchgrass (PvTB1) and dicots, such as Arabidopsis (BRC1 and BRC2), pea (PsBRC1), and tomato (SlBRC1) indicate conserved role of this gene in negatively regulating axillary bud outgrowth across both the lineages of angiosperms (Takeda et al., 2003; Kebrom et al., 2006; Aguilar-Martínez et al., 2007; Ramsay et al., 2011; Braun et al., 2012; Nicolas et al., 2015; Xu et al., 2016). Duplication and differential expression of CYC2 genes have played a key role in the evolution of symmetry across different lineages of the angiosperms (Luo et al., 1996; Reeves and Olmstead, 2003; Specht and Howarth, 2015; Yang et al., 2015). CYC ortholog of rice, RETARDED PALEA 1 (REP1), also played a key role in regulating floral zygomorphy (Yuan et al., 2009). Whereas, CYC3 genes in Arabidopsis have been reported to play a minor role in branching in both vegetative and floral organs (Finlayson, 2007).
Genes belonging to CIN clade (TCP2–5, 10, 13, 17, and 24) of class II have been mainly implicated in regulating flowering time, floral organ development, leaf development and senescence, and morphogenesis of lateral organs (Nath et al., 2003; Palatnik et al., 2003; Koyama et al., 2007; Schommer et al., 2008; Ballester et al., 2015; Yang et al., 2015). Some of the more recent roles reported include regulation of secondary cell wall thickening in roots and floral organs of Arabidopsis (Wang H. et al., 2015) and ovule development in Phalaenopsis equestris (Lin et al., 2016). Although in angiosperms, only CYC/TB1 genes have been implicating in branching, a recent study in Physcomitrella patens revealed a role of CIN gene PpTCP5 in determining sporangia architecture by negatively regulating branching (Ortiz-Ramírez et al., 2016). These results indicate regulation of branching as an ancient role of class II TCPs.
Furthermore, members of both the classes are targeted by pathogens to manipulate host defense. An effector SECRETED AY-WB PROTEIN 11 (SAP11), produced by aster yellows phytoplasma, binds and destabilizes TCP4 thereby, leading to reduced jasmonic acid (JA) synthesis, increased plant susceptibility and survival rate of the insect vector (Sugio et al., 2011, 2014). TCP13, 14, and 19 of Arabidopsis are also directly targeted by pathogen effectors to elicit effector-triggered susceptibility. Whereas, TCP8, 14, and 15 interact with Suppressor Of rps4-RLD1 (SRFR1), a negative regulator of effector-triggered immunity to influence plant susceptibility (Kim et al., 2014). Recently, Zhang et al. (2016) showed that infection with viral pathogen, rice ragged stunt virus (RRSV) in rice leads to increased accumulation of miR319-targeted TCP genes, decreased JA levels and increased plant susceptibility. The biotrophic pathogens, however, may be benefited from the activation of JA-dependent responses. A recent study showed that Pseudomonas syringae type III effector, HopBB1 interacts with Arabidopsis TCP14 and targets it to proteasome-mediated degradation. Consequently, TCP14-regulated subset of JA response genes are de-repressed thereby, promoting pathogen virulence (Yang et al., 2017).
Binding Site and Mechanism of Action
TCP proteins modulate gene expression by directly binding to the regulatory regions of their target genes. Previous studies have reported overlapping but specific binding sites of classes I and II proteins. Viola et al. (2012) showed that presence of glycine or aspartic acid at positions 11 and 15 in classes I and II proteins, respectively, determines their binding preference. However, changes in residue composition at other positions can also influence the DNA-binding preferences of TCP proteins (Viola et al., 2011). For example, class I TCP protein, TCP11, has distinct DNA binding specificity due to presence of threonine residue at position 15, occupied by arginine in most of the other TCP proteins (Viola et al., 2011). Biochemical studies in Arabidopsis revealed that redox state of the cell can also influence binding ability of class I TCP proteins (Viola et al., 2013). Oxidation of a conserved cysteine residue at position 20 (cys-20) in these proteins leads to formation of intermolecular disulfide bonds and covalently linked homodimers that cannot bind target DNA. The effect of Arabidopsis TCP15 on anthocyanin accumulation is lost after prolonged exposure to high light intensity due to oxidation of cys-20 (Viola et al., 2016).
