ABSTRACT
Activation of the stress-responsive transcription factor NRF2 is the major line of defense to combat oxidative or electrophilic insults. Under basal conditions, NRF2 is continuously ubiquitylated by the KEAP1-CUL3-RBX1 E3 ubiquitin ligase complex and is targeted to the proteasome for degradation (the canonical mechanism). However, the path from the CUL3 complex to ultimate proteasomal degradation was previously unknown. p97 is a ubiquitin-targeted ATP-dependent segregase that extracts ubiquitylated client proteins from membranes, protein complexes, or chromatin and has an essential role in autophagy and the ubiquitin proteasome system (UPS). In this study, we show that p97 negatively regulates NRF2 through the canonical pathway by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex, with the aid of the heterodimeric cofactor UFD1/NPL4 and the UBA-UBX-containing protein UBXN7, for efficient proteasomal degradation. Given the role of NRF2 in chemoresistance and the surging interest in p97 inhibitors to treat cancers, our results indicate that dual p97/NRF2 inhibitors may offer a more potent and long-term avenue of p97-targeted treatment.
KEYWORDS: KEAP1, NRF2, autophagy, cancer chemoresistance, oxidative stress, p97, proteasome, protein quality control, ubiquitylation
INTRODUCTION
The ability of organisms to adapt to changes in the environment is critical to survival. There are many biological systems that can sense both subtle and catastrophic changes; one such system is the KEAP1-NRF2-ARE signaling pathway (1). Nuclear factor (erythroid-derived 2)-like 2 (NRF2) is a basic leucine zipper transcription factor that is activated in response to oxidative stress or stress caused by electrophilic xenobiotic agents (2). Under basal conditions, NRF2 is bound by Kelch-like ECH-associated protein 1 (KEAP1), which serves as a substrate adapter for the cullin-3 (CUL3) E3 ubiquitin (Ub) ligase complex (3–5). This leads to constant ubiquitylation and subsequent proteasomal degradation of NRF2, ensuring low basal levels of NRF2. Upon oxidative or electrophilic insults, KEAP1 can be modified at its sensory cysteines, leading to a conformational change that prevents NRF2 ubiquitylation (6, 7). As a result, the NRF2-KEAP1-CUL3 complex is stabilized, allowing newly synthesized NRF2 to accumulate in the cytosol (8). NRF2 then translocates to the nucleus, where it can bind to small Maf proteins and turn on the transcription of antioxidant response element (ARE)-controlled genes (9, 10). These genes encode stress response and energy metabolism proteins that work in concert to remove cellular stress and modulate intermediary metabolism (1, 11). This mode of NRF2 modulation is known as the canonical pathway (12).
Like many, if not all, protective pathways, the NRF2 pathway also contains a dark side. Uncontrolled or deregulated expression of NRF2 confers a survival advantage to cells that promotes cancer and confers drug resistance (13, 14). This dark side of NRF2 has been shown to be present in many cancers that bear mutations of NRF2 regulatory proteins or NRF2 itself (15). It was reported recently that the incidence of mutations in NRF2 or KEAP1 is comparable to that for previously well-recognized tumor suppressors or oncogenes, such as TP53, PTEN, and KRAS (16). Moreover, deregulation of NRF2 can result from autophagic dysfunction (17). When the autophagic pathway is compromised, p62 accumulates and sequesters KEAP1 in the autophagosomes, which leads to increased NRF2 levels and subsequent activation of NRF2 signaling (18–20). Unlike the canonical pathway, which has a mechanism to rapidly stop NRF2 signaling when cellular redox homeostasis is restored, this noncanonical path leads to prolonged NRF2 activation (21–24). The detrimental effects of persistent NRF2 activation have been shown to result in tissue damage in a variety of model systems, demonstrating the importance of tight NRF2 regulation (23, 25, 26).
p97 (or valosin-containing protein [VCP]) is an ATPase associated with various cellular activities (AAA+) chaperone that uses the energy of ATP binding and hydrolysis to segregate ubiquitylated polypeptides from other biomolecules (27, 28). This segregase function of p97 is central to a number of critical biological functions, including transcription factor regulation, ubiquitin proteasome system (UPS)-mediated degradation, and autophagosome maturation (29–33). These functions indicate that p97 may be involved in the regulation of NRF2, which would have important clinical implications for a p97 inhibitor that recently entered clinical trials (34, 35). If p97 were to regulate NRF2, then inhibition of p97 would cause an increase in NRF2 levels, possibly leading to enhanced drug resistance. Understanding this mechanism would help to combat this, for instance, by coadministration of an NRF2 inhibitor with a p97 inhibitor. For this reason, we studied the cross talk between p97 and NRF2. Our studies reveal that p97 negatively regulates NRF2 through the canonical pathway. The interaction between p97 and NRF2 is ubiquitin dependent and requires the ubiquitin-binding p97 heterodimeric cofactors NPL4 and UFD1. Additionally, UBXN7 is required to form the connection between CUL3 and p97. Adding mechanistic details to the understanding of the regulation of NRF2 offers new possibilities for drug design and potential synergistic drug development opportunities.
RESULTS
Reduction of the p97 level or activity leads to increased levels of NRF2 and its target genes.
