Abstract
Seasonal reproduction in many organisms requires detection of day length. This is achieved by integrating information on the light environment with an internal photoperiodic time‐keeping mechanism. Arabidopsis thaliana promotes flowering in response to long days (LDs), and CONSTANS (CO) transcription factor represents a photoperiodic timer whose stability is higher when plants are exposed to light under LDs. Here, we show that PSEUDO RESPONSE REGULATOR (PRR) proteins directly mediate this stabilization. PRRs interact with and stabilize CO at specific times during the day, thereby mediating its accumulation under LDs. PRR‐mediated stabilization increases binding of CO to the promoter of FLOWERING LOCUS T (FT), leading to enhanced FT transcription and early flowering under these conditions. PRRs were previously reported to contribute to timekeeping by regulating CO transcription through their roles in the circadian clock. We propose an additional role for PRRs in which they act upon CO protein to promote flowering, directly coupling information on light exposure to the timekeeper and allowing recognition of LDs.
Keywords: Arabidopsis, circadian clock, CONSTITUTIVE PHOTOMORPHOGENIC 1, photoperiodic flowering, PSEUDO RESPONSE REGULATOR
Subject Categories: Plant Biology; Post-translational Modifications, Proteolysis & Proteomics; Transcription
Introduction
Many organisms recognize seasonal changes in their environment by perceiving day length and utilize this information to control key steps in their life cycle, such as the onset of reproduction or diapause. These responses, referred to as photoperiodism, allow organisms to adapt to high latitude where seasonal climatic fluctuations involving significant temperature changes occur. Photoperiodic responses are generally conferred by a mechanism that allows organisms to measure the length of day or night that fluctuates predictably during the year. This process involves comparison of the light environment against an internal photoperiodic time‐keeping mechanism downstream of the circadian clock. In this system, the circadian clock provides an endogenous autonomous rhythm with a period length of ~24 h that regulates the photoperiodic timer.
Day length strongly influences the timing of floral initiation in many plants, allowing them to adapt to higher latitude and enabling successful reproduction. Molecular and genetic studies using the model species Arabidopsis thaliana, which flowers specifically in response to photoperiod under long days (LDs) of spring, provided knowledge on the mechanistic framework that couples information on light exposure with the photoperiodic time‐keeping mechanism and enables measurement of day length. The CONSTANS (CO) gene was originally isolated as a photoperiodic floral promoter and defined as the integrator of light and timing information. This is achieved through control of its transcription by the circadian clock and regulation of CO protein stability by light exposure (Andres & Coupland, 2012; Song et al, 2014). Accumulation of CO transcripts therefore forms a diurnal rhythm for timekeeping under LDs and SDs (Suarez‐Lopez et al, 2001). Under LDs CO transcription is controlled by a clock‐controlled blue light photoreceptor FLAVIN‐BINDING, KELCH REPEAT, F‐BOX1 (FKF1), clock‐controlled CYCLING DOF FACTOR (CDFs) transcription factors, and a clock protein GIGANTEA (GI). In this mechanism, a FKF1‐GI protein complex temporally promotes transcription of CO by binding to and initiating degradation of CDFs, suppressors of CO transcription, in a light‐dependent manner (Imaizumi et al, 2003, 2005; Sawa et al, 2007; Fornara et al, 2009). Through this regulation, transcripts of CO accumulate in the afternoon under LDs when plants are exposed to light. This coincidence between CO mRNA and light exposure allows stabilization of translated CO protein at these times, causing CO to accumulate under LDs and achieving recognition of LDs (Appendix Fig S1; Valverde et al, 2004). CO then directly promotes floral transition through its capacity to activate transcription of the “florigen” gene FLOWERING LOCUS T (FT) specifically under LDs (Andres & Coupland, 2012; Song et al, 2014). By contrast, under SDs, CO transcription occurs only in the dark, and under these conditions, CO protein does not accumulate (Valverde et al, 2004).
In contrast to current knowledge of the time‐keeping mechanism associated with patterns of CO mRNA accumulation, how information after light exposure is transferred to CO protein is still unclear. The blue light photoreceptors cryptochrome 1 (cry1) and cryptochrome 2 (cry2) and the far‐red photoreceptor phytochrome A (phyA) are required to stabilize CO protein (Valverde et al, 2004; Zuo et al, 2011). A protein complex of an E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and SUPPRESSOR OF PHYA1 (SPA1), which are repressors of A. thaliana photomorphogenesis, mediates between the photoreceptors and CO protein stabilization (Laubinger et al, 2006; Jang et al, 2008; Liu et al, 2008; Zuo et al, 2011; Lau & Deng, 2012; Sarid‐Krebs et al, 2015). During the night, the COP1/SPA1 protein complex physically interacts with CO in the nuclei to promote its proteasomal degradation, whereas during the day, the phyA and cry photoreceptors promote COP1 accumulation in the cytoplasm thus allowing CO to accumulate in the nucleus (Osterlund & Deng, 1998; Laubinger et al, 2006; Jang et al, 2008; Liu et al, 2008; Zuo et al, 2011; Sarid‐Krebs et al, 2015). Since the COP1/SPA1 complex degrades CO protein during the night, it strongly reduces CO accumulation under SDs (Jang et al, 2008; Liu et al, 2008). However, in contrast to its clear biological function in the perception of SDs, COP1 also interferes with the recognition of LDs where a residual activity in the light further reduces CO protein levels during the day, perhaps because photoreceptor activity under LDs is insufficient to completely suppress COP1 function (Jang et al, 2008). FKF1 might overcome this effect because it has been demonstrated that FKF1 stabilizes CO under LDs (Song et al, 2012). However, activity of FKF1 is strictly limited to the afternoon, whereas CO stabilization occurs at other times during the light period, including the morning (Valverde et al, 2004; Song et al, 2012). Finally, the ubiquitin ligase HOS1 and the APETALA2 (AP2) family protein TARGET OF EAT1 (TOE1), GI, and an E3 ubiquitin ligase ZEITLUPE (ZTL) negatively regulate CO protein abundance, although how their activities are related to light signaling during photoperiodic flowering has not been elucidated (Lazaro et al, 2012, 2015; Song et al, 2014; Zhang et al, 2015). Thus, the mechanisms that ensure CO accumulation in response to light exposure to establish LD recognition and floral transition are not fully understood.
In parallel to the studies of day‐length measurement in A. thaliana, quantitative trait locus (QTL) analyses of flowering time identified PSEUDO RESPONSE REGULATOR (PRR) genes in a wide range of crop species. Ppd‐H1, Ppd‐1, BOLTING TIME CONTROL 1 (BvBTC1), PRR37, and SbPRR37 encode PRR proteins in barley, wheat, beet, rice, and sorghum, respectively, and allelic variation at these genes confers natural diversity in flowering time (Turner et al, 2005; Pin et al, 2009; Murphy et al, 2011; Campoli et al, 2012; Shaw et al, 2012; Koo et al, 2013). In A. thaliana, PRR proteins are encoded by a family of five genes and are defined as central components of the circadian clock (TIMING OF CAB EXPRESSION 1 (TOC1), PRR3, PRR5, PRR7, and PRR9) (Strayer et al, 2000; Ito et al, 2003; Yamamoto et al, 2003; Murakami et al, 2004; Para et al, 2007). The abundance of their transcripts and proteins exhibit circadian rhythms, and under diurnal conditions, they peak in expression sequentially at 2–3 h intervals during the light period in the order PRR9, PRR7, PRR5, PRR3, and TOC1 (Matsushika et al, 2000; Fujiwara et al, 2008). PRR9, PRR7, PRR5, and TOC1 proteins are degraded during the night, so they mainly accumulate during the day when they repress transcription of genes encoding other clock components such as LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) (Mas et al, 2003b; Farré & Kay, 2007; Ito et al, 2007; Kiba et al, 2007; Nakamichi et al, 2010; Huang et al, 2012). Mutations in the PRR genes also delay flowering under LDs (Nakamichi et al, 2005; Ito et al, 2008), but double or triple mutants exhibit much stronger phenotypes suggesting functional redundancy between the genes (Nakamichi et al, 2005, p. 549; Ito et al, 2008). Despite the significance of the PRRs in flowering, how they contribute to this process remains unclear. In A. thaliana, they appear to influence the time‐keeping mechanism associated with CO transcription in the photoperiodic flowering pathway indirectly through their role in the circadian clock (Nakamichi et al, 2007; Ito et al, 2008). On the other hand, in crops such as barley and rice, their effects on CO transcription are relatively weak, whereas they strongly affect FT transcription (Turner et al, 2005; Campoli et al, 2012; Koo et al, 2013). Therefore, these PRRs are likely to have a more direct effect on the regulation of the photoperiodic flowering pathway.