Presence of co-regulators may be imperative for the regulatory activity of TCPs. For example, a WD repeat-containing protein, LIGHT-REGULATED WD1 (LWD1) acts as a coactivator of TCP20 and 22 in regulating expression of morning gene CIRCADIAN CLOCK ASSOCIATED1 (CCA1) in Arabidopsis (Wu et al., 2016). Although TCP20 and 22 can bind to regulatory element in CCA1 promoter, even in the absence of LWDs, overexpression of TCP20/22 in lwd1lwd2 double mutant fails to activate CCA1 expression (Wu et al., 2016). Whether concomitant binding of TCPs and LWDs leads to any shifts in conformational state of TCPs is yet to be determined.
Several TCPs act as modulators of hormone biosynthesis, transport and signal transduction (Lopez et al., 2015; Nicolas and Cubas, 2016). A recent review summarizes crosstalk between TCPs and, biosynthesis and signaling of hormones viz., gibberellins, cytokinins, ABA, JA, brassinosteroids, strigolactones, and auxins (Nicolas and Cubas, 2016).
Cell/tissue-type or developmental stage-specific expression of members of same/different class seems to assist them in fine tuning the hormone production and balance. For example, TCP20 of class I suppresses expression of LIPOXYGENASE2 (LOX2), a key enzyme involved in JA biosynthesis in young leaves, whereas, TCP4 of class II promotes LOX2 expression thereby, promoting JA biosynthesis and senescence in mature leaves (Danisman et al., 2012). The same gene, TCP4, however, suppresses LOX2 expression in floral tissues (Rubio-Somoza and Weigel, 2013).
The role of TCPs in regulation of hormone activity may be indirect by interacting with regulators of hormone biosynthesis and response as exemplified by interaction of OsTCP19 with ABA INSENSITIVE4 and of OsTB1 with OsMADS57 (Nicolas and Cubas, 2016). Alternatively, TCPs may directly bind to the promoters of key genes involved in hormone biosynthesis as exemplified by regulation of DWARF4 by TCP1 and, regulation of LOX2 by TCP4/20 (Nicolas and Cubas, 2016). A recent study showed that YUCCA5, an enzyme involved in auxin biosynthesis, is direct target of TCP4 (Challa et al., 2016).
TCP proteins also regulate transcription of the non-coding RNAs that in turn target genes involved in hormonal signaling. For example, TCP4 directly regulates miR167a that targets auxin response factors, ARF6 and 8, involved in JA biosynthesis (Nagpal et al., 2005; Wu et al., 2006).
Analysis of cross-family TF interactions showed that TCPs exhibit high range of connectivity with members of other TF families (Bemer et al., 2017). Synergistic interactions between members of different TF families binding to different cis-elements in the targeted genes imply a combinatorial effect on target gene expression (Figure 1). TCP21 (CHE) of Arabidopsis interacts with C2C2/CO-like family component of circadian clock, TIMING OF CAB EXPRESSION1 (TOC1) during circadian regulation (Pruneda-Paz et al., 2009). The direct interaction between an Arabidopsis DOF TF, DOF6, and TCP14 affects seed germination (Rueda-Romero et al., 2012). CIN-TCPs interact with LBD domain containing ASYMMETRIC LEAVES 2 (AS2) TF to suppress KNOX gene expression during leaf development in Arabidopsis (Li et al., 2012). Similarly, the ternary complex between TCP, MYB, and bHLH family TFs (TCP3-R2R3MYB-TT8) is involved in regulating flavonoid biosynthesis and auxin response (Li and Zachgo, 2013). An interaction between MADS-box protein OsMADS57 and OsTB1 has been shown to modulate tillering in rice (Guo et al., 2013). Whereas, the interaction between TCP14 of Arabidopsis with GRAS domain containing DELLA proteins in inflorescence apical meristems determines plant height (Daviere et al., 2014). Interaction between CUC family TFs, CUC2 and 3 and, TCP4, regulates age-dependent leaf complexity in Arabidopsis (Rubio-Somoza et al., 2014).
FIGURE 1.
