Abstract
Objectives
Ubiquitin E3 ligase-mediated protein degradation regulates osteoblast function. Itch, an E3 ligase, affects numerous cell functions by regulating ubiquitination and proteasomal degradation of related proteins. However, the Itch-related cellular and molecular mechanisms by which osteoblast differentiation and function are elevated during bone fracture repair are as yet unknown.
Methods
We examined the expression levels of E3 ligases and NF-κB members in callus samples during bone fracture repair by quantitative polymerase chain reaction (qPCR) and the total amount of ubiquitinated proteins by Western blot analysis in wild-type (WT) mice. The expression levels of osteoblast-associated genes in fracture callus from Itch knockout (KO) mice and their WT littermates were examined by qPCR. The effect of NF-κB on Itch expression in C2C12 osteoblast cells was determined by a chromatin immunoprecipitation (ChIP) assay.
Results
The expression levels of WW Domain Containing E3 Ubiquitin Protein Ligase 1 (Wwp1), SMAD Specific E3 Ubiquitin Protein Ligase 1 (Smurf1), SMAD Specific E3 Ubiquitin Protein Ligase 2 (Smurf2) and Itch were all significantly increased in the fracture callus of WT mice, which was associated with elevated expression of NF-κB members and total ubiquitinated proteins. Callus tissue isolated from Itch KO mice expressed higher levels of osteoblast-associated genes, including Runx2, a positive regulator of osteoblast differentiation, but osteoclast-associated genes were not increased. Both NF-κB RelA and RelB proteins were found to bind to the NF-κB binding site in the mouse Itch promoter.
Conclusions
Our findings indicate that Itch depletion may have a strong positive effect on osteoblast differentiation in fracture callus. Thus, ubiquitin E3 ligase Itch could be a potential target for enhancing bone fracture healing.
Cite this article: J. Liu, X. Li, H. Zhang, R. Gu, Z. Wang, Z. Gao, L. Xing. Ubiquitin E3 ligase Itch negatively regulates osteoblast function by promoting proteasome degradation of osteogenic proteins. Bone Joint Res 2017;6:154–161. DOI: 10.1302/2046-3758.63.BJR-2016-0237.R1.
Keywords: Itch, Osteoblasts, Bone formation, Fracture, E3 ligase
Article focus
Itch-related cellular and molecular mechanisms by which osteoblast differentiation and function are elevated during bone fracture repair are as yet unknown. The aim of this study is to determine whether Itch-altered osteoblast differentiation and function are elevated in the fracture callus of Itch KO mice.
Key messages
Our findings indicate that Itch depletion has a strong positive effect on osteoblast differentiation in fracture callus, indicating that Itch (or factors that regulate Itch-mediated cellular events) could be a potential target for enhancing bone fracture healing.
Strengths and limitations
A strength of our study is the demonstration of potential involvement of protein ubiquitination and ubiquitin E3 Itch in bone fracture healing. A limitation is the lack of in vivo indication of the requirement of Itch in bone fracture healing.
Introduction
Bone is a highly dynamic tissue which is able to continuously renew itself. Skeletal homeostasis determines bone mass in adults by achieving balance between bone resorption by osteoclasts and bone formation by osteoblasts. Protein ubiquitination is an important regulation for controlling cell function, and this mechanism has been implicated in the control of bone cell homeostasis.
Ubiquitination is a post-translational modification that has many functional implications. Ubiquitinated proteins undergo proteasomal or lysosomal degradation.1 There are several types of ubiquitin ligases, which are classified according to their structures and their mechanisms of action. WW domain-containing ubiquitin ligases are a subgroup of the homologues to the E6AP carboxyl terminus (HECT) family of ubiquitin E3 ligases, which promote protein ubiquitination by binding to a PPXY motif on target proteins. This class of E3 ligases consists of Nedd4-1, Nedd4-2, Itch, Smurf1, Smurf2, WWp1 and WWp2,2,3 and promotes ubiquitination and subsequent proteasomal or lysosomal degradation of target proteins.1,4 Thus far, the WW domain-containing ubiquitin ligases, Smurf1, Smurf2, Wwp1 and Wwp2, have been reported to be involved in bone cell regulation through modification of the stability of multiple proteins including the BMP-Smad-Runx2 protein,4,5 Smad3 and GSK3β,6 JunB,7 and Goosecoid.8
Itch is a ubiquitin E3 ligase and is another member of the WW domain-containing ubiquitin ligases.9 Itch KO mice in a C57BL/6J background develop a progressive autoimmune disease.10 We reported that young Itch KO mice have increased bone mass and osteoblast differentiation.11 However, the involvement of E3 ligases, including Itch,-mediated cellular events in fracture repair has not previously been studied.