Here, we identify novel mechanisms contributing to day‐length measurement and define unexpectedly direct roles for PRRs in photoperiodic flowering. We show that PRRs convey information on light exposure to the photoperiodic time‐keeping mechanism by interacting with and stabilizing CO during the day, thereby allowing CO to accumulate at higher levels under LDs. In A. thaliana, diversity in temporal accumulation patterns among PRRs throughout the day contributes to detection of long‐light periods and to CO accumulation in the morning and the evening specifically under LDs. Besides the function of PRRs in controlling the activity of the photoperiodic time‐keeping mechanism associated with CO transcription through their role the circadian clock, we find unexpectedly that PRRs also transfer information on light exposure and day length to CO at the post‐translational level. This level of regulation allows CO to optimally respond to LDs and accelerates the floral transition under these conditions.
Results
PRRs induce FT transcription independently of CO transcription
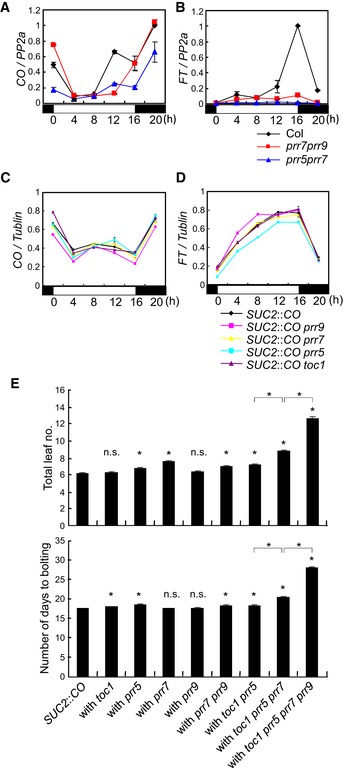
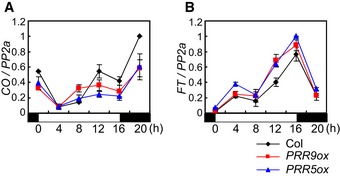
In A. thaliana toc1 prr5, prr5 prr7, and prr7 prr9 double mutants, the levels of CO mRNA are slightly reduced, contributing to late flowering (Nakamichi et al, 2007; Ito et al, 2008). However, in these mutants, transcript levels of FLOWERING LOCUS T (FT), which is a direct target of CO and encodes a florigen protein (Tiwari et al, 2010; Andres & Coupland, 2012; Song et al, 2012), are drastically reduced. We confirmed the effects of prr5 prr7 and prr7 prr9 mutations on CO and FT mRNAs under LD. CO mRNA in these mutants was lower than in wild‐type (WT) plants but still accumulated at some times, particularly 12 and 16 h after dawn, whereas FT mRNA was strongly reduced throughout the day (Fig EV1A and B). The effects of overexpression of PRR5 and PRR9 on CO and FT mRNA levels under LD were then tested. The abundance of CO mRNA was slightly lower in the overexpression plants compared to WT, especially at 12 and 16 h after dawn, whereas FT mRNA levels were inversely related to CO mRNA and elevated at these times (Fig 1A and B). These results are consistent with a hypothesis that PRRs upregulate FT mRNA level independently from affecting CO mRNA levels.
Figure EV1. Effects of prr mutations on FT expression and flowering time in WT and SUC2::CO backgrounds.
-
A, BExpression pattern of CO (A) and FT (B) mRNA in prr5 prr7 or prr7 prr9 double mutants under LD. Error bars indicate standard error within two biological replicates. In each replicate, two technical replicates were performed.
-
C, DEffect of single prr mutations on CO (C) and FT (D) mRNA accumulation in SUC2::CO plants under LD. Error bars indicate standard error within two technical replicates.
-
ETotal leaf number and bolting time of prr mutants in the SUC2::CO background grown under LDs. Approximately 40 plants of each genotype were grown under LDs. Error bars indicate standard error. Statistical significance between SUC2::CO and each prr mutant, and among multiple prr mutants was calculated using Student's t‐test; *P < 0.01; n.s. P > 0.01.
Figure 1. PRRs induce FT transcription independently from regulating CO transcription.
-
A, BExpression pattern of CO (A) and FT (B) mRNA in PRR5‐ or PRR9‐overexpressors under LD. The values of CO and FT mRNA levels were normalized to those of PP2a. For all data, error bars indicate standard error within two biological replicates and in each of these two technical replicates were performed.
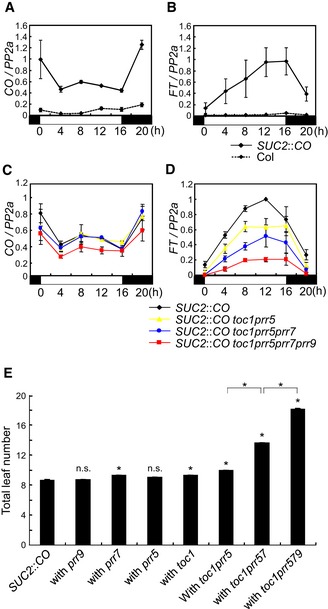
To further analyze the role of PRRs in FT transcription, the effects of prr mutations on FT expression were tested in a transgenic line expressing CO from the SUC2 promoter (An et al, 2004), thereby uncoupling CO transcription from impaired clock function in prr mutants. The SUC2 promoter expresses CO specifically in the phloem companion cells where it acts in WT plants to promote FT transcription (An et al, 2004). In these plants, FT expression was highly induced compared with WT due to increased accumulation of CO mRNA specifically in the phloem cells (Fig 2A and B). The prr mutations were introduced singly and in combination into SUC2::CO plants. Single prr5, prr7, prr9, or toc1 mutations in SUC2::CO background did not dramatically affect CO or FT expression (Fig EV1C and D). By contrast, combining prr mutations in SUC2::CO reduced FT mRNA levels without significantly affecting CO mRNA (Fig 2C and D). In toc1 prr5 SUC2::CO, the levels of FT mRNA were slightly reduced compared to SUC2::CO. In toc1 prr5 prr7 SUC2::CO plants, FT mRNA was further reduced, and this effect was even more enhanced in toc1 prr5 prr7 prr9 SUC2::CO plants (Fig 2C and D). The flowering times of these lines were also measured, and the toc1 prr5 prr7 SUC2::CO as well as the toc1 prr5 prr7 prr9 SUC2::CO line exhibited late‐flowering phenotypes compared to SUC2::CO, consistent with their reduced FT mRNA levels (Figs 2E and EV1E). Although FT mRNA levels were reduced in toc1 prr5 SUC2::CO plants compared to SUC2::CO, the mutant line was only mildly later flowering, presumably because FT levels are still so high in toc1 prr5 SUC2::CO that their effect on flowering is close to saturation. These results demonstrate that PRR proteins promote FT transcription independently from CO transcription.
