Abstract
Genome-wide association studies (GWAS) of multiple populations with distinctive genetic and lifestyle backgrounds are crucial to the understanding of Type 2 Diabetes Mellitus (T2DM) pathophysiology. We report a GWAS on the genetic basis of T2DM in a 3,286 Lebanese participants. More than 5,000,000 SNPs were directly genotyped or imputed using the 1000 Genomes Project reference panels. We identify genome-wide significant variants in two loci CDKAL1 and TCF7L2, independent of sex, age and BMI, with leading variants rs7766070 (OR = 1.39, P = 4.77 × 10−9) and rs34872471 (OR = 1.35, P = 1.01 × 10−8) respectively. The current study is the first GWAS to find genomic regions implicated in T2DM in the Lebanese population. The results support a central role of CDKAL1 and TCF7L2 in T2DM susceptibility in Southwest Asian populations and provide a plausible component for understanding molecular mechanisms involved in the disease.
Type 2 diabetes mellitus (T2DM) is a chronic metabolic disease with a complex pathogenesis defined by genetic predisposition and environmental factors1. In Lebanon, T2DM was evaluated on 8,050 Lebanese cases in 1990. Its prevalence and incidence, similar to the international averages, were 5.0% and 1.5 to 1.7% respectively2. In 1992, a study on 436 cases from all over the Lebanese territory gave prevalence of 7 to 8% for T2DM and 10 to 11% for impaired glucose tolerance3. In 2005, a study on 3,000 exclusively Lebanese individuals from Greater Beirut showed a prevalence of 11.3% which increased with older age4. The combined prevalence of previously and newly diagnosed T2DM was 15.8%. At that time, in the U.S., 6.3% of the population had T2DM: 4.5% diagnosed and 1.8% undiagnosed according to the 2004 National Diabetes Fact Sheet. These results suggest that the prevalence of T2DM in Greater Beirut is relatively high and is increasing among the Lebanese population.
Furthermore, according to the latest figures in the American National Diabetes Statistics Report, 29.1 million children and adults in the United States have diabetes, and 86 million people have prediabetes5. The pathogenesis of T2DM is closely associated with a positive family history, male gender, age over 45 years, overweight, hypertension, and abnormal lipid levels. In addition, the genetic contribution to T2DM is well recognized with a total of 91 established associated susceptibility loci6,7,8,9,10,11,12,13,14,15,16,17. The common variants in the reported loci however account for only a small proportion of the heritability of T2DM and the functional role of most of these variants remains far from clear. It is possible that a large number of highly-penetrant but rare T2DM susceptibility genetic variants remain to be identified. Additional genome-wide explorations including whole genome and exome sequencing in well-established groups of patients and controls may unravel these additional important genetic disease contributors which will undoubtedly help us understand the complex mechanisms involved in the development of T2DM.
The prevalence of T2DM is distinctly variable across populations and this variability adds to the disease complexity. This variability could be due to differences in lifestyle factors such as dietary habits, as well as behavioral patterns among populations. Multiple established T2DM susceptibility genetic loci have been identified from previous Genome Wide Association Studies (GWAS) in populations of European and Asian ethnicities17,18,19. It is however, equally important to replicate the behavior of these previously discovered associated variants in ethnically different populations and to identify novel predisposing genetic factors that may be stronger in their association than in previously studied populations.
To identify T2DM susceptibility loci in the Lebanese population, we performed a GWAS of more than 5,000,000 SNPs, which were directly genotyped or imputed using the 1000 Genomes Project reference panels. Our study enrolled 1,388 patients and 1,902 non-diabetic subjects with detailed cross-sectional demographic and clinical information. Previously, targeted T2DM susceptibility SNP replication studies have been conducted in the Lebanese population. However, this study seeks to establish a baseline association for T2DM in the Lebanese population and thus it is the first genome-wide scan to find genomic regions implicated in T2DM. We also make use of recent reference data from the 1000 Genomes Project, which was previously shown to identify novel and refined associations for type 2 diabetes mellitus20,21.
Results
The study population has a mean age of 62.9 (±11.1) (Table 1). The healthy control group has a mean age of 62.40 (±11.82), compared to 63.62 ± (9.98) for the T2DM patients. Sixty-three percent of the individuals were males with 40.05 percent suffering from T2DM. The mean BMI was of 28.1 (±4.6) according to standard measures. 52 samples that failed genotype call rate (<95%)27 and that showed relatedness (kinship coefficient range > 0.0442)28, were removed.
Table 1. Distribution in the surveyed population of age, gender, BMI, and coronary artery disease by T2DM diagnostic status.