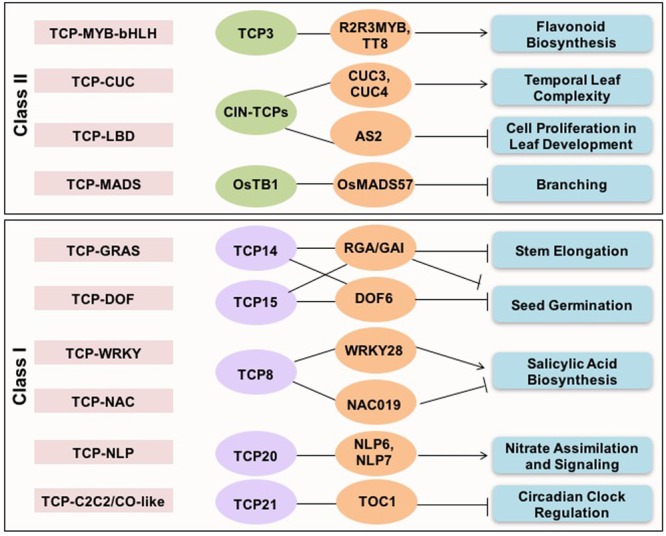
Cross-family transcription factor interactions shown by TCP proteins. The cross-family TF interactions exhibited by both classes I and II TCP proteins, and associated biological pathways are presented.
The choice of interaction partners also contributes to the functional diversity and specificity. For example, TCP8 may activate or repress ISOCHORISMATE SYNTHASE 1 (ICS1), a key gene involved in salicylic acid biosynthesis, by interacting with the transcriptional activators, WRKY28 and SAR DEFICIENT 1 or the NAC family repressor NAC109, respectively (Wang X. et al., 2015). Interactions between TCP20 and NIN-like TFs has been recently demonstrated to regulate nitrate assimilation and signaling (Guan et al., 2017). Overall, these studies highlight that TCP proteins are at the center of plant molecular networks and control diverse range of processes and signaling networks by recruiting specific interaction partners. Presence of intrinsically disordered region gives them extra flexibility to interact with diverse range of partners and make higher order complexes (Valsecchi et al., 2013).
Regulation of TCPs
The ability of TCPs to orchestrate plant response to both internal cues such as developmental signals and circadian rhythms; and diverse environmental factors such as light quality, nutrient availability, oxidative stress, etc., requires precise spatial and temporal control of their activity. Current research shows that the regulation of TCPs acts at several steps including transcription, mRNA stability, and post-translational modifications.
Regulation of gene expression includes a wide array of mechanisms. The spatial/temporal expression of TCP genes is directly associated with specific morphological phenotype or physiological response. For example, differential expression of GhCYC2 in Gerbera controls morphological differentiation of flower types along the radial axis of inflorescence (Broholm et al., 2008). Changes in the regulatory region of TB1 due to two transposable element insertions are responsible for its differential expression and domestication of maize (Zhou et al., 2011).
Alternative splicing also plays significant role in regulating gene expression. In potato, quality of light (R:FR) determines the ratio of two isoforms of a TCP gene BRC1a, only one of which is localized to nucleus and acts as a transcriptional activator (Nicolas et al., 2015). Transcriptional regulation by epigenetic mechanisms has also been demonstrated in TCPs. Differential methylation pattern in CYC orthologs resulted in differential expression of the gene causing dorsoventral asymmetry in flowers of Linaria vulgaris (Cubas et al., 1999b).
Role of non-coding microRNAs in post-transcriptional regulation of TCPs involved in flowering time and leaf morphogenesis is well-documented (Palatnik et al., 2003; Schommer et al., 2012; Spanudakis and Jackson, 2014). Both PCF5 and 6 of rice, involved in abiotic stress tolerance, are direct targets of miR319 (Luo et al., 2012; Wang et al., 2014). Downregulation of miR319-targeted TCP4, in response to sulfur dioxide exposure in Arabidopsis, reinforce the role of miRNAs in environmental regulation of TCPs (Li et al., 2016).
The final control comes at the level of post-translational modifications. These affect the activity and stability of the protein. Steiner et al. (2016) reported that regulation of TCP14 by SPINDLY, a Ser and Thr O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), prevents its proteolysis. Similarly, ubiquitin receptor proteins, DA1 and DA1-related proteins (DAR1 and DAR2), physically interact with TCP14 and 15, and affect their ubiquitination and stability (Peng et al., 2015). Ubiquitination sites have also been found on class I TCPs, TCP8 and 22, whereas, Ser-211 in TCP8 is phosphorylated (Valsecchi et al., 2013; Walton et al., 2016).