Figure 2. PRR genes contribute to SUC2::CO‐mediated FT transcription.
-
A, BComparison of CO (A) and FT (B) mRNA levels between WT and SUC2::CO.
-
C, DEffect of double, triple, and quadruple prr mutations on CO (C) and FT (D) mRNA in SUC2::CO background under LDs. The values of CO and FT mRNA levels were normalized to those of PP2a.
-
EEffect of prr mutations on flowering time in SUC2::CO background under LDs. For each line, 16 plants were used for flowering‐time measurement. Error bars indicate standard error. Statistical significance between SUC2::CO and each prr mutant, and among multiple prr mutants was calculated using Student's t‐test; *P < 0.01; n.s. P > 0.01.
PRRs stabilize CO protein under LD
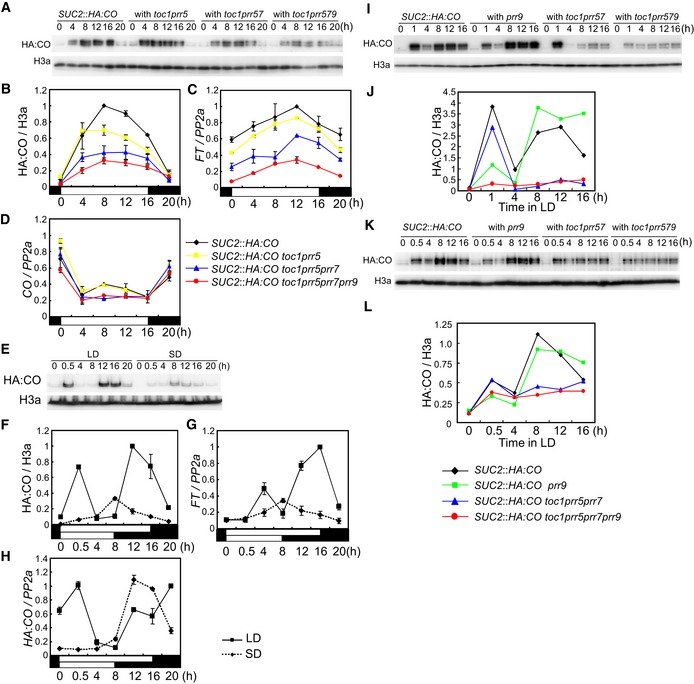
Based on the observation that prr mutations can reduce FT transcript levels independently from CO mRNA, we hypothesized that the prr mutations act in part at the post‐transcriptional level to reduce CO protein abundance. To test this idea, the prr mutations were introduced into SUC2::HA:CO, which allowed us to monitor HA:CO protein abundance in the prr mutants. In SUC2::HA:CO under LD, HA:CO protein accumulated during the day, but its levels were rapidly decreased in the night, although CO mRNA abundance remained high at this time (Fig 3A, B and D). This result is consistent with previous data demonstrating that in 35S::CO plants, where CO mRNA is constantly expressed, CO protein accumulates in the day and rapidly disappears in the night, due to light‐mediated stabilization of CO protein (Valverde et al, 2004; Song et al, 2012). During the light period under LD, HA:CO levels in toc1 prr5 SUC2::HA:CO were slightly reduced compared to SUC2::HA:CO and a further reduction of HA:CO protein was observed in toc1 prr5 prr7 SUC2::HA:CO and even more dramatically in toc1 prr5 prr7 prr9 SUC2::HA:CO mutants (Fig 3A and B). In these lines, FT mRNA levels were reduced and late flowering was also observed, consistent with the reduced HA:CO protein levels (Figs 3C and EV2F). CO mRNA levels were not significantly affected in these lines (Fig 3D). Taken together, these data indicate that the PRRs act redundantly to enhance CO protein accumulation under LDs.
Figure 3. PRR genes contribute to CO protein accumulation under LD.
-
A, BEffect of double, triple, and quadruple prr mutations on HA:CO protein accumulation in SUC2::HA:CO under LD.
-
C, DFT (C) and CO (D) mRNA expression under the same conditions as in (A, B).
-
E, FExpression pattern of HA:CO protein in pCO::HA:CO under LD and SD.
-
G, HFT (G) and HA:CO (H) mRNA expression in pCO::HA:CO under LD and SD.
-
I–LEffect of prr9 and toc1 prr5 prr7 mutations on HA:CO protein accumulation in SUC2::HA:CO background under LD. Samples collected early in the day were harvested at Zeitgeber time (ZT) 1 for (I and J) and at ZT 0.5 for (K and L).
Figure EV2. Stabilization and accumulation of HA:CO through PRR‐mediated inhibition of its proteasomal degradation, and its effect on flowering time.
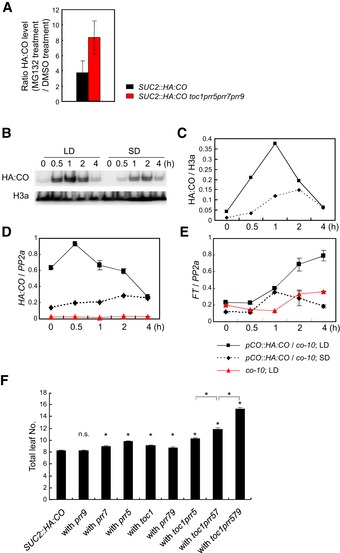
-
AEffect of MG132 treatment on recovery of HA:CO protein accumulation in toc1 prr5 prr7 prr9 SUC2::HA:CO. 10‐day‐old SUC2::HA:CO and toc1 prr5 prr7 prr9 SUC2::HA:CO were treated with either MG132 or DMSO at ZT5 and harvested at ZT8 under LD. The levels of HA:CO were normalized to those of H3a. The ratio of the values after MG132 treatment to those after DMSO treatment was calculated in each line, and fold change in HA:CO accumulation by the MG132 treatment was measured. Error bars indicate standard error within two biological replicates. In each replicate, two technical replicates were performed.
-
B, CAccumulation of HA:CO protein in pCO::HA:CO in the morning under LD and SD.
-
DAccumulation of HA:CO mRNA in pCO::HA:CO in the morning under LD and SD. Error bars indicate standard error within two technical replicates.
-
EAccumulation of FT mRNA in pCO::HA:CO in the morning under LD and SD, and FT mRNA accumulation in the morning requires CO. Error bars indicate standard error within two technical replicates.
-
FTotal leaf number of prr mutants in the SUC2::HA:CO background under LDs. 16 plants of each genotype were grown under LDs. Error bars indicate standard error. Statistical significance between SUC2::HA:CO and each prr mutant, and among multiple prr mutants was calculated using Student's t‐test; *P < 0.01; n.s. P > 0.01.
