Abstract
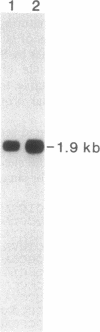
Many plants, as well as other organisms, accumulate betaine (N,N,N-trimethylglycine) as a nontoxic or protective osmolyte under saline or dry conditions. In plants, the last step in betaine synthesis is catalyzed by betaine-aldehyde dehydrogenase (BADH, EC 1.2.1.8), a nuclear-encoded chloroplastic enzyme. A cDNA clone for BADH (1812 base pairs) was selected from a lambda gt10 cDNA library derived from leaves of salt-stressed spinach (Spinacia oleracea L.). The library was screened with oligonucleotide probes corresponding to amino acid sequences of two peptides prepared from purified BADH. The authenticity of the clone was confirmed by nucleotide sequence analysis; this analysis demonstrated the presence of a 1491-base-pair open reading frame that contained sequences encoding 12 peptide fragments of BADH. The clone hybridized to a 1.9-kilobase mRNA from spinach leaves; this mRNA was more abundant in salt-stressed plants, consistent with the known salt induction of BADH activity. The amino acid sequence deduced from the BADH cDNA sequence showed substantial similarities to those for nonspecific aldehyde dehydrogenases (EC 1.2.1.3 and EC 1.2.1.5) from several sources, including absolute conservation of a decapeptide in the probable active site. Comparison of deduced and determined amino acid sequences indicated that the transit peptide may comprise only 7 or 8 residues, which is atypically short for precursors to stromal proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arakawa K., Takabe T., Sugiyama T., Akazawa T. Purification of betaine-aldehyde dehydrogenase from spinach leaves and preparation of its antibody. J Biochem. 1987 Jun;101(6):1485–1488. doi: 10.1093/oxfordjournals.jbchem.a122019. [DOI] [PubMed] [Google Scholar]
- Beach L. R., Spencer D., Randall P. J., Higgins T. J. Transcriptional and post-transcriptional regulation of storage protein gene expression in sulfur-deficient pea seeds. Nucleic Acids Res. 1985 Feb 11;13(3):999–1013. doi: 10.1093/nar/13.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry E. A., Trumpower B. L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987 Feb 15;161(1):1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- Dale R. M., Arrow A. A rapid single-stranded cloning, sequencing, insertion, and deletion strategy. Methods Enzymol. 1987;155:204–214. doi: 10.1016/0076-6879(87)55017-0. [DOI] [PubMed] [Google Scholar]
- Farrés J., Guan K. L., Weiner H. Primary structures of rat and bovine liver mitochondrial aldehyde dehydrogenases deduced from cDNA sequences. Eur J Biochem. 1989 Mar 1;180(1):67–74. doi: 10.1111/j.1432-1033.1989.tb14616.x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Goldberg A. M., McCaman R. E. Betaine aldehyde dehydrogenase: assay and partial purification. Biochim Biophys Acta. 1968 Aug 27;167(1):186–189. doi: 10.1016/0005-2744(68)90290-8. [DOI] [PubMed] [Google Scholar]
- Hanson A. D., May A. M., Grumet R., Bode J., Jamieson G. C., Rhodes D. Betaine synthesis in chenopods: Localization in chloroplasts. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3678–3682. doi: 10.1073/pnas.82.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. P., Colbert J. T., Lissemore J. L., Barker R. F., Quail P. H. Molecular cloning of cDNA for Avena phytochrome. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2332–2336. doi: 10.1073/pnas.81.8.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. C., Tani K., Fujiyoshi T., Kurachi K., Yoshida A. Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3771–3775. doi: 10.1073/pnas.82.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson J., von Bahr-Lindström H., Jeck R., Woenckhaus C., Jörnvall H. Mitochondrial aldehyde dehydrogenase from horse liver. Correlations of the same species variants for both the cytosolic and the mitochondrial forms of an enzyme. Eur J Biochem. 1988 Mar 15;172(3):527–533. doi: 10.1111/j.1432-1033.1988.tb13920.x. [DOI] [PubMed] [Google Scholar]