| Healthy controls | T2DM patients | Total | ||
|---|---|---|---|---|
| Sample size | 1,902 | 1,384 | 3,286 | |
| Age (SD) | 62.40 (11.82) | 63.62 (9.98) | 62.92 (11.1) | |
| Gender (%) | ||||
| Female | 597 (54) | 510 (46) | 1107 | |
| Male | 1305 (60) | 874 (40) | 2179 | |
| BMI (SD) | 28.07 (4.54) | 28.21 (4.74) | 28.13 (4.63) | |
| Coronary artery disease (%) | 1300 (56) | 1022 (44) | 2,322 |
Imputation
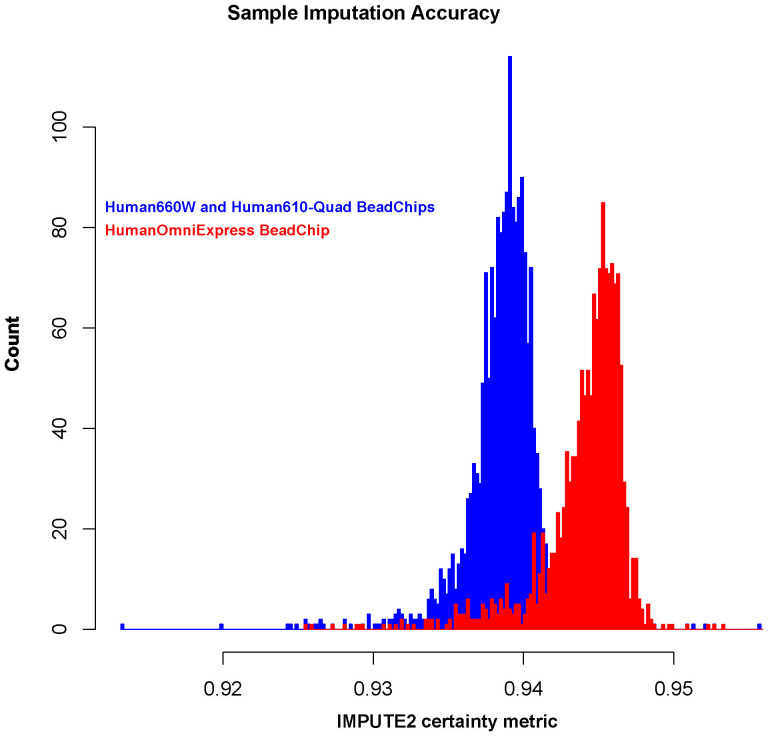
37995922 sites were imputed. The final set included 5891794 SNPs kept after QC (MAF > 0.05, AIMs removal). The accuracy of the genotype imputation depends on the array SNP coverage and the similarity of haplotypes between the study dataset and the reference panels. The Lebanese population was previously shown to be genetically very close to Europeans29 that are well represented in the 1000 Genomes Project references. This resulted in high imputation accuracy as shown by Impute2 certainty metric (Figure 1). Figure 1 shows biased imputation confidence depending on chip with the HumanOmniExpress (~700,000 SNPs) having better imputation accuracy compared to the Quad BeadChips (~550,000 SNPs). However, leading principal components reveals no bias regarding population coverage or stratification between chips (FigureS1).
Figure 1. Imputation accuracy.
Per-sample imputation confidence scores between true genotypes and genotypes predicted by imputation, averaged over imputation chunks. Accuracy increases with increased array SNP coverage.
Association with T2DM
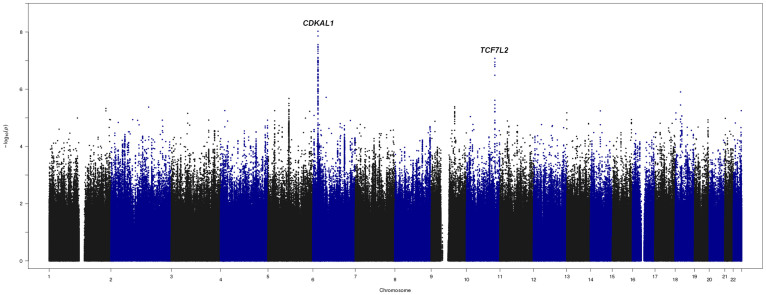
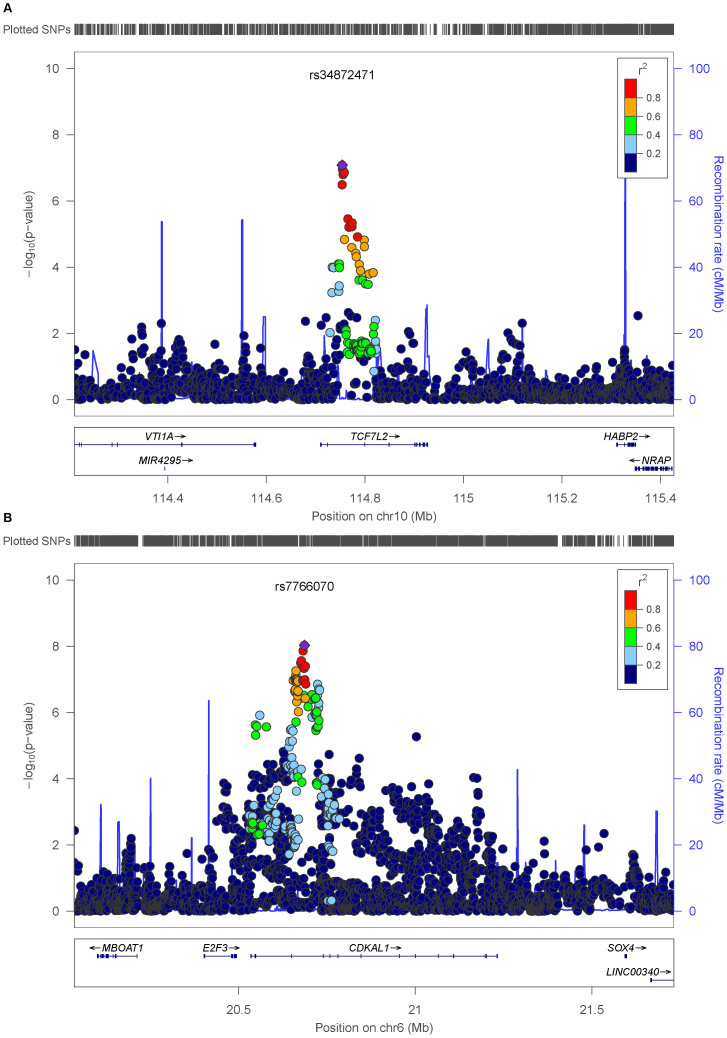
To map genetic loci associated with T2DM, genotyped and imputed data in 3,286 Lebanese individuals were tested for association with the disease (Figure 2). We identify variations located on chromosomes 4, 6, 9, 10, 12, and 18 that show trends of association with T2DM reaching P < 10−5 under an additive model (Supplementary Table 1). Seven variants located in CDKAL1 (cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1) reached genome-wide significance (P < 5 × 10−8) with lead signal rs7766070 (OR = 1.38, P = 9.37 × 10−9) (Figure 3, Table 2).