Key Challenges and Outlook
TCP genes appear to play central role in the biological signaling networks by interacting with many molecular and signaling components. These features not only make them ideal candidates to investigate the mechanism of combinatorial gene expression and hormonal crosstalk in plants, but also suggest them as promising targets for engineering crop plants. For this, a thorough understanding of their mechanism of action is imperative. Most of the functional genomic studies with TCPs are impeded by lack of three-dimensional structure, high level of genetic redundancy and lack of sufficient in vivo studies to identify in planta interaction partners and other regulatory components.
The theoretical predictions based on bHLH structure can be misleading. Deciphering three-dimensional structures of representative TCP proteins is of fundamental importance to gain mechanistic understanding of their functions. To cope with redundancy in TF genes, Hiratsu et al. (2003) developed a novel approach using a chimeric repressor gene-silencing technology (CRES-T), in which a TF is fused to the EAR-motif repression domain (SRDX) that dominantly represses the transcription of its target genes even in the presence of functionally redundant TFs (Mitsuda et al., 2011). Several authors have successfully used this technology to gain insights into TCP gene functions in Arabidopsis (Koyama et al., 2007; Guo et al., 2010; Aguilar-Martinez and Sinha, 2013). However, this technology cannot be used to decipher functions of essential genes. Danisman et al. (2012) used a bioinformatics approach to integrate data generated using pair-wise protein-protein interactions, phylogeny and expression profiling to predict functionally redundant TCP genes in Arabidopsis. Authors also validated one of the novel pairs, TCP19-TCP20, that functions redundantly in the leaf development. However, the interactions reported in their study are not immune to limitation of yeast two-hybrid technology. Due to high auto-activation capacity of class I TCP proteins, most of the connections were reported among class II TCP proteins. In planta studies during temporal stages of development and in response to pathogen infection or abiotic stresses would be required to precisely determine the interaction dynamics of TCP proteins.
Another interesting aspect of TCP genes is the predominant presence of introns in their UTRs (Francis et al., 2016). How these intron sequences influence gene expression, mRNA stability, or translational efficiency in TCPs remains unexplored.
Furthermore, although miR319-mediated regulation of CIN genes in both dicot and monocot species is well-documented, none of the TCP genes in Physocmitrella, Selaginella, and Marchantia polymorpha have a recognizable miR319 binding site (Axtell and Bowman, 2008; Schommer et al., 2012; Flores-Sandoval et al., 2016). Future studies will clarify if gain of miR319 targeting site has any role in the functional evolution of CIN genes in higher plants.
Furthermore, most of the earlier studies aimed at characterizing TCP gene functions focused on the model system, Arabidopsis. Although the TCP gene functions are now beginning to be elucidated in non-model systems as well (Table 1), this area of TCP research still needs momentum.
Table 1.
Teosinte Branched1/Cycloidea/Proliferating cell factors (TCP) proteins characterized from non-model systems and their roles.