In the current study, we have demonstrated high expression levels of E3 ligases, including Itch, and ubiquitinated proteins in fracture callus tissues. Itch KO mice have increased expression of osteoblast-associated genes in callus tissues that were isolated from an early stage of fracture repair. These callus tissues had increased expression levels of Runx2 mRNA, the essential transcription factor for osteoblast differentiation. Our findings suggest that Itch, or factors that regulate Itch-mediated cellular events, could be a potential target for enhancing bone fracture healing.
Materials and Methods
Animals
A total of ten 12-week-old wild-type C57BL/6J male mice were purchased from Jackson Laboratory (Bar Harbour, Maine). A total of five Itch KO mice on a C57BL/6J background were generated by breeding heterozygous female with heterozygous male mice. Homozygous mice for Itch deficiency (Itch KO mice) were genotyped using polymerase chain reaction (PCR) as we previously reported.11 The University Committee on Animal Resources at the University of Rochester has approved all procedures performed on the animals, as well as the housing conditions and the diet in this study.
Tibial fracture model
Open tibial fractures were created in WT and Itch KO mice (five pairs). In brief, an incision of 6 mm in length was made in the skin on the anterior side of the tibia after anaesthesia. A sterile 27 G x 1.25 inch needle was inserted into the marrow cavity of the tibia from the proximal end, temporarily withdrawn to facilitate transection of the tibia using a scalpel at midshaft, and then reinserted to stabilise the fracture. The incision was closed with 5-0 nylon sutures. Fractures were confirmed by radiograph. Callus tissues were harvested at days 3, 7, 10, 14, and 21 following fracture for RNA extraction. For examining the expression levels of total ubiquitinated proteins, the mice were treated with the proteasome inhibitor MG132 (1 mg/kg by intraperitoneal injection, EMD Millipore, Billerica, Massachusetts) 24 hours before they were killed at day 7 following fracture. Total ubiquitinated proteins were examined in callus tissues by Western blot analysis. The cortical bone tissues from the non-fractured legs were used as control.
Quantitative-PCR
Total RNA was extracted from callus tissues using TRIzol Reagent (Invitrogen, Carlsbad, California). cDNAs were synthesised using an iSCRIPT cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, California). Quantitative real-time (RT)-PCR amplifications were performed in the iCycler (Bio-Rad) real-time PCR machine using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instruction. The expression levels of ubiquitin E3 ligases (Itch, Wwp1, Smurf1 and Smur2), NF-κB members (RelA, RelB, p50, p52), osteoblast-related genes (Runx2 and alkaline phosphatase, ALP) and osteoclast-related genes (Receptor Activator of Nuclear Factor kappa-B, RANKL and osteoprotegerin, OPG) were examined with sequence specific primers (Table I).
Table I.
Sequence specific primers
| qPCR primers | Sequence |
|---|---|
| Itch | forward, 5’-TCACTTGGGCATAGGTCTCT-3’ |
| reverse, 5’-TGTGCCCAGACACTGAGTTA-3’ | |
| Wwp1 | forward, 5’-AGAATGGAGACCCTGCAACAAG-3’ |
| reverse, 5’-GCAGGAGTTGGGAACAACAGTA-3’ | |
| Smurf1 | forward, 5’-AGTTCGTGGCCAAATAGTGG-3’ |
| reverse, 5’-GTTCCTTCGTTCTCCAGCAG-3’ | |
| Smurf2 | forward, 5’-TGTAACACAGGCAAATCCCA-3’ |
| reverse, 5’-CGACGTGGGGACAGATAAAT-3’ | |
| RelA | forward, 5’-GCAGTATCCATAGCTTCCAG-3’ |
| reverse, 5’-GCACTGCATTCAAGTCATAGT-3’ | |
| RelB | forward, 5’-TACGACAAGAAGTCCACCA-3’ |
| reverse, 5’-CACATCAGCTTGAGAGAAGTC-3’ | |