Figure 2. Manhattan plot.
Plot shows results of T2DM GWA analysis assuming additive impact in 3286 Lebanese using >5,000,000 genotyped or imputed SNPs. The Y-axis corresponds to the significance of the association (−log10 p-values). The X-axis represents the physical location of the variant colored by chromosome.
Figure 3. Regional association plots for the TCF7L2 (A) and CDKAL1 (B) genes.
Each regional plot shows the chromosomal position (hg 19) of SNPs in the specific region against −log10 p values estimated assuming additive impact. The SNP with the highest association signal at each locus is shown as a purple star; the other SNPs are colored according to the extent of LD with that SNP. Estimated recombination rates from the 1000 Genomes Project European population (release March 2012) are shown as light blue lines.
Table 2. T2DM GWA results showing SNPs that reached genome-wide significance (<5 × 10−8) in the initial testing phase or after conditioning for sex, age, and BMI.
| Additive model | Adjusted for sex | Adjusted for age | Adjusted for BMI | Adjusted for sex, age, and BMI | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Imputation | Chr | gene | MAF | OR | 95%CI | p-value | OR | 95%CI | p-value | OR | 95%CI | p-value | OR | 95%CI | p-value | OR | 95%CI | p-value |
| rs7766070 | imputed | 6 | CDKAL1 | 0.29 | 1.38 | 1.24-1.54 | 9.37E-09 | 1.38 | 1.24-1.54 | 1.71E-08 | 1.38 | 1.24-1.54 | 4.64E-09 | 1.39 | 1.24-1.55 | 6.83E-09 | 1.39 | 1.25-1.55 | 4.77E-09 |
| rs9348441 | imputed | 6 | CDKAL1 | 0.29 | 1.38 | 1.24-1.53 | 1.38E-08 | 1.38 | 1.24-1.53 | 2.33E-08 | 1.38 | 1.24-1.53 | 7.16E-09 | 1.38 | 1.24-1.54 | 9.26E-09 | 1.39 | 1.24-1.55 | 6.38E-09 |
| rs35261542 | imputed | 6 | CDKAL1 | 0.29 | 1.38 | 1.23-1.53 | 2.75E-08 | 1.38 | 1.23-1.53 | 4.58E-08 | 1.38 | 1.23-1.54 | 1.56E-08 | 1.39 | 1.24-1.55 | 1.93E-08 | 1.39 | 1.24-1.55 | 1.45E-08 |
| rs7451008 | imputed | 6 | CDKAL1 | 0.29 | 1.37 | 1.23-1.53 | 3.18E-08 | 1.37 | 1.23-1.53 | 5.16E-08 | 1.37 | 1.23-1.53 | 1.91E-08 | 1.39 | 1.24-1.55 | 1.63E-08 | 1.39 | 1.24-1.55 | 1.31E-08 |
| rs1569699 | genotyped | 6 | CDKAL1 | 0.38 | 1.33 | 1.21-1.48 | 3.51E-08 | 1.33 | 1.21-1.48 | 7.11E-08 | 1.34 | 1.21-1.48 | 1.74E-08 | 1.35 | 1.22-1.49 | 1.85E-08 | 1.35 | 1.22-1.49 | 1.49E-08 |
| rs9368222 | imputed | 6 | CDKAL1 | 0.29 | 1.36 | 1.22-1.52 | 4.02E-08 | 1.36 | 1.22-1.52 | 6.46E-08 | 1.37 | 1.23-1.52 | 2.18E-08 | 1.37 | 1.23-1.53 | 2.49E-08 | 1.38 | 1.23-1.53 | 1.65E-08 |
| rs9350271 | imputed | 6 | CDKAL1 | 0.37 | 1.33 | 1.2-1.47 | 4.64E-08 | 1.33 | 1.2-1.47 | 9.50E-08 | 1.33 | 1.21-1.48 | 2.18E-08 | 1.34 | 1.21-1.49 | 3.40E-08 | 1.34 | 1.21-1.49 | 2.59E-08 |
| rs10946396 | imputed | 6 | CDKAL1 | 0.38 | 1.33 | 1.2-1.47 | 5.56E-08 | 1.33 | 1.2-1.47 | 1.04E-07 | 1.33 | 1.2-1.47 | 2.49E-08 | 1.34 | 1.21-1.48 | 5.23E-08 | 1.34 | 1.21-1.48 | 4.08E-08 |
| rs6456369 | imputed | 6 | CDKAL1 | 0.38 | 1.33 | 1.2-1.47 | 7.72E-08 | 1.33 | 1.2-1.47 | 1.36E-07 | 1.33 | 1.2-1.47 | 3.38E-08 | 1.33 | 1.2-1.48 | 7.33E-08 | 1.33 | 1.2-1.48 | 5.20E-08 |