When CO mRNA is expressed constitutively from the 35S promoter under LDs, CO protein peaks in abundance early in the morning and again in the afternoon (Valverde et al, 2004; Song et al, 2012). We tested this pattern in pCO::HA:CO lines in which HA:CO is expressed from the endogenous CO promoter. As in the 35S lines, the levels of HA:CO protein increased early in the morning and in the afternoon, and these peaks appeared more strongly under LD than SD (Figs 3E, F and H, and EV2B–D). The diurnal accumulation of FT mRNA was also similar to that of HA:CO protein (Figs 3G and EV2E), suggesting that both the morning and the evening peak of CO contribute to LD‐induction of flowering. In a co mutant induction of FT in the morning was impaired, indicating that FT transcription in the morning depends on CO activity (Fig EV2E; Kim et al, 2005). Next, we analyzed the temporal HA:CO protein accumulation in single and higher order prr mutants at different times during the day and compared those to the WT. In agreement with previous reports on PRR9 function in the morning, HA:CO accumulation was specifically reduced in the morning in prr9 SUC2::HA:CO plants (Figs 3I–L and EV3A), whereas the toc1 prr5 prr7 mutations strongly reduced the evening peak under LD (Fig 3I–L; Fujiwara et al, 2008). These data indicate that diversity in the timing of expression and activity of PRRs cause CO to accumulate at specific times during the day to generate the typical LD‐specific accumulation pattern. Previously, FKF1 was reported to stabilize CO protein specifically in the evening in 35S::CO plants (Song et al, 2012). However, in contrast to the dramatic reduction of HA:CO levels in toc1 prr5 prr7 prr9 SUC2::HA:CO, the fkf1 mutation did not strongly affect HA:CO levels in the same SUC2::HA:CO line (Fig EV3C–E). This result suggests that PRRs are more heavily involved than FKF1 in increasing CO protein levels when it is expressed from the SUC2 promoter in the phloem companion cells, the tissue in which CO promotes floral induction.
Figure EV3. Effect of prr9 and fkf1 mutations on HA:CO accumulation.
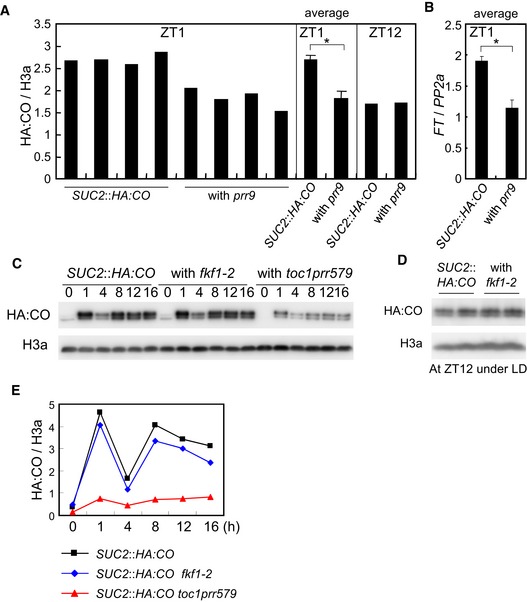
-
A, BAccumulation of HA:CO (A) and FT mRNA (B) in SUC2::HA:CO and prr9 SUC2::HA:CO in the morning. Each genotype was grown in four independent agar plates and harvested at ZT1 under LD. The levels of HA:CO and FT mRNA were analyzed in these samples. Error bars indicate standard error within these samples. Statistical significance was calculated using Student's t‐test, *P < 0.01. As the control, HA:CO levels among those lines grown in one plate for each and harvested at ZT12 is shown.
-
C–EEffect of the fkf1‐2 mutation on HA:CO accumulation in SUC2::HA:CO under LD. In (D), each genotype was grown on two independent agar plates and analyzed.
PRRs contribute to light‐mediated accumulation of CO
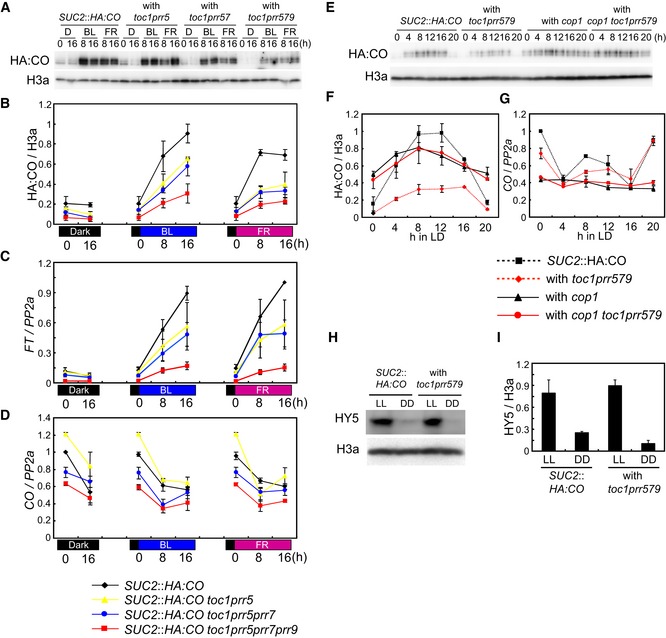
PRR proteins accumulate during the day and are degraded during the night (Mas et al, 2003b; Farré & Kay, 2007; Ito et al, 2007; Kiba et al, 2007; Fujiwara et al, 2008), so they could directly contribute to CO protein stabilization in light. CO is also known to be stabilized in plants exposed to blue light (BL) and far‐red light (FR) (Valverde et al, 2004). To understand whether CO protein abundance under different light regimes is dependent on PRR function, HA:CO protein accumulation was followed under BL and FR in SUC2::HA:CO plants and after introduction of prr mutations. In SUC2::HA:CO plants, the levels of HA:CO protein were low in the dark, but increased strongly in BL or FR independently of CO mRNA abundance (Fig 4A and B). The toc1 prr5 prr7 SUC2::HA:CO and toc1 prr5 prr7 prr9 SUC2::HA:CO lines showed strongly reduced HA:CO protein levels in both BL and FR when compared to SUC2::HA:CO (Fig 4A and B). Moreover, FT mRNA levels under both conditions correlated with the abundance of HA:CO protein, but not with CO mRNA (Fig 4C and D). Together with the observations that PRR proteins accumulate during the day, these results support the hypothesis that the PRR proteins contribute to light‐mediated accumulation of CO.
Figure 4. PRR genes are required for BL‐ and FR‐mediated CO protein accumulation.
-
A, BEffect of double, triple, and quadruple prr mutations on HA:CO protein accumulation in blue light (BL) and far‐red light (FR). Plants were grown under LDs and then transferred to darkness for 24 h. The plants were then transferred to continuous BL or FR at ZT 0. A population of plants was kept in darkness (D) as control. HA:CO protein was analyzed in each genotype at the illustrated times under BL, FR, or control D treatment.
-
CFT mRNA accumulation under the same conditions as in (A, B).
-
DCO mRNA accumulation under the same conditions as in (A, B).
-
E–GPRRs suppress ability of COP1 to degrade CO. (E, F) Effect of quadruple prr, cop1, and quintuple prr/cop1 mutations on CO protein abundance in SUC2::HA:CO background through a time course under LDs. (G) CO mRNA levels in genotypes used in (F).
-
H, IAbundance of HY5 protein in prr quadruple mutant in continuous light (LL) and continuous dark (DD). Plants were grown under LDs for 7 days, transferred to LL or DD at ZT 0 and kept for 40 h before harvesting.