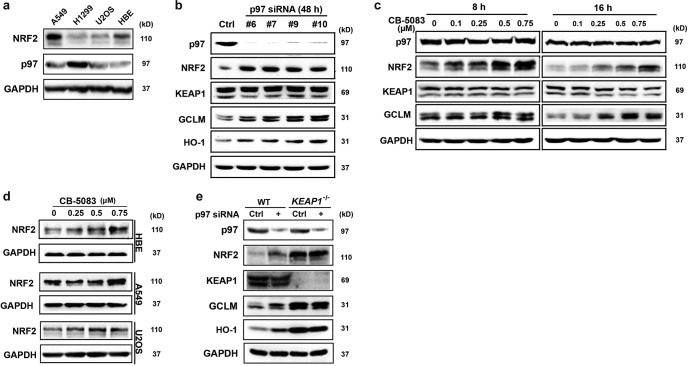
Because p97 is a ubiquitin-targeted AAA+ chaperone and also facilitates autophagosome maturation (27), it was hypothesized that p97 may be involved in NRF2 regulation. To test this, several cell lines, including a human bronchial epithelial cell line (HBE), non-small-cell lung cancer cell lines (H1299 and A549), and an osteosarcoma cell line (U2OS), were subjected to immunoblot analyses to compare the protein levels of p97. We found that H1299 cells had the highest level of p97 and the lowest level of NRF2, making them the most suitable cells for further research (Fig. 1a). Next, the expression of p97 was silenced in H1299 cells by use of four different small interfering RNAs (siRNAs) to exclude off-target effects of the p97 siRNAs; for further experiments, only p97 siRNA#7 was used, since differences among siRNAs were modest (Fig. 1b). As shown, increases in the levels of NRF2 and its target gene products, glutamate-cysteine ligase, modifier subunit (GCLM), and heme oxygenase 1 (HO-1), were observed when p97 expression was reduced (Fig. 1b). In contrast, no effects on KEAP1 protein levels were observed (Fig. 1b). Similar results were obtained with HBE cells (not shown); however, in A549 and U2OS cells, knockdown of p97 showed little effect on NRF2, GCLM, and HO-1 levels (data not shown). This was likely caused by low endogenous levels of p97, indicating inefficient p97 knockdown. To test if p97 activity is necessary to regulate NRF2, H1299 cells were treated with CB-5083, a potent and highly selective p97 inhibitor that is currently in clinical trials (34). Protein levels of NRF2 (peak induction at 4 to 8 h) and GCLM (peak induction at 16 h) increased in a dose-dependent manner when the activity of p97 was inhibited (Fig. 1c). Similarly, in A549, HBE, and U2OS cells treated with CB-5083, dose-dependent increases in NRF2 levels were observed (Fig. 1d). To explore the requirement of KEAP1 in the p97-mediated negative regulation of NRF2, H1299 KEAP1 knockout (KEAP1−/−) cells were generated using the clustered regularly interspaced short palindromic repeat (CRISPR) gene editing technique. The results showed that knockdown of p97 had no effect on NRF2, GCLM, or HO-1 levels in KEAP1−/− cells compared to those in wild-type (WT) H1299 cells (Fig. 1e). Collectively, these results indicate that p97 negatively regulates the NRF2 signaling pathway in a KEAP1-dependent manner in the cell lines tested.
FIG 1.
Reduction of the p97 level or activity leads to increased levels of NRF2 and its target genes. (a) Protein levels of NRF2 and p97 were examined by immunoblot analysis for two lung adenocarcinoma cell lines (A549 and H1299), a normal bronchial epithelial cell line (HBE), and an osteosarcoma cell line (U2OS). (b) H1299 cells were transfected with a scrambled siRNA (control [Ctrl]) or one of four different p97 siRNAs (#6, #7, #9, and #10; see Materials and Methods for details) for 48 h. Total cell lysates were used for immunoblot analysis with antibodies against the indicated proteins. In this panel, NRF2 appears as a single band because the protein lysates were resolved in 4 to 12% gradient gels. All other NRF2 bands appear as two bands because they were resolved in 7.5% gels. (c) H1299 cells were treated for 8 h (left) or 16 h (right) with the indicated doses of the p97 inhibitor CB-5083. Total cell lysates were subjected to immunoblot analyses. (d) HBE, A549, and U2OS cells were treated with the p97 inhibitor CB-5083 for 8 h. NRF2 expression was detected by immunoblot analysis. (e) H1299 KEAP1 knockout (KEAP1−/−) cells were generated using CRISPR-Cas9 gene editing. Wild-type (WT) and KEAP1−/− cells were transfected with Ctrl or p97 siRNA for 48 h. Total cell lysates were subjected to immunoblot analyses.
p97 interacts with NRF2 in a ubiquitin-dependent manner.
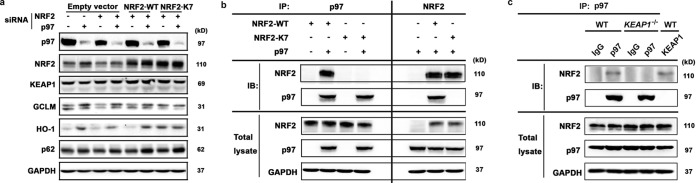
Given our observation that p97 regulates NRF2, we hypothesized that p97 physically interacts with ubiquitylated NRF2. To test this, H1299 cells were transfected with control siRNA, p97 siRNA, or NRF2 siRNA (to knock down endogenous NRF2 expression) and then cotransfected with vectors containing hemagglutinin (HA)-tagged wild-type NRF2 (NRF2-WT) or HA-tagged NRF2-K7 (a ubiquitylation-defective NRF2 mutant in which the seven ubiquitin-accepting lysines are replaced by alanines) (5). Immunoblot analysis was performed to detect the expression of NRF2 and its target genes (Fig. 2a). Even though the reduction of NRF2 was modest (∼50%), the expression levels of NRF2-WT and -K7 were sufficiently high to render the results significant. The data indicate that p97 decreases NRF2 levels and function only when the ubiquitin-receiving lysines in NRF2 are intact. Furthermore, to determine if p97 was in the complex with NRF2 and the requirement of NRF2 ubiquitylation, HA-tagged NRF2-WT or HA-tagged NRF2-K7 was coexpressed (with the levels adjusted to be equal) with FLAG-tagged p97. Reciprocal immunoprecipitation analyses indicated an interaction between p97 and NRF2-WT but not between p97 and NRF2-K7 (Fig. 2b). These results indicate that p97 interacts only with ubiquitylated NRF2, resulting in negative regulation of NRF2. Furthermore, to confirm that p97 was in the same complex as ubiquitylated NRF2, immunoprecipitation analyses of endogenous p97 and NRF2 were performed on WT and KEAP1−/− H1299 cells. The interaction between p97 and NRF2 was detected only in WT cells, not in KEAP1−/− cells, suggesting that p97 extracts ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex (Fig. 2c).