| rs6456368 | imputed | 6 | CDKAL1 | 0.31 | 1.35 | 1.21-1.5 | 8.23E-08 | 1.35 | 1.21-1.5 | 1.32E-07 | 1.35 | 1.21-1.5 | 3.88E-08 | 1.35 | 1.21-1.5 | 9.80E-08 | 1.35 | 1.21-1.5 | 6.91E-08 |
| rs7754840 | imputed | 6 | CDKAL1 | 0.31 | 1.34 | 1.21-1.49 | 9.67E-08 | 1.34 | 1.21-1.49 | 1.48E-07 | 1.34 | 1.21-1.49 | 4.45E-08 | 1.35 | 1.21-1.5 | 1.02E-07 | 1.35 | 1.21-1.5 | 6.71E-08 |
| rs4710940 | imputed | 6 | CDKAL1 | 0.38 | 1.32 | 1.2-1.46 | 9.77E-08 | 1.32 | 1.2-1.46 | 1.83E-07 | 1.33 | 1.2-1.47 | 4.35E-08 | 1.33 | 1.2-1.47 | 8.27E-08 | 1.33 | 1.2-1.48 | 6.28E-08 |
| rs35456723 | imputed | 6 | CDKAL1 | 0.3 | 1.35 | 1.21-1.5 | 1.02E-07 | 1.35 | 1.21-1.5 | 1.66E-07 | 1.35 | 1.21-1.5 | 4.86E-08 | 1.35 | 1.21-1.5 | 1.03E-07 | 1.35 | 1.21-1.5 | 7.44E-08 |
| rs9295474 | imputed | 6 | CDKAL1 | 0.31 | 1.35 | 1.21-1.5 | 1.07E-07 | 1.35 | 1.21-1.5 | 1.69E-07 | 1.35 | 1.21-1.5 | 5.19E-08 | 1.36 | 1.22-1.51 | 6.71E-08 | 1.36 | 1.22-1.51 | 4.51E-08 |
| rs7756992 | genotyped | 6 | CDKAL1 | 0.3 | 1.34 | 1.21-1.49 | 1.08E-07 | 1.34 | 1.21-1.49 | 1.78E-07 | 1.34 | 1.21-1.5 | 5.14E-08 | 1.35 | 1.21-1.51 | 6.63E-08 | 1.35 | 1.22-1.51 | 4.33E-08 |
| rs6456367 | imputed | 6 | CDKAL1 | 0.31 | 1.34 | 1.21-1.49 | 1.10E-07 | 1.34 | 1.21-1.49 | 1.71E-07 | 1.34 | 1.21-1.49 | 4.94E-08 | 1.35 | 1.21-1.5 | 1.09E-07 | 1.35 | 1.21-1.5 | 7.07E-08 |
| chr6:20660689:D | imputed | 6 | CDKAL1 | 0.37 | 1.33 | 1.2-1.47 | 1.11E-07 | 1.33 | 1.2-1.47 | 1.96E-07 | 1.33 | 1.2-1.47 | 4.96E-08 | 1.33 | 1.2-1.48 | 1.03E-07 | 1.33 | 1.2-1.48 | 7.54E-08 |
| rs7752780 | imputed | 6 | CDKAL1 | 0.31 | 1.34 | 1.21-1.49 | 1.11E-07 | 1.34 | 1.21-1.49 | 1.71E-07 | 1.34 | 1.21-1.49 | 4.93E-08 | 1.34 | 1.21-1.5 | 1.15E-07 | 1.35 | 1.21-1.5 | 7.31E-08 |
| rs34872471 | imputed | 10 | TCF7L2 | 0.4 | 1.32 | 1.19-1.46 | 8.30E-08 | 1.32 | 1.19-1.46 | 1.51E-07 | 1.32 | 1.2-1.46 | 4.80E-08 | 1.35 | 1.22-1.49 | 9.91E-09 | 1.35 | 1.22-1.5 | 1.01E-08 |
| rs35198068 | imputed | 10 | TCF7L2 | 0.4 | 1.31 | 1.19-1.45 | 1.12E-07 | 1.31 | 1.19-1.45 | 2.03E-07 | 1.32 | 1.19-1.46 | 6.61E-08 | 1.34 | 1.21-1.49 | 1.38E-08 | 1.35 | 1.22-1.49 | 1.42E-08 |
| rs7903146 | genotyped | 10 | TCF7L2 | 0.4 | 1.31 | 1.19-1.45 | 1.39E-07 | 1.31 | 1.19-1.45 | 2.44E-07 | 1.31 | 1.19-1.45 | 8.38E-08 | 1.34 | 1.21-1.48 | 1.74E-08 | 1.34 | 1.21-1.49 | 1.79E-08 |
| rs4506565 | imputed | 10 | TCF7L2 | 0.41 | 1.31 | 1.18-1.44 | 1.60E-07 | 1.31 | 1.18-1.44 | 2.70E-07 | 1.31 | 1.19-1.45 | 9.64E-08 | 1.34 | 1.21-1.48 | 2.00E-08 | 1.34 | 1.21-1.48 | 1.96E-08 |
| rs7901695 | genotyped | 10 | TCF7L2 | 0.41 | 1.3 | 1.18-1.43 | 3.23E-07 | 1.3 | 1.18-1.43 | 5.53E-07 | 1.3 | 1.18-1.44 | 2.03E-07 | 1.33 | 1.2-1.47 | 4.35E-08 | 1.33 | 1.2-1.47 | 4.50E-08 |
Adjustment for sex, age, and BMI (Table 2) identified 16 additional genome-wide significant variants in CDKAL1 and TCF7L2 (Transcription factor 7-like 2) and showed that association of these two loci with T2DM appear to be independent from age and BMI. These variants are also added to Table S1 along with imputation scores.