| p50 | forward, 5’-ACCAAAACAGAGGGGATTT-3’ |
| reverse, 5’-CCATTTGTGACCAACTGAA-3’ | |
| p52 | forward, 5’-CTGTCAAGATCTGTAACTATGAGG-3’ |
| reverse, 5’-ATGTCCTTGGGTCCTACAG-3’ | |
| ALP | forward, 5’-CTTGCTGGTGGAAGGAGGCAGG-3’ |
| reverse, 5’-CACGTCTTCTCCACCGTGGGTC-3’ | |
| Runx2 | forward, 5’-CAAGAAGGCTCTGGCGTTTA-3’ |
| reverse, 5’-TGCAGCCTTAAATGACTCGG-3’ | |
| RANKL | forward, 5’-CAGAAGGAACTGCAACACAT-3’ |
| reverse, 5’-CAGAGTGACTTTATGGGAACC-3’ | |
| OPG | forward, 5’-AATTGGCTGAGTGTTTTGGT-3’ |
| reverse, 5’-TGATGTTTCCACAGCTTCAG-3’ | |
| GAPDH | forward, 5’-GGTCGGTGTGAACGGATTTG-3’ |
| reverse, 5’-ATGAGCCCTTCCACAATG-3’ |
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified on the same plates and used to normalise the data. Each sample was prepared in triplicate and each experiment was repeated at least once. The relative abundance of each gene was calculated by subtracting the cycle threshold (CT) value of each sample for an individual gene from the corresponding CT value of GAPDH (ΔCT). ΔΔCT were obtained by subtracting the ΔCT of the reference point. These values were then raised to the power of 2 (2ΔΔCT) to yield fold expression relative to the reference point.
Western blot analysis
Whole-cell lysates were prepared from fracture callus tissues or C2C12 cells, an osteoblast/myoblast precursor cell line that was purchased from ATCC (Manassas, Virginia). Homogenised fracture callus tissues with liquid nitrogen or cells were lysed with mammalian protein extraction reagent (Pierce Chemical, Dallas, Texas) containing a protease inhibitor mixture (Roche Applied Science, Indianapolis, Indiana). Whole cell lysates (10 μg protein/lane) were loaded in 10% Sodium Dodecyl Sulfate (SDS)-PAGE gels, transferred to a nitrocellulose membrane, and immunoblotted with antibodies to β-actin, ubiquitin (Santa Cruz Biotechnology) and Itch (BD Biosciences, San Jose, California). Bands were visualized using ECL chemiluminescence (Amersham Biosciences, Piscataway, New Jersey).
Chromatin immunoprecipitation (ChIP) assay
The C2C12 cells were infected with retroviral virus encoding green fluorescent protein (GFP) control, NF-κB RelA or RelB for 24 hours. Infection efficiency was confirmed by a fluorescent microscopy. ChIP assay was performed with the MAGnify Chromatin Immunoprecipitation System (Invitrogen) according to the manufacturer’s instructions. 1x106 cells in 500 µl PBS were fixed with 1% formaldehyde for 15 minutes. Cells were then lysed in 50 µl Lysis Buffer containing protease inhibitor and sonicated on ice eight times with a 20-seconds-on, 20-seconds-off cycle at high power using the Bioruptor UCD-200 sonicator (Diagenode, Denville, New Jersey) to shear chromatin into 200 bp to 500 bp fragments. 10 µl of chromatin was diluted to 100 µl with dilution buffer. 10 µl was used as an Input. Antibodies against RelA and RelB or control rabbit IgG (Invitrogen) were used in the immunoprecipitation step. Antibodies were coupled to the Dynabeads (ThermoFisher Scientific, Waltham, Massachusetts) and then incubated with 100 µl diluted chromatin for two hours at 40°C. After adequate washing, the immuoprecipiated (IP) chromatin was reversed for the crosslinking with Crosslinking buffer containing Proteinase K for 15 minutes at 55°C. The purified DNA fragments were determined by quantitative PCR using the primers localised at the binding sites (Table II). Control primers localised at irrelevant down-streaming sites were used as a negative control. Amplification efficiency was calculated by 100x2e (Average Input - Average IP), where the Input Control was named as Input recovery.
Table II.