FIG 2.
p97 interacts with NRF2 in a ubiquitin-dependent manner. (a) H1299 cells were first transfected with p97 siRNA, NRF2 siRNA (to knock down endogenous NRF2), or both siRNAs for 24 h and then transfected with a plasmid containing either HA-tagged NRF2-WT or HA-tagged NRF2-K7 for another 24 h. Total cell lysates were subjected to immunoblot analysis. (b) H1299 cells were transfected with a plasmid containing FLAG-tagged p97 and a plasmid for either HA-tagged NRF2-WT or HA-tagged NRF2-K7 for 24 h. Different amounts of NRF2-WT and NRF2-K7 were used in the presence or absence of p97 to ensure equal expression of total NRF2. Cell lysates were used for reciprocal immunoprecipitation (IP) and immunoblot (IB) analyses. (Left) p97 was immunoprecipitated with an anti-FLAG antibody, and NRF2 was detected using an anti-HA antibody. (Right) NRF2-WT or NRF2-K7 was immunoprecipitated using an anti-HA antibody, and p97 was detected using an anti-FLAG antibody. (Bottom) An aliquot of total cell lysate was used for immunoblot analysis with an anti-FLAG (p97) or anti-HA (NRF2) antibody. (c) H1299 WT and KEAP1−/− cells were treated with 10 μM MG132 for 4 h to block degradation of ubiquitylated NRF2 before harvest. Cell lysates were subjected to immunoprecipitation analyses with anti-p97 or anti-NRF2 antibodies. Isotype IgG was used as a negative control, and anti-KEAP1 antibody was used as a positive control.
p97 facilitates ubiquitin-mediated degradation of NRF2.
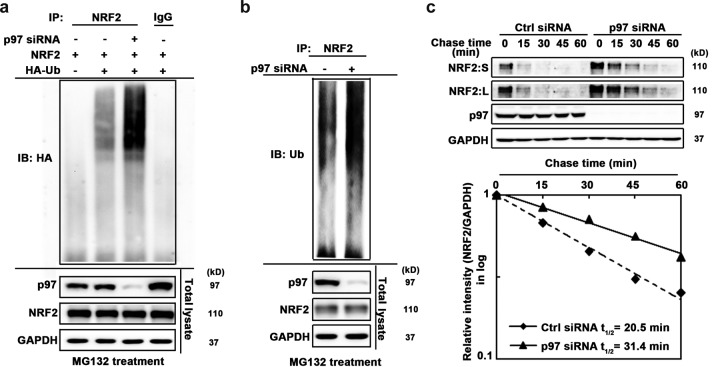
Based on the known function of p97 and our observation that p97 interacts with ubiquitylated NRF2, we hypothesized that p97 functions as a macromolecular machine that extracts ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex to deliver it to the 26S proteasome for degradation. To test this possibility, the effect of p97 knockdown on ubiquitylated NRF2 levels was examined in detail. H1299 cells were transfected with HA-ubiquitin (HA-Ub) and NRF2 along with p97 siRNA and then treated with MG132 for 4 h before harvest to block the degradation of ubiquitylated NRF2. Immunoblot analysis with total cell lysate confirmed the p97 knockdown and the blockage of NRF2 degradation by MG132 (Fig. 3a, bottom panel). Next, NRF2 was immunoprecipitated, and ubiquitylated NRF2 was detected by immunoblot analysis with an anti-HA antibody. A much higher level of ubiquitylated NRF2 was observed in the p97 siRNA-transfected cells (Fig. 3a), suggesting that p97 modulates the level of ubiquitylated NRF2. To further confirm this and to ensure that the effects observed were not an artifact of exogenous protein expression, the same experiment was carried out on endogenous NRF2 and ubiquitin. Again, an increase in ubiquitylated NRF2 was clearly present when cells were transfected with p97 siRNA (Fig. 3b). Finally, to test the role of p97 in enhancing the degradation of NRF2, the half-life of NRF2 was measured. H1299 cells were transfected with control siRNA or p97 siRNA and then treated with cycloheximide for the indicated times. The relative amounts of NRF2 at different time points were calculated from the immunoblot analysis and plotted to determine the half-life of NRF2. In p97 siRNA-transfected cells, the half-life of NRF2 increased from 20.5 min to 31.4 min (Fig. 3c). Collectively, these data suggest that p97 facilitates the degradation of NRF2.