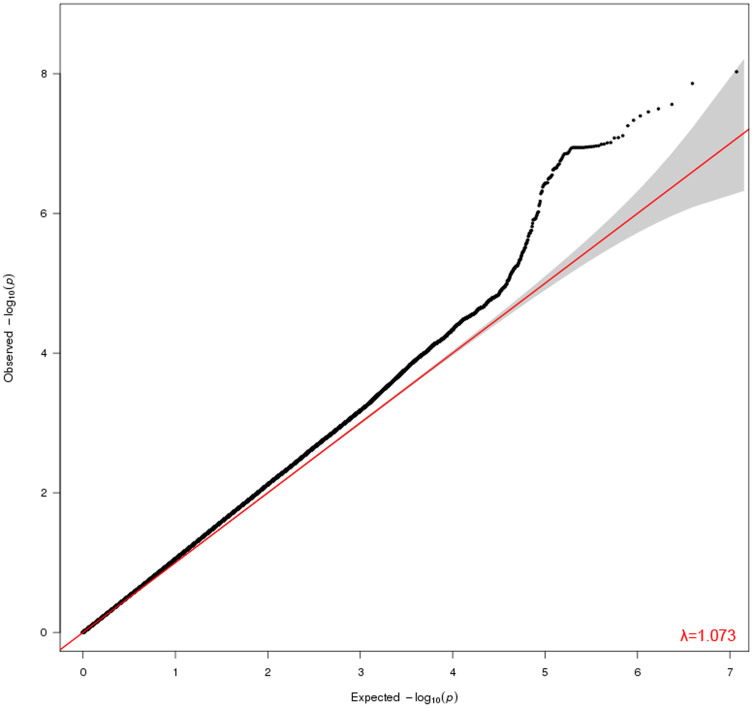
The genomic control inflation factor (λ) which compares observed association statistics against the expected distribution, was 1.073 suggesting possible but marginal over-dispersion of the association statistics (Figure 4)30.
Figure 4. Quantile-Quantile plot of the GWAS results.
Plot compares observed −log10 p values of the tested SNPs on the vertical axis to expected −log10 p values under the null hypothesis on the horizontal axis. The genomic control ratio was 1.073, indicating the lack of strong effect of systematic error such as population stratification.
SNPs of interest that showed trend of association but did not reach genome-wide significance were 12 variants on chromosome 9 (Supplementary Table 1), overlapping a region with microRNA 3910-1 and 3910-2 downstream the gene ROR2 (receptor tyrosine kinase-like orphan receptor 2).
Three of the SNPs, rs7766070, rs9368222, and rs34872471, identified in Table 2 were significant in DIAGRAM consortium6,7,9,10,11,15,17. One other, rs1044083315, was identified but not genome wide significant (Table S2), but which may be of particular interest in Middle Eastern populations31. Table S2 lists DIAGRAM consortium study SNPs along with comparisons with our results and imputation confidence scores.
Discussion
T2DM global prevalence has been steadily increasing over the past 50 years and is today considered a major international health concern. In addition to environmental factors such as diet and lifestyle, genetic susceptibility appears to have substantial role in the etiology of T2DM. Furthermore, T2DM risk alleles show significant disparity in frequencies across worldwide populations. Studies analyzing distribution patterns of genetic diseases found that T2DM risk alleles demonstrated the most extreme differentiation32, with population frequencies decreasing from Sub-Saharan Africa, and through Europe to East Asia32,33.
The current work contributes to the ongoing worldwide effort to identify the genetic factors that affect the risk of T2DM. We provide analysis in a novel population that is highly admixed with ancestral components from the Middle East, Europe, Central Asia, and sub-Saharan Africa29. The risk alleles identified in the Lebanese population could assist in understanding the disparity in T2DM prevalence across worldwide populations.
We report the first genome-wide scan for genetic susceptibility to T2DM in Lebanese subjects. We demonstrate strong association of T2DM with CDKAL1 and TCF7L2 genes previously shown to predispose to this condition15,18,19,24,26,34,35,36,37. A large number of genetic variants have recently been identified to be associated with T2DM in GWASs in Europeans6,10,15,17,19,25. The elucidation of the genetic mechanisms responsible for the heterogeneous condition of T2DM in different ethnicities may give important contributions to better understand the complexity of this disease. While some associations are consistent across different ethnic groups38,39, some, like the TCF7L2 variants, have been found to be heterogeneous6,25. Our study confirmed genome-wide association of 23 different variants across the CDKAL1 and TCF7L2 genes previously reported to play a role in T2DM in different populations23,40,41. This association remained significant after adjustment for sex, age, and BMI, suggesting that these risk factors are not likely to be major contributors to the observed association.