Primers for genotyping
| Primer type | Sequence |
|---|---|
| Genotyping primers | |
| Itch WT | 5’-ATCGTCTACTCACCCCACATAAGG-3’ |
| Itch KO | 5’-AAGAAGCAGCAGAGACAACGAGTG-3’ |
| Common: | 5’-TCTATGCTCTGTTGTCTCCCATGC-3’ |
| ChIP primers | |
| NF-κB binding site 1 | forward, 5’-GCAGAAATGTCCCAAAGA-3’ |
| reverse, 5’-TGGAAAGCCAGCAAAGC-3’ | |
| NF-κB binding site 2 | forward, 5’-TATGGAGATTATTAGGCTGG TG-3’ |
| reverse, 5’-GAGCGGGGTCTACAAAGT-3’ | |
| Irrelevant site | forward, 5’-GTTCCTACAGTCTAGTTGCATTCA-3’ |
| reverse, 5’-GATGTGGAGGTCAGAGGAC-3’ |
Statistical analysis
All results are given as mean and standard deviation (sd). Comparisons between the two groups were analysed using the two-tailed unpaired Student’s t-test. One-way analysis of variance (ANOVA) and Dunnett’s post hoc multiple comparisons were used for comparisons among three or more groups. A p-value of less than 0.05 was considered statistically significant.
Results
Increased expression of E3 ligases and NF-κB members in callus tissues during bone fracture repair in WT mice
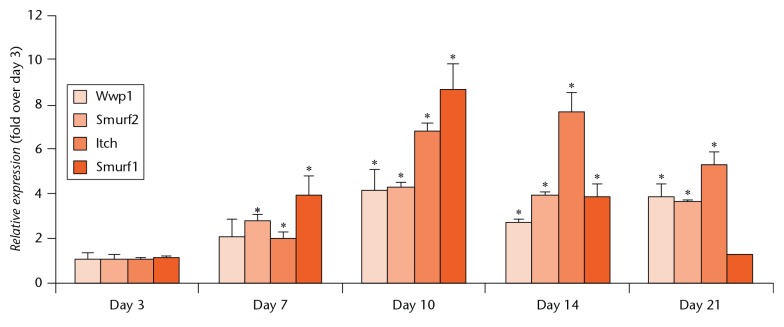
The WW domain-containing ubiquitin E3 ligases, including Wwp1, Smurf1 and Itch, regulates the stability of the positive osteoblast regulators Runx2, JunB and CXCR4.4,12,13 To determine if these E3 ligases are involved in fracture repair, we examined their expression levels in callus tissues from WT C57BL/6J mice which had undergone surgical tibia bone fracture. The expression levels of Smurf1, Smurf2 and Itch were all significantly increased, starting at day 7 following fracture, Wwp1 expression increased significantly, starting at day 10, and reached maximum expression at ten days following fracture (Itch expression reaching the maximum at 14 days) (Fig. 1).
Fig. 1.
Increased expression of multiple Nedd4 subclass of E3 ligases in fracture callus. Three-month-old WT C57 BL/6J male mice underwent a surgical tibia fracture and were killed at different time points post-fracture. The expression levels of E3 ligases in callus tissue were examined by qPCR. Values are mean and standard deviation of three mice. The relative expression was calculated as fold change using the expression level at day 3 as 1. *p<0.05 versus samples from day three.
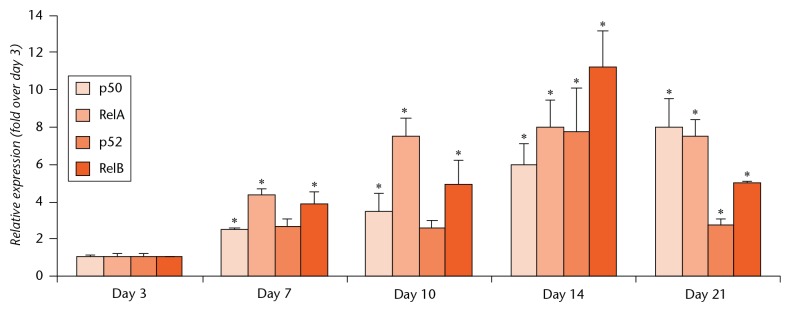
We have previously reported that NF-κB members are positive regulars of E3 ligase Itch in osteoclast lineage cells.14 Thus, we examined expression of NF-κB members at mRNA level extracted from callus tissues at different time points post-fracture. The expression levels of all NF-κB members, including p50, RelA, p52 and RelB, were significantly increased, starting at 7 days after fracture and reaching maximum expression at 14 days (Fig. 2), a similar time point to that of when E3 ligase levels were elevated.
Fig. 2.