In A. thaliana light‐signaling pathways, COP1 acts as a negative regulator to suppress photomorphogenesis by targeting proteins such as the transcription factor ELONGATED HYPOCOTYL 5 (HY5) for proteasomal degradation during the dark (Osterlund et al, 2000). In the light, phys and crys suppress COP1 function, allowing HY5 to accumulate and promote photomorphogenesis (Osterlund et al, 2000). The photoreceptor‐COP1 module also controls CO protein abundance, where phyA, cry1, and cry2 suppress the activity of COP1 to degrade CO thereby allowing CO to accumulate during the day (Zuo et al, 2011; Sarid‐Krebs et al, 2015). As PRRs stabilize CO protein during the day, we tested whether they might also act by suppressing COP1 activity at this time. HA:CO protein abundance was therefore compared in the toc1 prr5 prr7 prr9 SUC2::HA:CO line and a cop1 toc1 prr5 prr7 prr9 SUC2::HA:CO line that we constructed. In agreement with our previous observation, HA:CO protein levels were strongly reduced in the toc1 prr5 prr7 prr9 SUC2::HA:CO line but were increased by introduction of the cop1 mutation in the cop1 toc1 prr5 prr7 prr9 SUC2::HA:CO line (Fig 4E and F). Interestingly, the quintuple mutant cop1 toc1 prr5 prr7 prr9 SUC2::HA:CO contained similar amounts of HA:CO protein to cop1 SUC2::HA:CO controls (Fig 4E and F), demonstrating that the cop1 mutation is epistatic to the prr mutations with respect to CO protein abundance. Also, the higher abundance of HA:CO protein in the presence of the cop1 mutation could not be explained by increased levels of CO mRNA (Fig 4G). These results demonstrate that reduced CO abundance in prr mutants requires COP1 activity, and suggest that PRRs stabilize CO by suppressing COP1‐mediated degradation of CO in the light. We also tested whether PRRs contribute to HA:CO accumulation by suppressing its proteasome‐mediated degradation. SUC2::HA:CO and toc1 prr5 prr7 prr9 SUC2::HA:CO plants were treated with the proteasome inhibitor MG132, and the increase in HA:CO accumulation after MG132 treatment was compared between genotypes. In the toc1 prr5 prr7 prr9 SUC2::HA:CO line, HA:CO levels increased to a greater extent after MG132 treatment than in SUC2::HA:CO plants (Fig EV2A), supporting the idea that PRRs suppress proteasomal degradation of CO.
The abundance of HY5 protein was then compared in SUC2::HA:CO and toc1 prr5 prr7 prr9 SUC2::HA:CO plants, because if the PRRs generally suppress COP1 activity then in the prr quadruple mutant background the level of HY5 should be reduced due to increased COP1 function. Consistent with this hypothesis, impairing the function of several PRRs causes a long‐hypocotyl phenotype in A. thaliana similar to that of hy5 mutants (Fig EV4A; Yamashino et al, 2008). Unexpectedly, however, the levels of HY5 protein were not reduced in the light in the toc1 prr5 prr7 prr9 SUC2::HA:CO quadruple mutant (Fig 4H and I), although under the same conditions the levels of HA:CO protein were decreased. This result suggests that PRRs do not regulate all of the activities of COP1, but that they interfere more specifically with its function to target CO for proteasomal degradation. Furthermore, we also found that the long‐hypocotyl phenotype of toc1 prr5 prr7 prr9 is observed under LDs and SDs but not in continuous light (Fig EV4A), although COP1 overexpression was reported to cause a long hypocotyl even under this condition (Torii et al, 1998). These data also suggest that the reduction of CO protein and the long‐hypocotyl phenotype in the prr quadruple mutant are not caused by a general increase in COP1 activity. The long hypocotyl of the prr quadruple mutant may rather be due to impaired circadian‐clock function, causing hyper‐accumulation of transcripts of a clock‐controlled gene, such as PHYTOCHROME INTERACTING FACTOR 4 (PIF4), whose mRNA was increased in abundance in this background and encodes a transcription factor involved in promotion of hypocotyl growth in shade and dark (Fig EV4B; Franklin, 2008; Nakamichi et al, 2009). Accumulation of PIF4 protein requires shade or darkness (Nozue et al, 2007; Leivar et al, 2008; Lorrain et al, 2008), consistent with our data that the long‐hypocotyl phenotype in the prr mutant was observed only under LD and SD conditions, which include periods of darkness.
Figure EV4. Hypocotyl growth in the toc1 prr5 prr7 prr9 mutant under various day lengths and PIF4 mRNA level in the same background under LD.
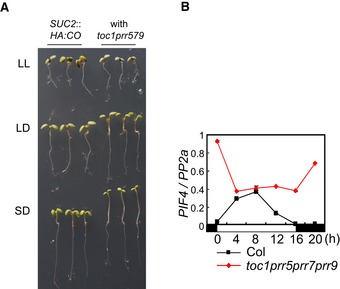
- The prr quadruple mutant exhibits long‐hypocotyl phenotypes under SDs and LDs but not in LL. Plants were grown on agar plates without sucrose for 7 days under LDs, SDs, or LL.
- Transcripts of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) highly accumulate in the prr quadruple mutant. Col and toc1 prr5 prr7 prr9 were grown under LDs. Error bars indicate standard error within two technical replicates.
PRRs stabilize CO independently from altering expression of other clock‐controlled genes
Circadian‐clock function is strongly impaired in the triple mutant prr5 prr7 prr9 (Nakamichi et al, 2005), suggesting that the observed effect of the quadruple toc1 prr5 prr7 prr9 mutant on HA:CO protein level could be due to altered expression of other clock‐regulated genes that affect CO accumulation. For example, the mRNA level of the clock‐controlled gene FKF1 was reduced in the quadruple mutant (Appendix Fig S2C). However, the abundance of HA:CO protein in fkf1 SUC2::HA:CO plants was similar to SUC2::HA:CO and clearly more than toc1 prr5 prr7 prr9 SUC2::HA:CO (Fig EV3C–E), indicating that the reduced HA:CO protein level in toc1 prr5 prr7 prr9 SUC2::HA:CO plants is not explained by reduced FKF1 activity. Moreover, in contrast to the late‐flowering phenotype of toc1 prr5 prr7 prr9 SUC2::HA:CO plants, the fkf1 SUC2::HA:CO line flowered at the same time as SUC2::HA:CO (Appendix Fig S2F). Also, although fkf1 mutation was reported not to affect the morning peak of HA:CO protein (Song et al, 2012), combining the four prr mutations greatly reduced CO protein and FT mRNA abundance in the morning (Fig 3I–L and Appendix Fig S2A, B, D and E). Finally, in the prr9 mutant where the circadian clock is not strongly affected in light/dark cycles (Appendix Fig S2G; Ito et al, 2003; Farré et al, 2005), HA:CO protein abundance was strongly reduced in the morning when PRR9 is expressed in WT plants (Figs 3I–L and EV3A). Another clock‐controlled gene TOE1 was reported to reduce CO protein abundance under LD (Zhang et al, 2015), and we found that its mRNA levels were increased in toc1 prr5 prr7 prr9 SUC2::HA:CO, suggesting that this might contribute to the observed reduction in CO protein abundance (Appendix Fig S2H and I). However, in prr9 SUC2::HA:CO where CO protein level is reduced specifically in the morning, TOE1 mRNA level was not increased at this time (Appendix Fig S2H and I). Its mRNA level was, however, increased in toc1 prr5 prr7 SUC2::HA:CO in the morning where these prr mutations do not affect the abundance of CO protein (Appendix Fig S2H and I). These results indicate that changes in TOE1 mRNA levels in the prr mutants are not linked to CO protein levels. Taken together, we conclude that the reduction of CO protein levels in prr mutants is not an indirect effect of altered expression of clock‐output genes such as FKF1 or TOE1, but rather reflects a more direct effect of PRRs on CO.