- Jones D. E., Jr, Brennan M. D., Hempel J., Lindahl R. Cloning and complete nucleotide sequence of a full-length cDNA encoding a catalytically functional tumor-associated aldehyde dehydrogenase. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1782–1786. doi: 10.1073/pnas.85.6.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Meulenberg C. H., Kingma J., Witholt B. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J Biol Chem. 1989 Apr 5;264(10):5442–5451. [PubMed] [Google Scholar]
- Landfald B., Strøm A. R. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J Bacteriol. 1986 Mar;165(3):849–855. doi: 10.1128/jb.165.3.849-855.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rudulier D., Strom A. R., Dandekar A. M., Smith L. T., Valentine R. C. Molecular biology of osmoregulation. Science. 1984 Jun 8;224(4653):1064–1068. doi: 10.1126/science.224.4653.1064. [DOI] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski C. B., Olson S. W., Piepenbrock M., Schmitt J. M., Bohnert H. J. Time Course of mRNA Induction Elicited by Salt Stress in the Common Ice Plant (Mesembryanthemum crystallinum). Plant Physiol. 1989 Mar;89(3):811–816. doi: 10.1104/pp.89.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J., Chua N. H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988 Aug;7(8):2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S. M., Moreau R. A., Yu C., Huang A. H. Betaine accumulation and betaine-aldehyde dehydrogenase in spinach leaves. Plant Physiol. 1981 Jun;67(6):1105–1108. doi: 10.1104/pp.67.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateman J. A., Doy C. H., Olsen J. E., Norris U., Creaser E. H., Hynes M. Regulation of alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (AldDH) in Aspergillus nidulans. Proc R Soc Lond B Biol Sci. 1983 Feb 22;217(1208):243–264. doi: 10.1098/rspb.1983.0009. [DOI] [PubMed] [Google Scholar]
- Pickett M., Gwynne D. I., Buxton F. P., Elliott R., Davies R. W., Lockington R. A., Scazzocchio C., Sealy-Lewis H. M. Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene. 1987;51(2-3):217–226. doi: 10.1016/0378-1119(87)90310-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröppel-Meier G., Kaiser W. M. Ion Homeostasis in Chloroplasts under Salinity and Mineral Deficiency : I. Solute Concentrations in Leaves and Chloroplasts from Spinach Plants under NaCl or NaNO(3) Salinity. Plant Physiol. 1988 Aug;87(4):822–827. doi: 10.1104/pp.87.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. K., Nelson D. E., Kuhn D., Hasegawa P. M., Bressan R. A. Molecular Cloning of Osmotin and Regulation of Its Expression by ABA and Adaptation to Low Water Potential. Plant Physiol. 1989 Jul;90(3):1096–1101. doi: 10.1104/pp.90.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero G. N. Protons, osmolytes, and fitness of internal milieu for protein function. Am J Physiol. 1986 Aug;251(2 Pt 2):R197–R213. doi: 10.1152/ajpregu.1986.251.2.R197. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Walker J. C., Howard E. A., Dennis E. S., Peacock W. J. DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6624–6628. doi: 10.1073/pnas.84.19.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel P., Weretilnyk E. A., Hanson A. D. Betaine aldehyde oxidation by spinach chloroplasts. Plant Physiol. 1986 Nov;82(3):753–759. doi: 10.1104/pp.82.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weretilnyk E. A., Hanson A. D. Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity. Arch Biochem Biophys. 1989 May 15;271(1):56–63. doi: 10.1016/0003-9861(89)90255-5. [DOI] [PubMed] [Google Scholar]
- Weretilnyk E. A., Hanson A. D. Betaine aldehyde dehydrogenase polymorphism in spinach: genetic and biochemical characterization. Biochem Genet. 1988 Feb;26(1-2):143–151. doi: 10.1007/BF00555495. [DOI] [PubMed] [Google Scholar]
- de Jong W. W., Zweers A., Cohen L. H. Influence of single amino acid substitutions on electrophoretic mobility of sodium dodecyl sulfate-protein complexes. Biochem Biophys Res Commun. 1978 May 30;82(2):532–539. doi: 10.1016/0006-291x(78)90907-5. [DOI] [PubMed] [Google Scholar]
- von Bahr-Lindström H., Hempel J., Jörnvall H. The cytoplasmic isoenzyme of horse liver aldehyde dehydrogenase. Relationship to the corresponding human isoenzyme. Eur J Biochem. 1984 May 15;141(1):37–42. doi: 10.1111/j.1432-1033.1984.tb08152.x. [DOI] [PubMed] [Google Scholar]