Increased expression of NF-κB members in fracture callus. Samples were obtained from the callus tissue as in Figure 1. The expression levels of NF-κB members were determined by quantitative polymerase chain reaction (qPCR). Values are mean and standard deviation of three mice. The relative expression was calculated as fold change using the expression level at day 3 as 1. *p<0.05 versus samples from day 3.
Increased amount of total ubiquitinated proteins in fracture callus tissues
We sought to determine if the total number of ubiquitinated (Ub)-proteins in callus tissues is increased in the presence of proteasome inhibitor MG132. The total amount of Ub-proteins in callus tissues from the fractured legs was increased compared with cortical bone tissues from non-fractured legs (Fig. 3).
Fig. 3.
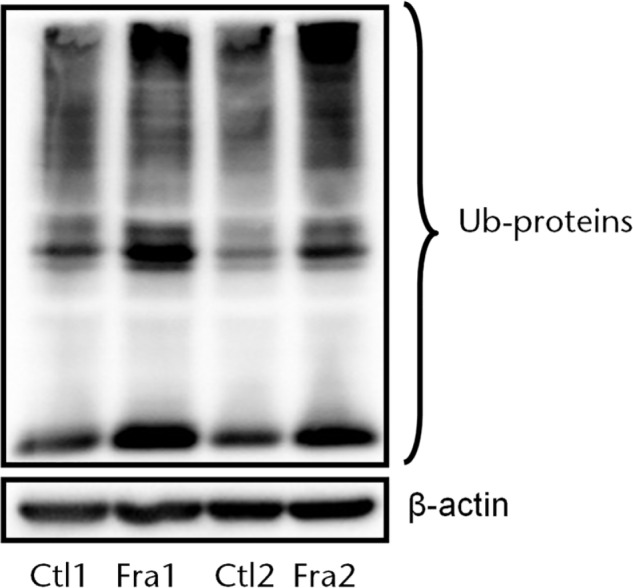
Increased total ubiquitinated proteins in fracture callus. WT mice received bone fracture surgery as in Figure 1. Callus tissue from fractured legs was harvested at day 7 after mice received MG132 for 24 hours. Total ubiquitinated (Ub) proteins were determined by Western blot analysis using anti-ubiquitin antibody. The cortical bone samples from the non-fractured legs were used as a control (Ctl).
NF-κB upregulates Itch expression in osteoblast progenitor cells
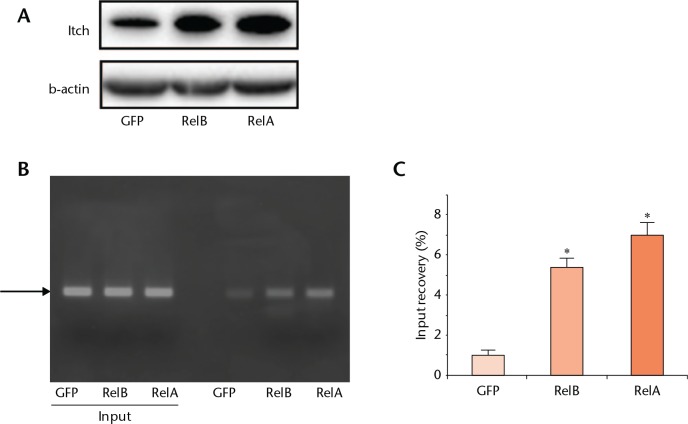
We have demonstrated that NF-κB RelA mediated RANKL induces Itch expression in osteoclasts by directly binding to NF-κB sites in the itch promoter.14 We wanted to know if this also occurs in osteoblasts. We have also previously identified two putative NF-κB binding sites at the -3473/-3465 and -3183/-3175 location of the murine Itch promoter (National Centre for Biotechnology Information RefSeq accession number NC_000068) using TFSEARCH software (version 1.3, National Institute of Advanced Industrial Science and Technology, Tokyo).14 We overexpressed RelA and RelB in C2C12 cells and found that both of them could increase Itch expression (Fig. 4a) and then performed ChIP assays using an anti- NF-κB RelA or RelB antibody to pull down protein-chromatin complexes and two primer sets around these NF-κB binding sites. Results showed RelA and RelB bindings to the NF-κB binding sites of the Itch promoter (Figs 4b and 4c).
Fig. 4.