PRR proteins interact with CO
The above data support a direct effect of PRRs on HA:CO protein stabilization and therefore suggest that PRRs might physically interact with CO. To test this possibility fluorescence resonance energy transfer (FRET) and in vivo co‐immunoprecipitation assays were performed. Plasmids carrying 35S::CO:YFP and 35S::PRR:CFP were transfected into A. thaliana protoplasts, and FRET between YFP and CFP was measured through acceptor photobleaching. All PRR:CFP proteins were localized in nuclei, and they were also co‐localized with CO:YFP (Fig 5A). In the FRET assay, high FRET signals between CO:YFP and each PRR:CFP protein were detected (Figs 5A and B, and EV5B). As negative controls, plasmids containing transgenes that express CFP or YFP protein fused to AGAMOUS LIKE 16 (AGL16), a member of the MADS‐box transcription factor family that was reported to localize to the nucleus, were transfected into protoplasts (Parenicova et al, 2003; Hu et al, 2014). Significantly lower FRET signals were observed between each PRR:CFP and AGL16:YFP or between AGL16:CFP and CO:YFP than between PRR:CFP and CO:YFP (Figs 5B and EV5B). The interaction between PRRs and CO was also tested in Nicotiana benthamiana leaves. Agrobacterium strains carrying 35S::PRR:CFP and 35S::CO:YFP constructs were infiltrated into N. benthamiana leaves and FRET between CFP and YFP measured. High FRET signals between CO:YFP and each PRR:CFP protein were detected (Fig EV5C and D). By contrast, significantly lower FRET signals were observed between each PRR:CFP and YFP alone, or between CFP alone and CO:YFP (Fig EV5D). Results obtained by the FRET analyses were then further tested by performing in vivo co‐immunoprecipitation assays. Thus, PRR:GFP and HA:CO proteins were transiently expressed in N. benthamiana leaves and co‐immunoprecipitation performed. Agrobacterium strains carrying 35S::PRR:GFP and 35S::CO:HA constructs were infiltrated into tobacco leaves. PRR:GFP was immunoprecipitated with an antibody against GFP and for each PRR a HA:CO co‐immunoprecipitation could be observed (Fig 5C). By contrast, HA:CO was not precipitated by the GFP antibody when 35S:HA:CO was infiltrated alone or with 35S::TRB3:YFP, which expresses a fusion of TELOMERE REPEAT BINDING 3 fused to YFP (Figs 5C and EV5A; Schrumpfova et al, 2014; Zhou et al, 2016). In summary, these results indicate that each PRR can physically interact with CO in planta.
Figure 5. PRR proteins physically interact with CO protein in planta .
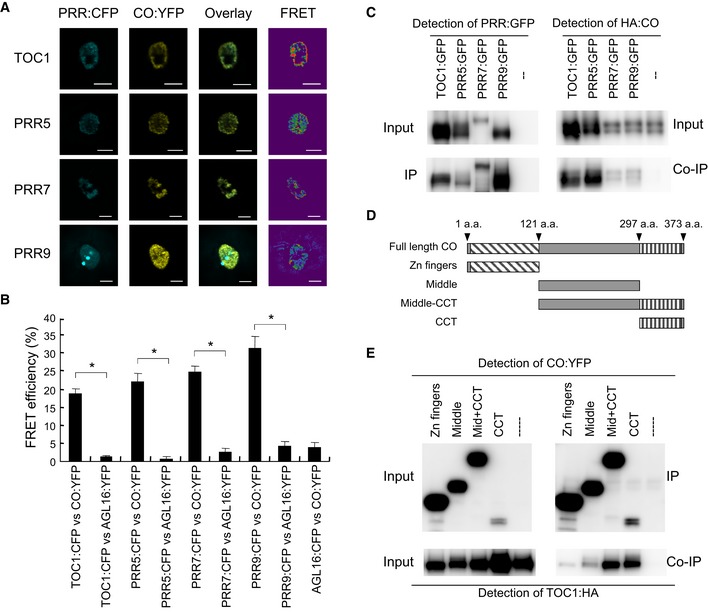
-
A, BFluorescence resonance energy transfer (FRET) analysis between PRR:CFP and CO:YFP. PRR:CFP and CO:YFP were co‐expressed in A. thaliana protoplasts, and FRET analyses were performed with 20 cells. Statistical significance was calculated using Student's t‐test. *P < 0.01. Error bars indicate standard error within the 20 samples. Scale bars indicate 10 μm.
-
CIn vivo immunoprecipitation assay with PRR:GFP and CO:HA protein. PRR:GFP and CO:HA were co‐expressed in N. benthamiana, and co‐immunoprecipitation assays performed.
-
DThe schematic diagram of truncated CO proteins used for co‐IP in (E).
-
ETOC1 preferentially binds to the CCT domain in CO. TOC1:HA was co‐infiltrated with YFP fused to truncated versions of CO protein shown in (D) in N. benthamiana, and co‐immunoprecipitation was performed.
Figure EV5. PRRs interact with CO in vivo .
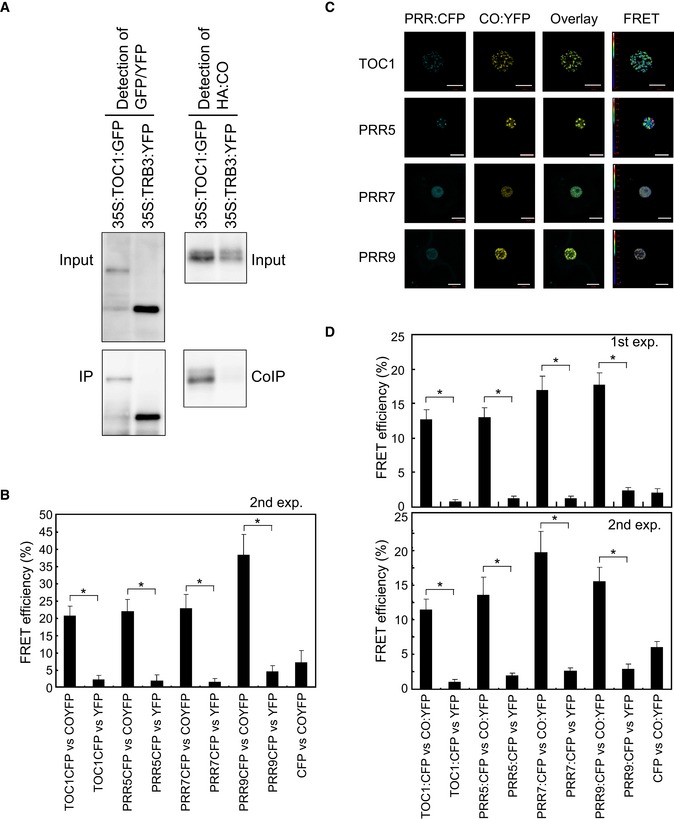
-
ACo‐IP experiment in N. benthamiana, TRB3 protein was used as the negative control in the experiment shown in Fig 5. Agrobacterium carrying either 35S::TOC1:GFP or 35S::TRB3:YFP were co‐infiltrated with that carrying 35S::HA:CO in N. benthamiana leaves. TOC1:GFP and TRB3:YFP proteins were immunoprecipitated with the GFP antibody, and co‐immunoprecipitation of HA:CO protein was checked.
-
BFRET between PRR:CFP and CO:YFP in A. thaliana protoplasts. This is a second experiment of the FRET shown in Fig 5A and B. For each construct combination, FRET in 10 protoplasts was analyzed.
-
C, DFRET analysis between PRR:CFP and CO:YFP. PRR:CFP and CO:YFP were co‐expressed in N. benthamiana by Agroinfiltration, and FRET analyses were performed. FRET in 30 cells for each construct combination was analyzed for the first experiment. For the second experiment, 15 cells for each combination were analyzed. Scale bars indicate 10 μm.