| Species | Gene | Function | Reference | |
|---|---|---|---|---|
| Dicots | Brassica rapa | BrpTCP4 | miR319a-regulated, regulates transition from round to cylindrical head shape | Mao et al., 2014 |
| BrTCP24 | Suppresses growth of plant cells in Chinese cabbage | Gao et al., 2016 | ||
| Cucumis melon | CmTCP1 | Involved in development of tendrils from lateral shoots | Mizuno et al., 2015 | |
| Cucumis sativus | TEN | Causal gene for rare variation of tendril-less phenotype | Wang S. et al., 2015 | |
| Gerbera hybrida | GhCYC2 | A gradient of GhCYC2 expression correlates with flower type specification along inflorescence axis | Broholm et al., 2008 | |
| Gossypium hirsutum | GhTCP14 | Regulates auxin-mediated development of cotton fiber cells | Wang et al., 2013 | |
| Ipomoea nil | InTCP4 | miR319-regulated, affect floral initiation, flower development and cotyledon senescence | Glazińska et al., 2014 | |
| Pisum sativum | PsBRC1 | Regulates shoot branching putatively in response to cytokinin and strigolactone signaling | Braun et al., 2012 | |
| Solanum lycopersicon | LA (LANCEOLATE) | miR319-regulated, involved in leaf margin development and compound leaf formation | Ori et al., 2007 | |
| SlBRC1b | Suppresses shoot branching | Martín-Trillo et al., 2011 | ||
| SlTCP14-2 | Target of pathogen effector CRN12_997 of Phytophthora capsici and prevents plant defense | Stam et al., 2013 | ||
| Solanum tuberosum | BRC1a | Involved in controlling lateral branching | Nicolas et al., 2015 | |
| Monocots | Hordeum vulgare | INTERMEDIUM-C | Regulate tillering and fertility of lateral spikelets | Ramsay et al., 2011 |
| Oryza sativa | FC1 (FINE CULM1) | Ortholog of maize TB1 and mutants exhibit reduced plant height and increased tillering | Takeda et al., 2003 | |
| REP1 (RETARDED PALEA1) | Controls palea development and floral zygomorphy | Yuan et al., 2009 | ||
| OsTCP5 | Controls mesocotyl elongation in rice | Hu et al., 2014 | ||
| OsTCP19 | Involved in salinity and drought tolerance | Mukhopadhyay and Tyagi, 2015 | ||
| OsTCP21 | Involved in cold stress tolerance and plant defense response against rice ragged stunt virus (RRSV) | Wang et al., 2014; Zhang et al., 2016 | ||
| PCF2 | Involved in salt stress tolerance | Almeida et al., 2017 | ||
| PCF5 | Involved in drought and salinity stress tolerance | Luo et al., 2012 | ||
| PCF6 | Involved in cold tolerance | Wang et al., 2014 | ||
| Petunia hybrida | PhTCP3 | Regulates branching through strigolactone signaling | Revel et al., 2015 | |
| Phalaenopsis equestris | PePCF10 | Involved in leaf and ovule development | Lin et al., 2016 | |
| PeCIN8 | Regulates ovule, leaf and petal development | Lin et al., 2016 | ||
| Sorghum bicolor | SbTB1 | Negatively regulates tillering by suppressing bud outgrowth | Kebrom et al., 2006 | |
| Switchgrass | PvTB1 | Negatively regulates tillering | Xu et al., 2016 | |
| Zea mays | BAD1 | Regulates inflorescence architecture by affecting lateral branch angle | Bai et al., 2012 | |
| TB1 | Negatively regulates tillering and promotes formation of female inflorescence | Doebley et al., 1997 | ||
| Bryophytes | Physcomitrella patens | PpTCP5 | Negatively regulates sporophyte branching | Ortiz-Ramírez et al., 2016 |
Author Contributions
ND and RS conceptualized, prepared the framework and drafted the review. VB collected the data from the literature and helped in drafting the manuscript. MS contributed in preparing the framework and revising the article. All authors read and approved the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. We acknowledge the financial assistance in the form of Ramalingaswami fellowship and project grant by Department of Biotechnology, Government of India, and Start-Up grant from UGC through UGC-FRP scheme.