After adjusting for age and BMI, most variants in CDKAL1 and TCF7L2 showed stronger association to T2DM, indicating that these genes confer susceptibility to T2DM independently from the classical clinical risks. Functional studies of these two genes show their implication in down-regulating blood glucose level. CDKAL1 encodes a 65 kD protein of unknown function expressed in pancreatic islet and skeletal muscle26. CDKAL1 shows homology with CDK5 regulatory subunit associated protein 1 (CDK5RAP1), an inhibitor of CDK5 activation. CDK5 plays a role in the regulation of pancreatic beta cell function by down-regulating insulin expression42,43. The TCL7L2 gene product is a high mobility group (HMG) box-containing transcription factor implicated in blood glucose homeostasis. TCL7L2 acts through regulation of proglucagon through repression of the proglucagon gene in enteroendocrine cells via the Wnt signaling pathway44.
T2DM is a socio-economic burden in Lebanon and genetic studies aim at posing important public health questions in regard to strategies for diagnosis, treatment, and prevention of the disease. Previous replication studies examining a total of 29 SNPs in the Lebanese population have identified variants associated with T2DM in COL8A1, KCNQ1, ALX4, HNF145 (n = 2071), CDKAL150 (n = 1422), and TCF7L246,47 (n = 1610).
Our genome-wide analysis of more than 5,000,000 SNPs in the largest T2DM study to date in the region has only identified variations in the CDKAL1 and TCF7L2 genes to be genome wide significantly associated with the disease.
Lebanon represents a Levantine region with higher European affinity than other parts of the Middle East48, and therefore potentially fills in the map between SNPs identified in European populations contrasted with other Middle Eastern populations. Further, since T2DM is an emerging issue within the Middle East, these populations may differentially highlight some variants distinct from other regions. Even accepting relatively low power compared to that available in meta-studies, the moderately small number of replicated GWS significant SNPs in our study is less than expected, especially given the affinity of Lebanon with other European populations48.
Our results contribute to ongoing efforts to identify and confirm risk alleles associated with T2DM. Mapping disease alleles in different populations can provide important perspectives in disease diagnosis and steer functional studies to provide better understanding of the pathogenesis of T2DM.
Methods
Study Participants
The Lebanese study participants were all of Lebanese origin and were recruited to this study in three phases. A first recruitment campaign, conducted with the collaboration of the Lebanese University Medical Center, led to the recruitment of 506 subjects from the suburbs of Beirut, the capital of Lebanon. In a second campaign conducted in North Lebanon, 492 subjects were successfully recruited. The 2,292 remaining participants were recruited as part of the Functional Genomic Diagnostic Tools for Coronary Artery Disease project initiative (FGENTCARD) from the Rafic Hariri University Hospital, and the “Centre Hospitalier du Nord” in Lebanon49,50. Research and methods were carried out in compliance with the Helsinki Declaration and with the approval of the LAU institutional review board and local ethics committees on human research (Reference number SMPZ08072010-4). All participants signed an informed consent and data and blood samples were obtained from each individual. By taking part in the two recruitment campaigns, participants (1) answered a detailed questionnaire, (2) gave a blood sample for DNA analysis and (3) gave a blood sample for HbA1C, fasting blood glucose (FBS), and lipid profile measures after 12 hours fasting. For the remaining FGENTCARD 2,292 participants, a detailed questionnaire was duly filled, a blood sample was obtained for DNA and metabolites analysis and annotations were coded from medical charts for data such as laboratory tests, prescribed medications, and presence of clinical conditions. Table 1 describes clinical and demographic characteristics of the study participants49. Previously reported51 analysis of these subjects showed association with recruitment for questionnaire variables, such as “exercise level,” “smoking usage”. However, blood panel associations showed very consistent odds ratios for T2DM and coronary artery disease risk independently of recruitment.
Selection of Patients and Controls
For the 998 participants selected through the two recruitment campaigns, an HbA1C of 48 mmol/mol (6.5%) was used as the cut-off point for diagnosing T2DM in line with the World Health Organization definition of type 2 diabetes mellitus diagnosis52. For the 2,292 participants from the FGENTCARD project, T2DM was diagnosed by an ascertained physician supported by subjects HbA1C levels and or their two-hour plasma glucose concentration after an oral glucose tolerance test as documented in their medical records. For the control dataset, 85.23% of the participants were ≥50 years old and have no history of T2DM. Body mass index (BMI) was calculated according to standard measurements. Selection criteria for DNA analysis were based on the complete availability of the genotyping data and the T2DM-relevant characteristics, resulting in the selection of 3,286 subjects.
Genotyping and imputation
DNA was extracted using a standard phenol-chloroform extraction procedure. Samples were genotyped using Illumina HumanOmniExpress-12V1-1 Multi-use (623 controls, 839 cases, ~700,000 SNPs) and Illumina Human610-660W Quad BeadChips (1279 controls, 545 cases, ~550,000 SNPs). Plink27 was used for data management and quality control keeping 3,286 samples with call rate ≥95%, SNPs call rate ≥98%, Hardy-Weinberg p > 10−6, MAF ≥ 1%. Genotype data was converted to NCBI genome build 37 using LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Chromosomes were phased with SHAPEIT53 using the 1000 Genomes Phase I haplotypes54 provided on the IMPUTE2 website (1000 Genomes Phase I integrated haplotypes (produced using SHAPEIT2), September 2013) based on sequence data for 1,092 TGP samples from release 20110521. IMPUTE255 was employed to impute genotypes in blocks of ~5 Mb using all populations in the 1000 Genomes Phase I data. Accuracy of imputation was measured using IMPUTE2's information metric “info”.