NF-κB upregulates Itch expression in osteoblasts. C2C12, an osteoblast/myoblast cell line, was infected with GFP control, NF-κB RelA or RelB virus for 24 hours. a) Expression levels of Itch protein were determined by Western blot analysis. b) Chromatin immunoprecipitation (ChIP) assays were performed on immunocomplexes that were pulled down with anti-RelA, anti-RelB antibody or Immunoglobulin G (IgG). Precipitated DNA was measured by polymerase chain reaction (PCR) and quantitative PCR using sequence-specific primers. Values are mean (sd) of determinates in triplicate. * p < 0.05 versus green fluorescent protein (GFP)-infected samples.
Increased expression of osteoblast-related genes in callus tissues from Itch KO mice following facture
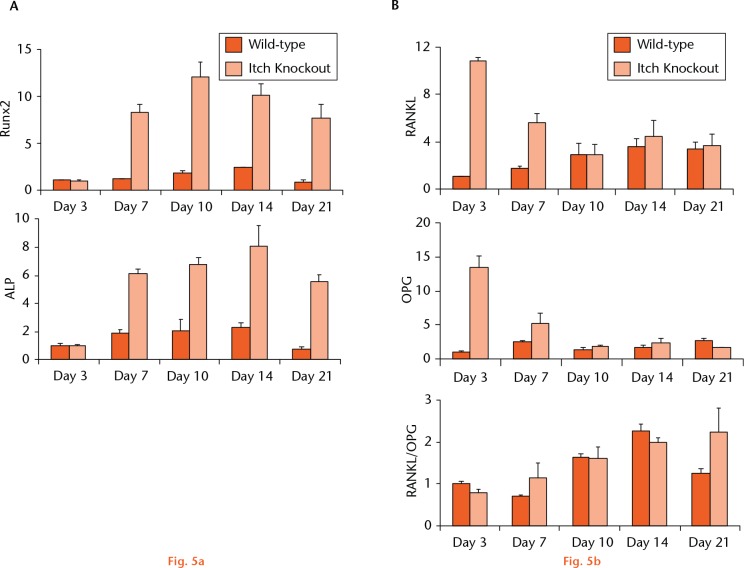
Itch functions as an E3 ligase to promote ubiquitination of targeting proteins, including the osteoblast positive regulators, thereby negatively regulates osteoblast differentiation.11 Thus, in the absence of Itch, osteoblast differentiation should be increased. To determine if this is the case at the bone fracture site, we examined the expression levels of osteoblast-associated genes (Runx2, ALP) and genes associated with bone remodelling, RANKL and OPG in fracture callus of Itch KO mice at different time points post-fracture. Runx2 and ALP are two positive osteoblast regulators which are regulated partially through ubiquitination and proteasome degradation in osteoblasts.7,15 It was seen that RANKL and OPG play critical roles in osteoclast formation and bone remodelling. The expression levels of ALP and Runx2 were significantly increased in fracture callus of Itch KO mice compared to WT, starting from day 7 post-fracture. The ratio of RANKL/OPG was not changed, although their expression levels were also elevated in Itch KO callus tissues (Fig. 5).
Altered expression patterns for genes associated with bone formation and remodelling in fracture callus of Itch KO mice. Itch KO and WT control mice underwent a surgical fracture and were killed at different time points post-fracture. The expression levels of genes related to osteoblast a) and osteoclast b) differentiation were examined by quantitative polymerase chain reaction (qPCR). Values are mean and standard deviation of triplicate experiments. The relative expression was calculated as fold change using the expression level at day 3 as 1.
Discussion
In this study, we examined the potential involvement of E3 ligase Itch in fracture repair using WT and global Itch knockout (KO) mice. We demonstrated elevated expression levels of WW domain-containing ubiquitin E3 ligases including Itch at early stages of fracture repair. Elevated Itch expression was associated with increased expression of NF-κB members. NF-κB upregulated Itch expression by binding to the NF-κB binding site in the Itch promoter in an osteoblast progenitor cell line. Callus tissues from Itch KO mice expressed high levels of osteoblast-positive regulator Runx2 and ALP. These findings suggest that during the early stage of fracture repair, inflammation (NF-κB) may increase expression of multiple WW domain-containing ubiquitin E3 ligases including Itch, which may limit osteoblast differentiation. Thus, Itch or factors that regulate Itch, or other WW domain-containing E3 ligase-mediated cellular events, could be a potential target for enhancing bone fracture healing.