References
- Aggarwal P., Das Gupta M., Joseph A. P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22 1174–1189. 10.1105/tpc.109.066647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez J. A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 458–472. 10.1105/tpc.106.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A., Sinha N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4:406 10.3389/fpls.2013.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida D. M., Gregorio G. B., Oliveira M. M., Saibo N. J. (2017). Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 93 61–77. 10.1007/s11103-016-0547-7 [DOI] [PubMed] [Google Scholar]
- Axtell M. J., Bowman J. L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13 343–349. 10.1016/j.tplants.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Bai F., Reinheimer R., Durantini D., Kellogg E. A., Schmidt R. J. (2012). TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. U.S.A. 109 12225–12230. 10.1073/pnas.1202439109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester P., Navarrete-Gomez M., Carbonero P., Onate-Sanchez L., Ferrandiz C. (2015). Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plant. 155 21–32. 10.1111/ppl.12327 [DOI] [PubMed] [Google Scholar]
- Bemer M., Van Dijk A. D., Immink R. G., Angenent G. C. (2017). Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci. 22 66–80. 10.1016/j.tplants.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Braun N., De Saint Germain A., Pillot J.-P., Boutet-Mercey S., Dalmais M., Antoniadi I., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158 225–238. 10.1104/pp.111.182725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm S. K., Tähtiharju S., Laitinen R. A., Albert V. A., Teeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U.S.A. 105 9117–9122. 10.1073/pnas.0801359105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa K. R., Aggarwal P., Nath U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28 2117–2130. 10.1105/tpc.16.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen Y., Ding A., Chen H., Xia F., Wang W., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15 1–14. 10.4238/gmr.15027728 [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999a). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 215–222. [DOI] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999b). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 157–161. [DOI] [PubMed] [Google Scholar]
- Danisman S., Van Der Wal F., Dhondt S., Waites R., De Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviere J. M., Wild M., Regnault T., Baumberger N., Eisler H., Genschik P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24 1923–1928. 10.1016/j.cub.2014.07.012 [DOI] [PubMed] [Google Scholar]
- De Paolo S., Gaudio L., Aceto S. (2015). Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 5:16265 10.1038/srep16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Finlayson S. A. (2007). Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 48 667–677. 10.1093/pcp/pcm044 [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E., Dierschke T., Fisher T. J., Bowman J. L. (2016). Efficient and inducible use of artificial MicroRNAs in Marchantia polymorpha. Plant Cell Physiol. 57 281–290. 10.1093/pcp/pcv068 [DOI] [PubMed] [Google Scholar]
- Francis A., Dhaka N., Bakshi M., Jung K. H., Sharma M. K., Sharma R. (2016). Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci. Rep. 6:38488 10.1038/srep38488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang F., Zhang Y., Lifeng L., Li H., Li L., et al. (2016). BrTCP24 Gene Useful for Controlling Growth of Cabbage and Application Thereof. US 9353380 B2. [Google Scholar]
- Glazińska P., Wilmowicz E., Wojciechowski W., Frankowski K., Kopcewicz J. (2014). Impact of InMIR319 and light on the expression of InTCP4 gene involved in the development of Ipomoea nil plants. Acta Physiol. Plant 36 29–43. 10.1007/s11738-013-1384-9 [DOI] [Google Scholar]
- Guan P., Ripoll J.-J., Wang R., Vuong L., Bailey-Steinitz L. J., Ye D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A 114 2419–2424. 10.1073/pnas.1615676114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Xu Y., Liu H., Mao Z., Zhang C., Ma Y., et al. (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4 1566 10.1038/ncomms2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E. B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22 1161–1173. 10.1105/tpc.109.069203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Howarth D. G., Donoghue M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103 9101–9106. 10.1073/pnas.0602827103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yamauchi T., Yang J., Jikumaru Y., Tsuchida-Mayama T., Ichikawa H., et al. (2014). Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 55 30–41. 10.1093/pcp/pct150 [DOI] [PubMed] [Google Scholar]
- Kebrom T. H., Burson B. L., Finlayson S. A. (2006). Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140 1109–1117. 10.1104/pp.105.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Son G. H., Bhattacharjee S., Kim H. J., Nam J. C., Nguyen P. D., et al. (2014). The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 78 978–989. 10.1111/tpj.12527 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484. 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xue M., Yi H. (2016). Uncovering microRNA-mediated response to SO2 stress in Arabidopsis thaliana by deep sequencing. J. Hazard. Mater. 316 178–185. 10.1016/j.jhazmat.2016.05.014 [DOI] [PubMed] [Google Scholar]