FIG 3.
p97 facilitates ubiquitin-mediated degradation of NRF2. (a) Ubiquitylation of ectopically expressed NRF2. H1299 cells were first transfected with p97 siRNA for 24 h and then transfected with expression vectors for FLAG-NRF2 and HA-ubiquitin (HA-Ub) for an additional 24 h. Before harvest, cells were treated with 10 μM MG132 for 4 h to block degradation of ubiquitylated NRF2. NRF2 was immunoprecipitated using an anti-FLAG antibody, and immunoprecipitated NRF2 was subjected to immunoblot analysis with an anti-HA antibody. (Bottom) An aliquot of total cell lysate was used for immunoblot analysis. (b) Ubiquitylation of endogenous NRF2. H1299 cells were transfected with p97 siRNA for 48 h. NRF2 was immunoprecipitated with an anti-NRF2 antibody, and immunoprecipitated NRF2 was subjected to immunoblot analysis with an antiubiquitin (anti-Ub) antibody for detection of endogenous ubiquitylated NRF2. (Bottom) An aliquot of total cell lysate was used for immunoblot analysis. (c) H1299 cells were transfected with a scrambled siRNA (Ctrl siRNA) or p97 siRNA for 48 h. Cells were treated with cycloheximide for the indicated time before harvest. (Top) Total cell lysates were subjected to immunoblot analysis. (Bottom) NRF2 and GAPDH levels were determined by densitometry, and the level of NRF2 relative to that of GAPDH was plotted as a function of time to determine the half-life of NRF2. NRF2:S, short exposure; NRF2:L, long exposure.
p97 is involved in the canonical NRF2 pathway.
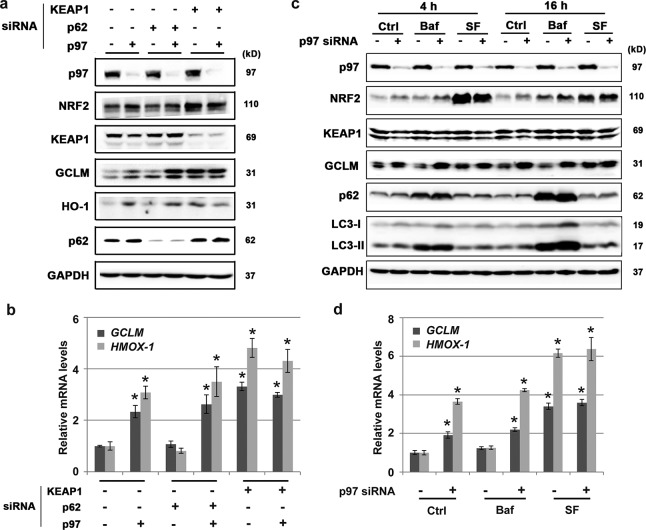
As discussed above, NRF2 can be modulated in a canonical, UPS-mediated manner or in a noncanonical, autophagy-mediated manner. Because p97 has been implicated in both of these pathways, we sought to differentiate how p97 modulates NRF2. First, KEAP1 expression was knocked down, and as expected, NRF2 levels increased, since KEAP1-mediated ubiquitylation of NRF2 was abolished. Moreover, no further increase in NRF2 was seen when both KEAP1 and p97 expression was knocked down (Fig. 4a). Since KEAP1 is an essential component of both the canonical and noncanonical regulation of NRF2, p62 expression was also knocked down to test if p97 worked through the noncanonical pathway. siRNA-mediated knockdown of p62 had no effect on the inverse relationship between p97 and NRF2 (Fig. 4a). These results are consistent with the mRNA expression levels of the NRF2 downstream genes GCLM and HMOX-1 (the gene that encodes HO-1) as analyzed by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 4b). These data suggest that the negative effects of p97 on NRF2 are KEAP1 dependent and operate independently of the p62-mediated noncanonical autophagy pathway. Consistent with this, blocking autophagy by use of bafilomycin (Baf) failed to alter the effects of p97 knockdown on NRF2 upregulation (Fig. 4c). As shown, when p97 was knocked down, an increase of NRF2 was observed in control (Ctrl) cells, with a further increase in the NRF2 level in Baf-treated cells, at both the 4-h and 16-h time points (Fig. 4c). The same trend was observed in the analysis of mRNA levels of GCLM and HMOX-1 by qRT-PCR (Fig. 4d). These results further support the canonical pathway as the intersection between p97 and NRF2. Additionally, the canonical activator sulforaphane (SF) was used to confirm that the effect of p97 on NRF2 is through the canonical pathway. SF increased the level of NRF2, but there was no further increase when p97 was knocked down (Fig. 4b). This is most likely due to the observations that SF blocks KEAP1-mediated ubiquitylation of NRF2 (5) and that nonubiquitylated NRF2 is not a substrate of p97. These data also indicate that p97 works in the canonical NRF2 pathway.
FIG 4.
p97 is involved in the canonical NRF2 pathway. (a and b) H1299 cells were transfected with the indicated siRNAs for 48 h. Total cell lysates were subjected to immunoblot analysis (a) and qRT-PCR analysis of GCLM and HMOX-1 (the gene encoding HO-1) (b). (c and d) H1299 cells were transfected with p97 siRNA for 48 h, followed by a 4-h or 16-h treatment with the autophagy blocker bafilomycin (Baf) (100 nM for 4 h or 50 nM for 16 h) or the canonical NRF2 activator sulforaphane (SF) (5 μM for 4 h or 2.5 μM for 8 h). Total cell lysates were subjected to immunoblot analysis (c) and qRT-PCR analysis of GCLM and HMOX-1 (for the 16-h treatment only) (d). Results were obtained from three independent experiments. *, P < 0.05 compared to control.
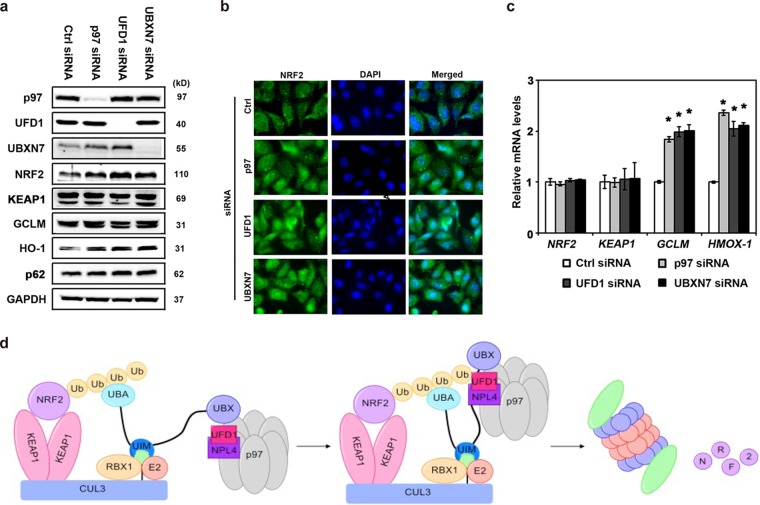
Decreased levels of the p97 cofactors UFD1 and NPL4 and the UBA-UBX protein UBXN7 lead to increased expression of NRF2 and NRF2 target genes.