Statistical analysis
GWAS can be confounded by population stratification. The most common approach to correct for population stratification is to include the principal components from the PCA as covariates. However, there is no solid method to determine how many components to include and selection is usually subjective with inconclusive results. Correction for stratification becomes even more complicated in populations like the Lebanese where genetic structure is recent and not driven by the usual factors of distance and geography29. Furthermore, previous studies have shown that historical migrations to Lebanon have potentially created disease-associated population structures that are similar to those in the source populations22,56. In this study, we use insights from our previous studies on population stratifications in the Lebanese to avoid type 1 errors in the GWAS.
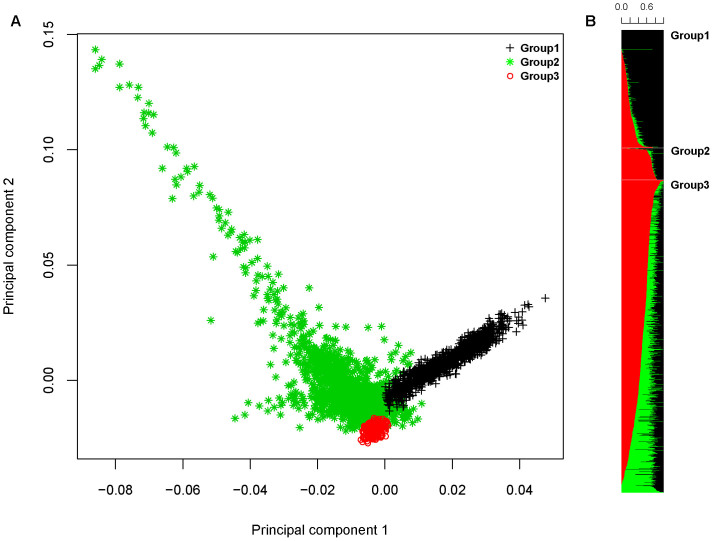
We apply a method that uses available 1000 Genomes Project reference panels to impute a resolved ancestry dataset29 as well as the current T2DM cohort to the same SNP coverage resolution. We next use two model-based clustering methods to unambiguously classify subjects into groups (Figure 5):
1- mclust (http://www.stat.washington.edu/mclust/) uses model-based hierarchical clustering and Bayesian Information Criterion to classify samples into related groups.
2- ADMIXTURE57 assigns individuals in a model-based manner into ancestral populations using maximum likelihood estimates.
Figure 5. Population structure in the Lebanese.
A) Principal component analysis shows an orthogonal population structure. Model-based clustering using mclust identifies three groups similar to previously reported stratification in the Lebanese. B) Estimation of individual ancestry using ADMIXTURE at K = 3 shows three groups with disparate proportions of ancestral allele frequencies.
The two methods identified similar population structures and assigned samples into three ancestral groups (Figure 5) similar to previous ancestry reports on the Lebanese29. We next proceed by extracting 192,310 ancestry informative markers (AIMs) between the groups and exclude it from the GWA analysis. We have also tested our method by using the identified ancestry groups as covariates in the GWAS and obtained identical results.
Association tests were performed using SNPTEST v2.458, employing frequentist association tests and taking into account uncertainty in imputed genotypes (calling threshold 0.9). At each SNP, an additive, dominant, or recessive model was tested versus a model of no association. Association tests were also conditioned on age, sex, and BMI to test for a genetic effect over and above that explained by these covariates. Chip type and enrollment were also both included in a separate analysis; results showed correlated but no genome-wide significant SNPs. Further, a PCA painted by chip type (Figure S1) shows no significant bias. LocusZoom59 was used to plot regional association results from the genome-wide association scans.
Supplementary Material
Supplementary Info
Acknowledgments
We thank the patients for agreeing to participate in the study. We thank Dr. Bouchra Douaihy and Mrs. Hana Sbeite for helping in the campaigns. We thank the dispensaire de la maison maronite de soins et d'hébergement- Ain El Remmaneh, Ehden Governmental Hospital - Ehden, Caritas Liban - Dahr el Ain, Lebanese Red Cross - Amioun, Saint Joseph dispensary - Zgharta, and dispensary of Kfarhabou –Donnieh for hosting the campaigns. This publication was made possible by a grant from the Qatar National Research Fund under its National Priorities Research program (NPRP) award number NPRP 09-215-3-049. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Qatar National Research Fund.
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the experiments: M.G.S., M.H., H.E.S., D.E.P. and P.A.Z. Performed the statistical analysis: M.H. Analyzed the data: M.G.S., M.H., A.K.S., D.E.P., H.E.S. and P.A.Z. Contributed reagents/materials/genotyping: Y.A.S., Y.A., K.H., J.R., F.M., D.G. and H.E.S. Wrote the paper: M.G.S., M.H., D.E.P. and P.A.Z. with help from all co-authors.
References
- Stumvoll M., Goldstein B. J. & van Haeften T. W. Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365, 1333–46 (2005). [DOI] [PubMed] [Google Scholar]
- Hirbli K. I., Aboujaoude J. H., Ghorra F. S. & Barakat-el-Khoury W. M. [Prevalence and incidence of diabetes mellitus in Lebanon]. Diabete Metab 16, 479–83 (1990). [PubMed] [Google Scholar]
- Hirbli K. I., Gerges T. A., Karam V. J. & Saikaly J. A. [The estimation of the prevalence of diabetes mellitus in Lebanon]. J Med Liban 40, 22–30 (1992). [PubMed] [Google Scholar]
- Hirbli K. I. et al. Prevalence of diabetes in greater Beirut. Diabetes Care 28, 1262 (2005). [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. Atlanta, GA: US Department of Health and Human Services. (2014) (Date of access:03/10/2014). Available from: http://www.cdc.gov/diabetes/pubs/statsreport14/national-diabetes-report-web.pdf.