To determine the domain that mediates the physical interaction between CO and the PRRs, a series of in vivo immunoprecipitation assays were performed on truncated variants of CO fused to YFP, which were co‐expressed with TOC1:HA in N. benthamiana (Fig 5D and E). TOC1:HA was most strongly co‐immunoprecipitated with the YFP fusion protein containing the CCT domain of CO, indicating that TOC1:HA preferentially binds to this region of CO (Fig 5E). This domain was previously reported to be involved in mediating the binding of CO to DNA through its cognate binding site, defined as the CO response element (CORE) in the FT promoter (Tiwari et al, 2010). Moreover, the CCT domain was also defined as the region with which COP1 physically interacts (Jang et al, 2008; Liu et al, 2008).
Stabilization by the PRRs increases CO binding to FT
CO binds to a canonical CORE in the FT promoter, thereby enabling chromatin looping and enhanced FT transcription (Adrian et al, 2010; Tiwari et al, 2010; Song et al, 2012; Cao et al, 2014). Since prr mutations strongly reduced the levels of HA:CO protein and FT transcripts in SUC2::HA:CO plants, we tested whether lower HA:CO protein levels in toc1 prr5 prr7 prr9 SUC2::HA:CO resulted in reduced binding of HA:CO to COREs at FT. To this end, a scan of the sequence of the entire intergenic up‐ and downstream region as well as the coding region of FT identified five canonical COREs. Chromatin immunoprecipitation (ChIP) was then performed in SUC2::HA:CO and toc1 prr5 prr7 prr9 SUC2::HA:CO plants to test binding of HA:CO at each of the 5 putative CO binding sites. In agreement with previous results, HA:CO was found to be strongly enriched at CORE1/2 in SUC2::HA:CO plants, whereas in toc1 prr5 prr7 prr9 SUC2::HA:CO quadruple mutant binding of HA:CO was strongly reduced, demonstrating a correlation between HA:CO protein abundance and its ability to bind to the CORE at FT (Fig 6A and B). Moreover, the ChIP experiment identified a previously unrecognized binding site for CO, CORE3, which is located in the second intron of FT and a reduction in enrichment of HA:CO was also detected at this CORE in toc1 prr5 prr7 prr9 SUC2::HA:CO plants (Fig 6A and B). In addition, ChIP experiments were performed using 35S::TOC1 SUC2::HA:CO plants, and HA:CO was found to be more strongly enriched at the COREs of FT in these plants compared to SUC2::HA:CO plants (Fig EV6). Although the effect of TOC1 overexpression on HA:CO accumulation was not tested, these ChIP experiments are also consistent with our proposal that CO protein levels as determined by PRR activity are related to the amount of CO that is bound to COREs at FT.
Figure 6. Occupancy of CO protein at the FT promoter is reduced in prr quadruple mutants.
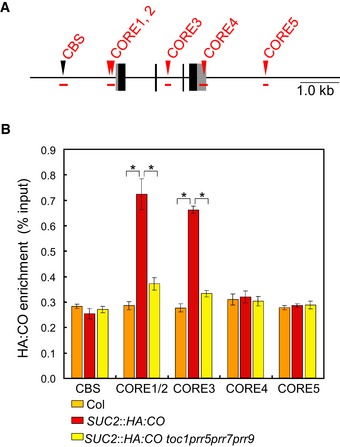
- The location of each DNA fragment in the FT gene tested in ChIP assays. The amplicons are shown as red horizontal bars. The position of each predicted CO response element (CORE) is shown as red triangle.
- ChIP of CORE‐containing amplicons within the FT gene with HA:CO protein. SUC2::HA:CO, toc1 prr5 prr7 prr9 SUC2::HA:CO, and Col were grown under LDs and harvested at ZT12. Enrichment of the CORE segments (CORE1‐5 shown in A) was tested in each line. Error bars indicate standard error within three biological replicates. In each replicate, three technical replicates were performed. Statistical significance was obtained with Student's t‐test, *P < 0.01.
Figure EV6. Overexpression of TOC1 increases binding of CO to FT .
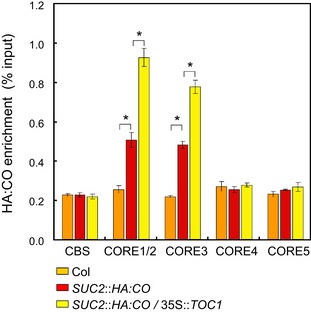
ChIP with 35S::TOC1 SUC2::HA:CO at CORE sequences within the FT gene. The plants were harvested at ZT12 under LD. HA:CO protein was immunoprecipitated from these lines, and the enrichment of the fragments was measured by qPCR. Error bars indicate standard error among three biological replicates. In each replicate, three technical replicates were performed. Statistical significance differences were obtained with Student's t‐test, *P < 0.01.
Discussion
Perception of changes in day length enables plants to precisely determine the timing of the floral transition in their life cycle. In this process, a light‐signaling pathway that transfers information on the light environment to a photoperiodic time‐keeping mechanism is crucial. However, despite its significance, how information on light exposure is conveyed to the timing mechanism is not fully understood. In this study, we demonstrate that in A. thaliana, PRRs enable measurement of LDs by transferring information on light exposure to CO protein. This is achieved through the roles of PRRs in binding and stabilizing CO during the day to allow it to accumulate specifically under LDs. PRR paralogues contribute to detection of long‐light periods through their diverged temporal expression patterns, resulting in CO accumulation in the morning and the evening specifically under LDs and thereby inducing FT transcription at these times. We propose that PRRs stabilize CO protein to promote FT transcription and flowering, conveying information on day length to the photoperiodic time‐keeping mechanism and allowing accurate monitoring of LDs.
Novel roles for PRRs in controlling photoperiodic flowering
PRRs were originally identified as components of the circadian clock in A. thaliana and generally proposed to contribute to day‐length measurement through control of the time‐keeping mechanism associated with CO transcription (Strayer et al, 2000; Yanovsky & Kay, 2002; Ito et al, 2003, 2008; Yamamoto et al, 2003; Murakami et al, 2004; Nakamichi et al, 2007; Para et al, 2007). By contrast, we defined a previously unidentified role of these circadian‐clock components in regulating CO activity, where they interact with and stabilize CO protein during the day, ensuring its accumulation under LDs. Therefore, specific circadian‐clock components not only transfer temporal information to a photoperiodic time‐keeping mechanism but also convey information on light exposure to the time‐keeping mechanism, establishing measurement of day length.