Next, we sought to identify the critical p97 cofactors involved in this regulatory process. Because NRF2 is regulated in a ubiquitin-dependent manner, it was reasoned that the heterodimeric pair UFD1/NPL4 was one likely candidate, as this pair has been shown to direct p97 to ubiquitylated proteins to be extracted (36). Indeed, knockdown of UFD1 increased the protein levels of NRF2, GCLM, and HO-1, as determined by immunoblot analysis (Fig. 5a), and increased protein levels and nuclear translocation of NRF2, as shown by immunofluorescence assay (Fig. 5b). Since NRF2 is regulated by a CUL3 E3 complex and a proteomic investigation from the Deshaies group indicated that UBA-UBX proteins are associated with the p97-cullin E3 ligase network (37, 38), a series of siRNAs targeting UBA-UBX proteins was tested for effects on NRF2. Knockdown of UBXN7 (a UBA-UBX protein) led to increases of NRF2, GCLM, and HO-1 by both immunoblot analysis (Fig. 5a) and immunofluorescence assay (Fig. 5b). In contrast, knockdown of UBXN7 had no effects on KEAP1 or p62 protein levels. Furthermore, in the presence of siRNA targeting the p97, UFD1, or UBXN7 gene, the mRNA levels of GCLM and HMOX-1 were increased relative to those in the presence of control siRNA (Fig. 5c). In contrast, NRF2 and KEAP1 mRNA levels were not affected, which is consistent with the fact that NRF2 is typically regulated posttranslationally at the level of protein stability.
FIG 5.
Decreased levels of p97, UFD1/NPL4, and UBXN7 increase NRF2 signaling. (a to c) H1299 cells were transfected with the indicated siRNA for 48 h. (a) Total cell lysates were subjected to immunoblot analysis. (b) Cells were grown on glass coverslips for 48 h and then were subjected to indirect immunofluorescence analysis using an anti-NRF2 antibody. Hoechst 33258 was included to label nuclei. (c) Total RNA was extracted and reverse transcribed, and equal amounts of cDNA were used for qRT-PCR. *, P < 0.05 compared to Ctrl siRNA. The experiments were repeated three times, each with duplicate samples. (d) Model of the p97-mediated extraction of NRF2 from the KEAP1-CUL3-RBX1 complex for proteasomal degradation in the canonical NRF2 pathway. NRF2 is recruited to the KEAP1-CUL3-RBX1 E3 ligase complex by KEAP1, leading to ubiquitylation of NRF2, which is segregated from the complex by p97. UBXN7 acts as a scaffold protein that brings together all the components, which we propose to take place by the following model. The UIM domain binds to NEDDylated (NEDD8) CUL3 (in green), the UBA domain binds to ubiquitylated NRF2, and the UBX domain recruits p97 through interaction with the p97 N-terminal domain. The UFD1/NPL4 heterodimer likely binds to both the p97 N-terminal domain and NRF2, bringing these two proteins into close proximity. This allows p97 to act as an ATP-driven motor to extract ubiquitylated NRF2 from the KEAP1-CUL3 complex to deliver it to the 26S proteasome for degradation.
DISCUSSION
Careful regulation of oxidative stress is critical to organismal well-being. The dietary and chemical modulation of the KEAP1-NRF2-ARE pathway to boost cellular defense mechanisms and confer protection against various diseases has been well documented (39–41). Typically, this takes the form of small electrophilic modifiers, such as sulforaphane, which covalently modify the sensory cysteines of KEAP1, preventing ubiquitylation of NRF2 and leading to activation of the NRF2-mediated cellular protective response (42). More recently, it was revealed that activation of NRF2 is not always positive. When this activation goes unchecked, NRF2 can confer a survival advantage to cells and lead to excessive growth (43). This dark side of NRF2 actually suggests that, in certain contexts, NRF2 inhibitors might be of benefit to cancer treatment, as supported by in vitro and in vivo evidence (44, 45). These examples argue for a continued need to dissect the details of the NRF2 pathway to develop new treatments and refine existing ones.
In the present study, we identified a critical link between NRF2, p97, and proteasomal degradation. siRNA-mediated knockdown of p97 and chemical inhibition of p97 activity led to increases in NRF2 levels. Further exploration of this connection by use of siRNAs showed that p97 employs the heterodimeric cofactor complex UFD1/NPL4. The proteins of this complex are the best-studied p97 cofactors and have been shown to target p97 to ubiquitylated substrates (46). Further, UBXN7 was also shown to be required for p97-mediated NRF2 regulation. Previous work has shown an interaction between p97, UBXN7, CUL2, and HIF1α (47). However, UBXN7 showed an effect opposite that of p97, i.e., UBXN7 knockdown decreased HIF1α levels (37). The significance of this complex biology remains unclear. In the present case, UBXN7 is inversely correlated with NRF2 levels, giving the first clear link between a UBA-UBX protein, p97, a CUL-containing E3 ligase, and a transcription factor. The structural details of this complex require further investigation, including the precise mode of interaction between p97, each cofactor, and the CUL3 complex, as well as the need for both a UBA domain and UFD1/NPL4, both of which have been demonstrated to bind to ubiquitin. In addition, our data clearly demonstrate that p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. However, it is currently undefined if p97 is able to extract ubiquitylated NRF2 from other E3s, such as the βTrCP-CUL1 complex, the endoplasmic reticulum (ER) membrane-associated E3 HRD1, and possibly other E3s that have yet to be identified (48–50).