Bone mass is dependent on the balance between bone formation by osteoblasts and bone resorption by osteoclasts.16-18 The differentiation and function of these cells are regulated by a number of factors, including the ubiquitin–proteasome pathway. In this pathway, the WW domain-containing ubiquitin ligase family that is composed of Smurf1, Smurf2, Wwp1, and Itch, and NEDD4 plays an important role in regulating bone cell functions.19 The WW domain-containing ubiquitin ligase family defines substrate specificity by recognising the target protein and then degradation in the 26S proteasome.20,22 For example, Smurf1 directly interacts with Runx2 and induces its degradation.19 The inflammatory cytokine tumour necrosis factor alpha (TNFα) increases Smurf1 expression in osteoblasts,5 leading to the degradation of Smad1 and Runx2, and subsequently impaired osteoblastogenesis.19 The ubiquitin–proteasome pathway is also involved in osteoclastogenesis by directly controlling important signalling pathways such as the nuclear factor kappa-B (NF-κB) pathway.22 Smurf2 transgenic mice in which the Smurf2 transgene is driven by the Col2a1 promoter develop an osteoarthritis-like phenotype.23 Wwp1 plays an important role in chronic inflammation-induced osteoblast inhibition and regulates JunB degradation.12 Itch regulates the stability of multiple proteins via ubiquitination and subsequent proteasomal or lysosomal degradation.24 In this study, we demonstrated for the first time that the expression levels of multiple WW domain-containing ubiquitin ligase members are significantly increased in callus tissues at an early stage of bone fracture, which is associated with elevated expression of NF-κB members. More importantly, we demonstrated that the total amount of ubiquitinated proteins in fracture calluses was also increased compared with samples from non-fractured bones. These data suggest that the ubiquitin–proteasome pathway may participate in the fracture repair process.
In osteoclasts, we reported that Itch inhibits RANKL-induced osteoclast formation and limits TRAF6 de-ubiquitination by binding the de-ubiquitinating enzyme cylindromatosis. Itch transcription is directly regulated by RANKL during osteoclast differentiation via NF-κB.14 In the current study, we demonstrated that NF-κB members also promote Itch expression in osteoblasts, indicating that NF-κB activation may affect protein ubiquitination and degradation in osteoblast lineage cells via Itch and perhaps other WW domain-containing E3 ligases.
Itch deficiency causes progressive autoimmune diseases10,25 and increased osteoclastogenesis.14 The molecular mechanisms by which Itch deficiency leads to autoimmune disease and multi-organ inflammation have been linked to persistent activation of the Jun amino terminal kinase (JNK) and NF-κB signal pathways in T cells and macrophages in response to inflammatory cytokines.26 Itch KO T cells have activated JNK signal protein kinase cascade and accelerated degradation of c-Jun and JunB proteins.27 Itch limits TNF-induced NF-κB activation by facilitating A20-mediated ubiquitination and degradation of the adaptor protein, Receptor-interacting protein (RIP), in T cells and macrophages.28,29 Itch is also required for negative regulation of TNF- and lipopolysaccharide (LPS)-mediated TNF receptor-associated factor 6 (TRAF6) ubiquitination induced by RING finger protein 11 (RNF11).30 We demonstrated increased expression of ALP and Runx2 mRNA in the fracture callus of Itch KO mice, but there is no change in the RANKL/OPG ratio. ALP and Runx2 levels are closely related to the degree of osteoblast differentiation. Increased expression of ALP and Runx2 in the callus tissue of Itch KO mice suggests that Itch-mediated osteoblast change may play a dominant role in the fracture repair process. However, this hypothesis needs to be tested in mice carrying osteoblast-specific depletion of Itch.
In summary, we demonstrated increased expression levels of WW domain-containing ubiquitin ligases in callus tissues compared to non-fracture bone, which is accompanied by increased amount of Ub-proteins. We also detected higher levels of osteoblast-associated genes in callus tissues from Itch KO mice than in samples from WT mice. These data suggest for the first time that the ubiquitin–proteasome pathway may participate in the bone fracture repair process. However, whether factors or drugs that modulate the protein ubiquitination and degradation can be used to promote fracture repair is something that needs to be examined.
Footnotes
Author Contributions: J. Liu: Study design, Study conduct, Data collection, Data analysis, Data interpretation, Drafting manuscript.