- Li S. (2015). The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal. Behav. 10:e1044192 10.1080/15592324.2015.1044192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76 901–913. 10.1111/tpj.12348 [DOI] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H., Dong A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71 99–107. 10.1111/j.1365-313X.2012.04973.x [DOI] [PubMed] [Google Scholar]
- Lin Y.-F., Chen Y.-Y., Hsiao Y.-Y., Shen C.-Y., Hsu J.-L., Yeh C.-M., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67 5051–5066. 10.1093/jxb/erw273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. A., Sun Y., Blair P. B., Mukhtar M. S. (2015). TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 20 238–245. 10.1016/j.tplants.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincet C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Luo H., Li D., Zhou M., Hu Q. (2012). Methods and compositions for enhanced resistance to abiotic stress in plants. US 20130117882 A1. [Google Scholar]
- Ma J., Liu F., Wang Q., Wang K., Jones D. C., Zhang B. (2016). Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 6:21535 10.1038/srep21535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manassero N. G., Viola I. L., Welchen E., Gonzalez D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. Concepts 4 111–127. 10.1515/bmc-2012-0051 [DOI] [PubMed] [Google Scholar]
- Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. (2014). MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164 710–720. 10.1104/pp.113.228007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E. G., Serra F., Marcel F., Rodríguez-Buey M. L., Schmitz G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67 701–714. 10.1111/j.1365-313X.2011.04629.x [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Matsui K., Ikeda M., Nakata M., Oshima Y., Nagatoshi Y., et al. (2011). “CRES-T, an effective gene silencing system utilizing chimeric repressors,” in Plant Transcription Factors: Methods and Protocols eds Yuan L., Perry S. E. (Totowa, NJ: Humana Press; ) 87–105. [DOI] [PubMed] [Google Scholar]
- Mizuno S., Sonoda M., Tamura Y., Nishino E., Suzuki H., Sato T., et al. (2015). Chiba Tendril-Less locus determines tendril organ identity in melon (Cucumis melo L.) and potentially encodes a tendril-specific TCP homolog. J Plant Res. 128 941–951. 10.1007/s10265-015-0747-2 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P., Tyagi A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5:9998 10.1038/srep09998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C. M., Weber H., Ploense S. E., Barkawi L. S., Guilfoyle T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118. 10.1242/dev.01955 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Herve C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65 23–33. 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- Nicolas M., Cubas P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33 33–41. 10.1016/j.pbi.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Nicolas M., Rodríguez-Buey M. L., Franco-Zorrilla J. M., Cubas P. (2015). A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr. Biol. 25 1799–1809. 10.1016/j.cub.2015.05.053 [DOI] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez C., Hernandez-Coronado M., Thamm A., Catarino B., Wang M., Dolan L., et al. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9 205–220. 10.1016/j.molp.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Peng Y., Chen L., Lu Y., Wu Y., Dumenil J., Zhu Z., et al. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27 649–662. 10.1105/tpc.114.132274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J. L., Breton G., Paser A., Kay S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323 1481–1485. 10.1126/science.1167206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L., Comadran J., Druka A., Marshall D. F., Thomas W. T., Macaulay M., et al. (2011). INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43 169–172. 10.1038/ng.745 [DOI] [PubMed] [Google Scholar]
- Reeves P. A., Olmstead R. G. (2003). Evolution of the TCP gene family in Asteridae: cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol. Biol. Evol. 20 1997–2009. 10.1093/molbev/msg211 [DOI] [PubMed] [Google Scholar]
- Revel S., Bart J. J., Luo Z., Oplaat C., Susan E. L., Mark W. W., et al. (2015). Environmental control of branching in petunia. Plant Physiol. 168 735–751. 10.1104/pp.15.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2013). Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 9:e1003374 10.1371/journal.pgen.1003374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Zhou C. M., Confraria A., Martinho C., Von Born P., Baena-Gonzalez E., et al. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24 2714–2719. 10.1016/j.cub.2014.09.058 [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P., Barrero-Sicilia C., Gomez-Cadenas A., Carbonero P., Onate-Sanchez L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds interacts with TCP14. J. Exp. Bot. 63 1937–1949. 10.1093/jxb/err388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Bresso E. G., Spinelli S. V., Palatnik J. F. (2012). Role of microRNA miR319 in plant development. Signal. Commun. Plants 15 29–47. 10.1007/978-3-642-27384-1_2 [DOI] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chetelat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanudakis E., Jackson S. (2014). The role of microRNAs in the control of flowering time. J. Exp. Bot. 65 365–380. 10.1093/jxb/ert453 [DOI] [PubMed] [Google Scholar]