Importantly, a series of genetic p97 variants have been linked to a multisystem disorder called inclusion body myopathy associated with Paget's disease of bone and frontotemporal dementia (IBMPFD) (51). In this autosomal dominant lethal disorder, mutations in p97 lead to compromised autophagosome maturation. Blocked autophagy was shown to lead to increased NRF2 levels through the noncanonical p62-dependent pathway (18). Therefore, we tested if p97 downregulated NRF2 through the autophagic pathway. Using a series of genetic and chemical approaches, we were able to establish that loss of p97 function does not lead to increased NRF2 through the autophagic route.
We thus put forth the following model for the canonical regulation of NRF2 by p97. NRF2 is recruited to the KEAP1-CUL3-RBX1 E3 ligase complex by KEAP1, and this leads to ubiquitylation of NRF2, which is segregated from the complex by p97. UBXN7 acts as a scaffold protein that brings together all the components, which we propose to take place as follows: the UIM domain binds to NEDDylated (NEDD8) CUL3, the UBA domain binds to ubiquitylated NRF2, and the UBX domain recruits p97 through interaction with the p97 N-terminal domain. The UFD1/NPL4 heterodimer likely binds to both the p97 N-terminal domain and NRF2, bringing these two proteins into close proximity. This allows p97 to act as an ATP-driven motor to extract ubiquitylated NRF2 from the KEAP1-CUL3 complex to deliver it to the 26S proteasome for degradation (Fig. 5d).
This is the first example of cross talk between NRF2 and p97 and is also the first example of UBXN7 working in concert with p97 to directly regulate a transcription factor. This process has important implications for chemotherapy (52). Currently, the compound CB-5083 is in clinical trials for treatment of a variety of malignancies, but as we have shown, this compound increases the level of NRF2 in multiple cancer cell lines. Upregulation of NRF2 may very well lead to increased drug resistance by decreasing the intracellular concentration of CB-5083 through increased detoxification and excretion and by enhancing cellular defense mechanisms (damage repair, redox homeostasis, and energy balance) (1). Accordingly, a recent study found that multiple myeloma cells resistant to proteasome inhibitors significantly upregulated proteins involved in redox homeostasis, protein folding and destruction, and metabolic regulation (53). Moreover, the protein with the highest induction was the P-glycoprotein multidrug resistance-associated protein 1 (MDR1; also known as ABCB1), encoded by an NRF2 target gene, further supporting the notion that drug-induced NRF2 upregulation leads to cancer cell chemoresistance (53, 54). Here we put forth the possibility that treatment with p97 or proteasome inhibitors will be more efficacious if it is coupled with an NRF2 inhibitor, such as brusatol (45), to overcome resistance.
MATERIALS AND METHODS
Chemicals, antibodies, and cell culture.
Bafilomycin (Baf) was purchased from Sigma, CB-5083 (p97 inhibitor) was a generous gift from the Deshaies lab, and sulforaphane (SF) was purchased from Santa Cruz Biotechnology. Primary antibodies against NRF2, KEAP1, p97, GCLM, HO-1, p62, UFD1, UBXN7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology. Antibody against the hemagglutinin (HA) epitope was obtained from Trevigen. Antibody against LC3 was obtained from Sigma. The Alexa Fluor 488-conjugated secondary antibody was obtained from Invitrogen. HBE cells were obtained from Dieter Gruenert, whose lab generated and characterized them; the rest of the cells were purchased from the American Type Culture Collection (ATCC), where they were tested and authenticated by short tandem repeat (STR) analysis. All cells were maintained at 37°C in a humidified incubator containing 5% CO2. H1299 and A549 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Atlanta Biological) and 0.1% gentamicin (Invitrogen). HBE cells were maintained in minimal essential medium (MEM) supplemented with 10% FBS, 1% l-glutamine, and 0.01% gentamicin. U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 1% l-glutamine, and 0.01% gentamicin.
Generation of H1299 KEAP1−/− cells.
H1299 KEAP1 knockout (KEAP1−/−) cells were generated using CRISPR-Cas9-mediated gene editing (55, 56). Two single guide RNA (sgRNA) sequences targeted coding sequences near the promoter region of KEAP1 (sgRNA-A, 5′-AGCGTGCCCCGTAACCGCAT-3′; and sgRNA-B, 5′-GATCTACACCGCGGGCGGCT-3′). These sgRNAs were cloned into the pSpCas9(BB)-2A-GFP plasmid. H1299 cells were cotransfected with 1 μg of the pSpCas9(BB)-2A-GFP plasmid carrying sgRNA-A and 1 μg of the pSpCas9(BB)-2A-GFP plasmid carrying sgRNA-B. Green fluorescent protein (GFP)-positive cells were isolated using fluorescence-activated cell sorting (FACS) and plated at low confluence for colony isolation. Individual clones were expanded, and the genomic DNA for KEAP1 was sequenced. Finally, the KEAP1 knockout was further confirmed by examining protein expression of KEAP1 and NRF2 by immunoblotting.
Construction of recombinant DNA molecules.
The p97 expression vectors were constructed by cloning a PCR-generated fragment into the pCMV-HA vector (Invitrogen) or pFlag-CMV (Sigma). The sequences were confirmed by direct nucleotide sequencing. NRF2-WT and NRF2-K7 were described previously (5).