- Cho Y. S. et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet 44, 67–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42, 105–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A. et al. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner J. S. et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 43, 984–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet 19, 2706–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44, 991–1005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X. O. et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet 6, e1001127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F. J. et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 6, e1000847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42, 579–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T. et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet 42, 864–8 (2010). [DOI] [PubMed] [Google Scholar]
- Zeggini E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40, 638–45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki H. et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 40, 1098–102 (2008). [DOI] [PubMed] [Google Scholar]
- Hara K. et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet 23, 239–46 (2013). [DOI] [PubMed] [Google Scholar]
- Huang J., Ellinghaus D., Franke A., Howie B. & Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet 20, 801–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. Y-chromosome R-M343 African lineages and sickle cell disease reveal structured assimilation in Lebanon. J Hum Genet 56, 29–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S. et al. TCF7L2 gene polymorphisms and type 2 diabetes risk: a comprehensive and updated meta-analysis involving 121, 174 subjects. Mutagenesis 28, 25–37 (2013). [DOI] [PubMed] [Google Scholar]
- Saxena R. et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet 90, 410–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R. et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes 62, 1746–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. Genome-wide diversity in the levant reveals recent structuring by culture. PLoS Genet 9, e1003316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K. & Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 60, 155–66 (2001). [DOI] [PubMed] [Google Scholar]
- Al-Daghri N. M. et al. Association between type 2 diabetes mellitus-related SNP variants and obesity traits in a Saudi population. Mol Biol Rep 41, 1731–40 (2014). [DOI] [PubMed] [Google Scholar]
- Chen R. et al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLoS Genet 8, e1002621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona E. et al. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. PLoS Genet 9, e1003447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T. W. T. C. C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y. et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet 44, 302–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. R. et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet 8, e1002741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancakova A. et al. Single-nucleotide polymorphism rs7754840 of CDKAL1 is associated with impaired insulin secretion in nondiabetic offspring of type 2 diabetic subjects and in a large sample of men with normal glucose tolerance. J Clin Endocrinol Metab 93, 1924–30 (2008). [DOI] [PubMed] [Google Scholar]
- Haiman C. A. et al. Consistent directions of effect for established type 2 diabetes risk variants across populations: the population architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes 61, 1642–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R. et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes 58, 478–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y. et al. Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. J Clin Endocrinol Metab 93, 3136–41 (2008). [DOI] [PubMed] [Google Scholar]
- Palmer N. D. et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 60, 662–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda M., Rukstalis J. M. & Habener J. F. Inhibition of cyclin-dependent kinase 5 activity protects pancreatic beta cells from glucotoxicity. J Biol Chem 281, 28858–64 (2006). [DOI] [PubMed] [Google Scholar]
- Wei F. Y. et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med 11, 1104–8 (2005). [DOI] [PubMed] [Google Scholar]
- Yi F., Brubaker P. L. & Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem 280, 1457–64 (2005). [DOI] [PubMed] [Google Scholar]
- Almawi W. Y. et al. A replication study of 19 GWAS-validated type 2 diabetes at-risk variants in the Lebanese population. Diabetes Res Clin Pract 102, 117–22 (2013). [DOI] [PubMed] [Google Scholar]
- Nemr R. et al. Replication study of common variants in CDKAL1 and CDKN2A/2B genes associated with type 2 diabetes in Lebanese Arab population. Diabetes Res Clin Pract 95, e37–40 (2012). [DOI] [PubMed] [Google Scholar]
- Nemr R. et al. Transcription factor-7-like 2 gene variants are strongly associated with type 2 diabetes in Lebanese subjects. Diabetes Res Clin Pract 98, e23–7 (2012). [DOI] [PubMed] [Google Scholar]
- Badro D. A. et al. Y-chromosome and mtDNA genetics reveal significant contrasts in affinities of modern Middle Eastern populations with European and African populations. PLoS One 8, e54616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saade S. et al. Large scale association analysis identifies three susceptibility loci for coronary artery disease. PLoS One 6, e29427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youhanna S. et al. Parental consanguinity and family history of coronary artery disease strongly predict early stenosis. Atherosclerosis 212, 559–63 (2010). [DOI] [PubMed] [Google Scholar]
- Platt D. E. et al. Circulating lipid levels and risk of coronary artery disease in a large group of patients undergoing coronary angiography. J Thromb Thrombolysis (2014) Advance online publication. 10.1007/s11239-014-1069-2. [DOI] [PubMed] [Google Scholar]
- Colagiuri S. Glycated haemoglobin (HbA1c) for the diagnosis of diabetes mellitus--practical implications. Diabetes Res Clin Pract 93, 312–3 (2011). [DOI] [PubMed] [Google Scholar]
- Delaneau O., Zagury J. F. & Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods 10, 5–6 (2013). [DOI] [PubMed] [Google Scholar]
- Abecasis G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P. & Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. mtDNA lineages reveal coronary artery disease-associated structures in the Lebanese population. Ann Hum Genet 76, 1–8 (2012). [DOI] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J. & Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J. & Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info