X. Li: Study design, Study conduct, Data collection, Data analysis, Data interpretation, Drafting manuscript.
H. Zhang: Study design, Study conduct, Data analysis, Data interpretation, Drafting manuscript.
R. Gu: Study design, Data interpretation, Drafting manuscript.
Z. Wang: Study design, Data interpretation, Drafting manuscript.
Z. Gao: Study design, Data interpretation, Drafting manuscript, Co-Corresponding author.
L. Xing: Study design, Data interpretation, Drafting manuscript, Co-Corresponding author.
ICMJE Conflicts of Interest: None declared
Funding Statement
This work was supported by grants from the National Institute of Health, United States (AR48697, AR63650, and AR053586) and from NYSTEM (CO-29548), USA, and from the National Natural Foundation of China (81202037).
References
- 1. Durrington HJ, Upton PD, Hoer S, et al. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J Biol Chem 2010;285:37641-37649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhi X, Chen C. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci 2012;69:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 2009;10:398-409. [DOI] [PubMed] [Google Scholar]
- 4. Guo R, Yamashita M, Zhang Q, et al. Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. J Biol Chem 2008;283:23084-23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaneki H, Guo R, Chen D, et al. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem 2006;281:4326-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu Q, Chen D, Zuscik MJ, O’Keefe RJ, Rosier RN. Overexpression of Smurf2 stimulates endochondral ossification through upregulation of beta-catenin. J Bone Miner Res 2008;23:552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao L, Huang J, Guo R, et al. Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. J Bone Miner Res 2010;25:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zou W, Chen X, Shim JH, et al. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat Cell Biol 2011;13:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melino G, Gallagher E, Aqeilan RI, et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ 2008;15:1103-1112. [DOI] [PubMed] [Google Scholar]
- 10. Perry WL, Hustad CM, Swing DA, et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet 1998;18:143-146. [DOI] [PubMed] [Google Scholar]
- 11. Zhang H, Xing L. Ubiquitin E3 ligase itch negatively regulates osteoblast differentiation from mesenchymal progenitor cells. Stem Cells 2013;31:1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao L, Huang J, Zhang H, et al. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells 2011;29:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhandari D, Robia SL, Marchese A. The E3 ubiquitin ligase atrophin interacting protein 4 binds directly to the chemokine receptor CXCR4 via a novel WW domain-mediated interaction. Mol Biol Cell 2009;20:1324-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H, Wu C, Matesic LE, et al. Ubiquitin E3 ligase Itch negatively regulates osteoclast formation by promoting de-ubiquitination of TNF receptor-associated factor 6. J Biol Chem 2013;288:22359-22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem 2003;278:27939-27944. [DOI] [PubMed] [Google Scholar]
- 16. Katagiri T, Takahashi N. Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis 2002;8:147-159. [DOI] [PubMed] [Google Scholar]
- 17. Kawamura N, Kugimiya F, Oshima Y, et al. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS One 2007;2:e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hie M, Tsukamoto I. Increased expression of the receptor for activation of NF-kappaB and decreased runt-related transcription factor 2 expression in bone of rats with streptozotocin-induced diabetes. Int J Mol Med 2010;26:611-618. [DOI] [PubMed] [Google Scholar]
- 19. Garrett IR, Chen D, Gutierrez G, et al. Selective inhibitors of the osteoblast proteasome stimulate bone formation in vivo and in vitro. J Clin Invest 2003;111:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A 2001;98:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sévère N, Dieudonné FX, Marie PJ. E3 ubiquitin ligase-mediated regulation of bone formation and tumorigenesis. Cell Death Dis 2013;4:e463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2010;2:a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu Q, Kim KO, Sampson ER, et al. Induction of an osteoarthritis-like phenotype and degradation of phosphorylated Smad3 by Smurf2 in transgenic mice. Arthritis Rheum 2008;58:3132-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heissmeyer V, Rao A. Itching to end NF-kappaB. Nat Immunol 2008;9:227-229. [DOI] [PubMed] [Google Scholar]
- 25. Lohr NJ, Molleston JP, Strauss KA, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet 2010;86:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang D, Elly C, Gao B, et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol 2002;3:281-287. [DOI] [PubMed] [Google Scholar]
- 27. Gao M, Labuda T, Xia Y, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 2004;306:271-275. [DOI] [PubMed] [Google Scholar]
- 28. Tao M, Scacheri PC, Marinis JM, et al. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol 2009;19:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shembade N, Harhaj NS, Parvatiyar K, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol 2008;9:254-262. [DOI] [PubMed] [Google Scholar]
- 30. Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J 2009;28:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]