Transfection of siRNA and cDNA.
Transfection of cDNA was performed using Lipofectamine 3000 (Invitrogen). Hiperfect (Qiagen) was used for transfection of small interfering RNA (siRNA). NRF2 siRNA (SI03246614; targets the 3′ untranslated region [UTR] to knock down endogenous NRF2 only), p97 siRNA (GS7415; contains siRNA#6, siRNA#7, siRNA#9, and siRNA#10), p97 siRNA#6 (SI03019618), p97 siRNA#7 (SI03019730), p97 siRNA#9 (S103149657), p97 siRNA#10 (S104350444), p62 siRNA (SI03116750), KEAP1 siRNA (SI03246439), UFD1 siRNA (GS7353; contains four siRNAs, namely, SI04251030, SI04138624, SI04132583, and SI03213133), UBXN7 siRNA (1027416; contains four siRNAs, namely, S100455364, S100455371, S100455378, and S100455385), and control siRNA (Ctrl; 1027281) were purchased from Qiagen. p97 siRNA#7 was used for all the data presented in this article.
mRNA extraction and real-time qRT-PCR.
Total mRNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using equal amounts of mRNA and a Transcriptor first-strand cDNA synthesis kit (Promega). The detailed primer sequences are shown in Table 1.
TABLE 1.
Primers used in this study
| Target gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| NRF2 | ACACGGTCCACAGCTCATC | TGTCAATCAAATCCATGTCCTG |
| KEAP1 | ACCACAACAGTGTGGAGAGGT | CGATCCTTCGTGTCAGCAT |
| GCLM | GACAAAACACAGTTGGAACAGC | CAGTCAAATCTGGTGGCATC |
| HMOX-1 | AACTTTCAGAAGGGCCAGGT | CTGGGCTCTCCTTGTTGC |
| GAPDH | CTGACTTCAACAGCGACACC | TGCTGTAGCCAAATTCGTTGT |
The real-time PCR (RT-PCR) conditions were as follows: one cycle of initial denaturation (95°C for 3 min), 40 cycles of amplification (95°C for 10 s, 60°C for 20 s, and 72°C for 5 s), melting curve analysis (95°C for 5 s, 65°C for 1 min, and 97°C continuously), and a cooling period (40°C for 30 s). Mean crossing point (Cp) values were determined and normalized to the respective Cp values for the GAPDH reference gene. Data are presented as fold changes in gene expression compared to that of the control siRNA group. The RT-PCR analyses were repeated in three independent experiments performed in duplicate. Data are shown as means ± standard deviations (SD).
Immunoblot analysis, ubiquitylation assay, and protein half-life.
Cells were harvested in sample buffer (62.5 mM Tris-HCl [pH 6.9], 3% SDS, 10% glycerol, 5% beta-mercaptoethanol, and 0.1% bromophenol blue). After sonication, cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and subjected to immunoblot analysis. For the endogenous ubiquitylation assay, H1299 cells were transfected with control or p97 siRNA for 48 h. For the exogenous ubiquitylation assay, H1299 cells were transfected with control or p97 siRNA for 24 h and then cotransfected with expression vectors for HA-tagged ubiquitin, NRF2, and KEAP1. After 24 h, cells were treated with 10 μM MG132 for 4 h. Cells were harvested in buffer containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), and 1 mM dithiothreitol (DTT) and immediately boiled. The lysates were then diluted 5-fold in buffer without SDS and incubated with an anti-NRF2 antibody. Immunoprecipitated proteins were analyzed by immunoblotting with an antibody against the HA epitope. To measure the half-life of NRF2, H1299 cells were transfected with either control or p97 siRNA for 48 h, and then 50 μM cycloheximide was added to block de novo protein synthesis. Total cell lysates were collected at different time points and subjected to immunoblot analysis with antibodies against NRF2, p97, and GAPDH. The relative intensities of the bands were quantified using a ChemiDoc CRS gel documentation system and Quantity One software from Bio-Rad (Hercules, CA).
Immunoprecipitation.
H1299 cells were transfected with empty vector or a vector expressing NRF2-WT, NRF2-K7, or FLAG-tagged p97. Cell lysates were collected at 24 h posttransfection in radioimmunoprecipitation assay (RIPA) buffer containing 10 mM sodium phosphate (pH 8.0), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS. DTT (1 mM), 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (PIC) (Sigma) were also added to the RIPA buffer. Cell lysates were incubated with 1 μg of antibody and 10 μl of protein A-agarose beads on a rotator at 4°C overnight. To detect protein expression in the total cell lysates, 10 μl of cell lysate in RIPA buffer was mixed with 10 μl of 2× sample buffer (62.5 mM Tris-HCl [pH 6.9], 3% SDS, 10% glycerol, 5% beta-mercaptoethanol, 0.1% bromophenol blue) and boiled for 5 min. The immunoprecipitated complexes were washed with RIPA buffer with DTT, PMSF, and PIC three times and eluted in sample buffer by boiling for 5 min. Samples were then resolved by PAGE and subjected to immunoblot analysis.
Immunofluorescence assay.
For indirect immunofluorescence assay, cells were grown on glass coverslips (Fisher Scientific) in 35-mm dishes (BD Biosciences). Cells were fixed in prechilled methanol for 20 min. NRF2 was detected using an anti-NRF2 antibody and an Alexa Fluor 488-conjugated anti-rabbit secondary antibody. Nuclei were stained using Hoechst 33258. All images were taken at room temperature with a Zeiss Observer.Z1 microscope using a 63× oil immersion Plan-Apochromat objective (Zeiss). Images were acquired with Slidebook 4.2.0.11 software (Intelligent Imaging Innovations, Inc.).
Statistical analysis.