- Specht C. D., Howarth D. G. (2015). Adaptation in flower form: a comparative evodevo approach. New Phytol. 206 74–90. 10.1111/nph.13198 [DOI] [PubMed] [Google Scholar]
- Stam R., Motion G., Boevink P. C., Huitema E. (2013). A conserved oomycete CRN effector targets and modulates tomato TCP14-2 to enhance virulence. bioRxiv. 10.1101/001248 [DOI] [PubMed] [Google Scholar]
- Steiner E., Livne S., Kobinson-Katz T., Tal L., Pri-Tal O., Mosquna A., et al. (2016). The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171 1485–1494. 10.1104/pp.16.00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Maclean A. M., Grieve V. M., Hogenhout S. A. (2011). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108 E1254–E1263. 10.1073/pnas.1105664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Maclean A. M., Hogenhout S. A. (2014). The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 202 838–848. 10.1111/nph.12721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanka M., Ashikari M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33 513–520. 10.1046/j.1365-313X.2003.01648.x [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Coscueta E. R., Gonzalez D. H. (2016). Expression of a repressor form of the Arabidopsis thaliana transcription factor TCP16 induces the formation of ectopic meristems. Plant Physiol. Biochem. 108 57–62. 10.1016/j.plaphy.2016.06.031 [DOI] [PubMed] [Google Scholar]
- Valsecchi I., Guittard-Crilat E., Maldiney R., Habricot Y., Lignon S., Lebrun R., et al. (2013). The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol. Biosyst. 9 2282–2295. 10.1039/c3mb70128j [DOI] [PubMed] [Google Scholar]
- Viola I. L., Camoirano A., Gonzalez D. H. (2016). Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 170 74–85. 10.1104/pp.15.01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I. L., Guttlein L. N., Gonzalez D. H. (2013). Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol. 162 1434–1447. 10.1104/pp.113.216416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I. L., Reinheimer R., Ripoll R., Manassero N. G., Gonzalez D. H. (2012). Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 287 347–356. 10.1074/jbc.M111.256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I. L., Uberti Manassero N. G., Ripoll R., Gonzalez D. H. (2011). The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 435 143–155. 10.1042/BJ20101019 [DOI] [PubMed] [Google Scholar]
- Walton A., Stes E., Cybulski N., Van Bel M., Inigo S., Durand A. N., et al. (2016). It’s time for some “site”-seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28 6–16. 10.1105/tpc.15.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Y., Li S.-S., Han G.-Z. (2015). Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167 872–886. 10.1104/pp.114.247403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Mao Y., Yang J., He Y. (2015). TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 6:436 10.3389/fpls.2015.00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yang X., Xu M., Lin X., Lin T., Qi J., et al. (2015). A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Mol. Plant 8 1795–1808. 10.1016/j.molp.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Wang X., Gao J., Zhu Z., Dong X., Wang X., Ren G., et al. (2015). TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 82 151–162. 10.1111/tpj.12803 [DOI] [PubMed] [Google Scholar]
- Wang M.-Y., Zhao P.-M., Cheng H.-Q., Han L.-B., Wu X.-M., Gao P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162 1669–1680. 10.1104/pp.113.215673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-T., Sun X.-L., Hoshino Y., Yu Y., Jia B., Sun Z.-W., et al. (2014). MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 9:e91357 10.1371/journal.pone.0091357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-F., Tsai H.-L., Joanito I., Wu Y.-C., Chang C.-W., Li Y.-H., et al. (2016). LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7:13181 10.1038/ncomms13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. F., Tian Q., Reed J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133 4211–4218. 10.1242/dev.02602 [DOI] [PubMed] [Google Scholar]
- Xu K., Wang Y., Shi L., Sun F., Liu S., Xi Y. (2016). PvTB1, a Teosinte Branched1 gene homolog, negatively regulates tillering in switchgrass. J. Plant Growth Regul 35 44–53. 10.1007/s00344-015-9505-x [DOI] [Google Scholar]
- Yang L., Teixeira P. J. P. L., Biswas S., Finkel O. M., He Y., Salas-Gonzalez I., et al. (2017). Pseudomonas syringae Type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21 156–168. 10.1016/j.chom.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhao X. G., Li C. Q., Liu J., Qiu Z. J., Dong Y., et al. (2015). Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family gesneriaceae. Plant Physiol. 169 2138–2151. 10.1104/pp.15.01181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Gao S., Xue D., Luo D., Li L., Ding S., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149 235–244. 10.1104/pp.108.128231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ding Z., Wu K., Yang L., Li Y., Yang Z., et al. (2016). Suppression of jasmonic acid-mediated defense by viral-inducible MicroRNA319 facilitates virus infection in rice. Mol. Plant 9 1302–1314. 10.1016/j.molp.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Yan J., Song R. (2011). Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res. 21 1267 10.1038/cr.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]