For qRT-PCR analyses, results are presented as means ± SD for at least three independent experiments performed in duplicate or triplicate. Statistical tests were performed using SPSS 10.0. Unpaired Student's t tests were used to compare the means for two groups. One-way analysis of variance was applied to compare the means for three or more groups. P values of <0.05 were deemed significant.
ACKNOWLEDGMENTS
This research was funded by grants ES023758 (E.C. and D.D.Z.), CA154377 (D.D.Z.), ES015010 (D.D.Z.), and ES006694 (a center grant).
We thank Raymond Deshaies for CB-5083.
S.T., P.L., G.L., H.C., and T.W. performed experiments. E.C. and M.R.D.L.V. wrote the manuscript. J.T., S.T., P.L., and G.L. prepared the figures. D.D.Z. and E.C. designed and supervised the study.
We have no competing financial interests to declare.
REFERENCES
- 1.Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD. 2015. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med 88:108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. 1995. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15:4184–4193. doi: 10.1128/MCB.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang DD, Hannink M. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird L, Lleres D, Swift S, Dinkova-Kostova AT. 2013. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc Natl Acad Sci U S A 110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 10.Katsuoka F, Yamamoto M. 2016. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene 586:197–205. doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova AT, Abramov AY. 2015. The emerging role of Nrf2 in mitochondrial function. Free Radic Biol Med 88:179–188. doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder B, Jiang T, Wu T, Tao S, Rojo de la Vega M, Tian W, Chapman E, Zhang DD. 2015. Molecular mechanisms of Nrf2 regulation and how these influence chemical modulation for disease intervention. Biochem Soc Trans 43:680–686. doi: 10.1042/BST20150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes JD, McMahon M. 2006. The double-edged sword of Nrf2: subversion of redox homeostasis during the evolution of cancer. Mol Cell 21:732–734. doi: 10.1016/j.molcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. 2008. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. 2008. Dual roles of Nrf2 in cancer. Pharmacol Res 58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. 2010. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 17.Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD. 2015. p62 links autophagy and Nrf2 signaling. Free Radic Biol Med 88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 20.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Zhang S, Chan JY, Zhang DD. 2007. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol 27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Wu T, Zhao F, Lau A, Birch CM, Zhang DD. 2011. KPNA6 (importin α7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol Cell Biol 31:1800–1811. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M. 2011. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD. 2013. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol 33:2436–2446. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. 2014. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol 61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, Yamamoto M, Tanaka K, Mizushima N, Komatsu M, Kopito RR. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol 191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 28.Xia D, Tang WK, Ye Y. 2016. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107:667–677. doi: 10.1016/S0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 30.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ndoja A, Cohen RE, Yao T. 2014. Ubiquitin signals proteolysis-independent stripping of transcription factors. Mol Cell 53:893–903. doi: 10.1016/j.molcel.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sha Z, Goldberg AL. 2014. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol 24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Wang Y, Li C, Shi Z, Hao Q, Wang W, Song X, Zhao Y, Jiao S, Zhou Z. 2015. The transitional endoplasmic reticulum ATPase p97 regulates the alternative nuclear factor NF-kappaB signaling via partial degradation of the NF-kappaB subunit p100. J Biol Chem 290:19558–19568. doi: 10.1074/jbc.M114.630061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou HJ, Wang J, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, Madriaga A, Kiss von Soly S, Kumar A, Parlati F, Yakes FM, Shawver L, Le Moigne R, Anderson DJ, Rolfe M, Wustrow D. 2015. Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083). J Med Chem 58:9480–9497. doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]
- 35.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. 2015. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G. 2012. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1alpha accumulation. BMC Biol 10:36. doi: 10.1186/1741-7007-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. 2012. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol 19:511–516. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surh YJ, Kundu JK, Na HK, Lee JS. 2005. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J Nutr 135:2993S–3001S. [DOI] [PubMed] [Google Scholar]
- 40.Kensler TW, Wakabayashi N, Biswal S. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 41.Su ZY, Shu L, Khor TO, Lee JH, Fuentes F, Kong AN. 2013. A perspective on dietary phytochemicals and cancer chemoprevention: oxidative stress, nrf2, and epigenomics. Top Curr Chem 329:133–162. doi: 10.1007/128_2012_340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kensler TW, Egner PA, Agyeman AS, Visvanathan K, Groopman JD, Chen JG, Chen TY, Fahey JW, Talalay P. 2013. Keap1-nrf2 signaling: a target for cancer prevention by sulforaphane. Top Curr Chem 329:163–177. doi: 10.1007/128_2012_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaramillo MC, Zhang DD. 2013. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson E, Feinstein E, Biswal S. 2008. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res 68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. 2011. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A 108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Y. 2006. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol 156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. 2008. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. 2011. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. 2013. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu T, Zhao F, Gao B, Tan C, Yagishita N, Nakajima T, Wong PK, Chapman E, Fang D, Zhang DD. 2014. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev 28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. 2004. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 52.Chapman E, Fry AN, Kang M. 2011. The complexities of p97 function in health and disease. Mol Biosyst 7:700–710. doi: 10.1039/C0MB00176G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soriano GP, Besse L, Li N, Kraus M, Besse A, Meeuwenoord N, Bader J, Everts B, den Dulk H, Overkleeft HS, Florea BI, Driessen C. 2016. Proteasome inhibitor-adapted myeloma cells are largely independent from proteasome activity and show complex proteomic changes, in particular in redox and energy metabolism. Leukemia 30:2198–2207. doi: 10.1038/leu.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeong HS, Ryoo IG, Kwak MK. 2015. Regulation of the expression of renal drug transporters in KEAP1-knockdown human tubular cells. Toxicol In Vitro 29:884–892. doi: 10.1016/j.tiv.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 55.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]