In WT plants, CO mRNA accumulates in the morning and in the evening, coinciding with exposure to light and stabilizing CO protein at these times specifically under LDs. Such a temporal accumulation of CO protein is also observed in 35S::CO or SUC2::HA:CO lines where CO mRNA is expressed at similar levels throughout the day and is not regulated by the circadian clock (Fig 3I–L and Appendix Fig S3A and B; Valverde et al, 2004; Song et al, 2012). Here, several lines of experimental evidence indicate that PRRs are involved in this diurnal accumulation pattern of CO. The quantitative effect of the prr9 mutation on HA:CO accumulation varied among experiments (Figs 3I–L and EV3A), but supported a role for PRR9 in conferring accumulation of CO specifically in the morning. Such a role was also consistent with the reduced FT mRNA levels observed in the morning in prr9 SUC2::HA:CO plants (Fig EV3B) and in the slightly late‐flowering phenotype previously described for prr9 mutants (Nakamichi et al, 2005). On the other hand, TOC1, PRR5, and PRR7 cause accumulation of CO in the evening, thereby illustrating how each PRR protein that acts in the central circadian‐clock mechanism can also directly convey temporal information during the day to the photoperiodic timer CO. These PRR activities coincide with the morning and evening accumulation of CO mRNA in WT plants and enhance CO protein accumulation at these times specifically under LDs allowing CO to bind more strongly to FT (Fig 7). This elegant system directly couples CO stability and function in photoperiodic flowering to cycling circadian‐clock components, ensuring that CO only binds to and activates FT transcription at specific times during the day dependent on day length. Since CO and PRRs contain similar CCT DNA‐binding domains (Matsushika et al, 2000; Gendron et al, 2012), PRRs might also bind along with CO to the FT promoter and contribute to the control of FT transcription, as was recently shown for the interaction between PRRs and PIF transcription factors on their target genes (Soy et al, 2016; Zhu et al, 2016). Morning FT expression is more pronounced when plants are exposed to shade (Wollenberg et al, 2008), suggesting that specific PRRs might also control this process by coupling a distinct flowering signal to CO protein stability or FT induction in the morning.
Figure 7. A model of day‐length recognition through CO and PRRs.
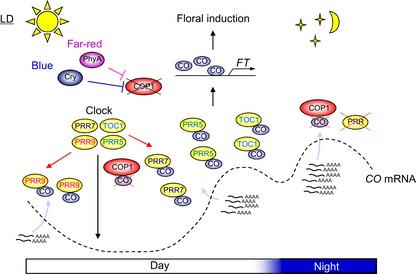
PRRs control activity of the photoperiodic time‐keeping mechanism through regulation of temporal abundance of CO mRNA under LDs and SDs. Under LDs, accumulation of CO transcripts coincides with exposure to light and PRRs function to stabilize CO protein during the day, thereby allowing it to accumulate under LDs and establishing recognition of LDs. In this mechanism, PRR9 mediates accumulation of CO protein in the morning, whereas TOC1, PRR5, and PRR7 mediate its accumulation in the evening. The mechanism of CO stabilization by PRRs is associated with their activities to suppress COP1 function to degrade CO. During the night under LDs, these stabilization mechanisms are lost because expression of PRRs is reduced, leading COP1 to target CO for degradation. CO protein stabilized by PRR during the day binds to the FT promoter to activate transcription of FT inducing flowering under LDs.
PRRs participate in light signaling that mediates photoperiodic flowering
The abundance of transcripts and proteins of PRRs exhibit circadian rhythms and under diurnal conditions they peak in expression sequentially at 2‐ to 3‐h intervals during the light period in the order PRR9, PRR7, PRR5, and TOC1 (Fujiwara et al, 2008). Overexpression of PRR5 slightly affects the peak time of some PRRs under LDs, but mutations in PRR genes do not affect the peak time of expression of other PRRs under these conditions (Appendix Fig S4A and B). These proteins are degraded during the night, so they mainly accumulate during the day when CO protein also accumulates (Mas et al, 2003b; Farré & Kay, 2007; Ito et al, 2007; Kiba et al, 2007). Through this regulation, PRRs allow detection of long‐light periods and contribute to recognition of LDs, interacting with and stabilizing CO protein during the day specifically under LDs.
COP1 is a key factor in controlling CO abundance by targeting it for degradation by the proteasome. Our genetic data using the quintuple mutant cop1 prr9 prr7 prr5 toc1 in the SUC2::HA:CO background support the idea that PRRs suppress COP1‐mediated degradation of CO during the day (Fig 4E and F). However, we also found that the levels of HY5 protein were not altered in toc1 prr5 prr7 prr9 mutant (Fig 4H and I), suggesting that PRRs do not participate in mechanisms that control either COP1 mRNA level, COP1 protein abundance or other mechanisms, including its light‐dependent nucleocytoplasmic partitioning, that broadly regulate COP1 activity (Osterlund & Deng, 1998; Laubinger et al, 2006; Zuo et al, 2011). The long‐hypocotyl phenotype of toc1 prr5 prr7 prr9 mutant might suggest that PRRs prevent the function of COP1 in the established light‐signaling pathway (Fig EV4A). However, this long‐hypocotyl phenotype may be due to increased accumulation of transcripts of the clock‐controlled gene PIF4, which encodes a transcription factor involved in shade‐ and dark‐mediated promotion of hypocotyl growth (Fig EV4B; Franklin, 2008; Nakamichi et al, 2009). Furthermore, PRRs physically bind to CO (Figs 5 and EV5), further decreasing the possibility that PRRs suppress general COP1 activity or participate in the mechanisms that mediate light‐dependent suppression of COP1 activity, such as promotion of its migration to the cytoplasm. Therefore, a more attractive hypothesis is that PRRs form a light‐signaling mechanism dedicated to photoperiodic flowering through their accumulation during the day, transferring information on light exposure to CO protein. The pattern of accumulation of PRR proteins in the light is in part mediated by their degradation in the dark. TOC1 and PRR5 proteins are targeted for degradation by ZTL, which was originally identified as a component of the circadian clock (Somers et al, 2000; Mas et al, 2003b; Kiba et al, 2007). It is still not known how PRR7 and PRR9 proteins are degraded during the night.
Taken together, CO protein accumulation is likely to be controlled by a general light‐signaling pathway involving COP1 and a dedicated signaling mechanism involving PRRs, which mainly function to degrade CO in the night and stabilize it in the day, respectively. However, COP1 clearly remains active at a reduced level during the day, triggering reduction of CO protein level at these times and lowering the abundance of CO under LDs (Jang et al, 2008). Our genetic data using the quintuple mutant cop1 prr9 prr7 prr5 toc1 in the SUC2::HA:CO background clearly support the idea that PRRs suppress COP1‐mediated degradation of CO during the day (Fig 4E and F). Therefore, although PRRs may not control general COP1 activity or participate in light‐dependent migration of COP1 to the cytoplasm, they may function in a more specific way to reduce COP1 activity on CO during the day and thereby limit this activity to darkness. In turn, this would ensure sufficient CO protein accumulation under LDs at the specific times that the PRRs are expressed and confer appropriate timing of floral induction under these conditions. The precise molecular mechanism by which PRRs suppress CO degradation by COP1 is still unclear. An attractive hypothesis is that since both PRR and COP1 proteins bind to CO protein, suppression of CO degradation by PRRs might occur at the level of protein–protein interaction. For example, PRRs might interact with CO to directly interfere with COP1 binding and thereby reduce CO degradation. Our co‐immunoprecipitation data showing that TOC1 preferentially binds to the CCT domain in CO (Fig 5D and E) is consistent with this hypothesis, since this domain is also responsible for interaction with COP1 (Jang et al, 2008; Liu et al, 2008). Alternatively, PRRs might act specifically in the phloem companion cells of the vasculature where CO is expressed to directly affect COP1 activity by mechanisms similar to those observed in the general light‐signaling pathways. According to this model, in our study the effect of toc1 prr5 prr7 prr9 mutations on HY5 levels in the vasculature would not be detected because HY5 is broadly expressed (Oyama et al, 1997). However, in this case, since PRRs are also broadly expressed in A. thaliana to control circadian‐clock functions (Para et al, 2007; Fujiwara et al, 2008), the activity of PRRs to directly control COP1 activity might be expected to occur in other tissues as well.
In addition to COP1 and FKF1 that were discussed above, HOS1 and TOE1 have also been implicated in regulating CO stability (Lazaro et al, 2012; Zhang et al, 2015). Although it is possible that HOS1 and TOE1 might also be regulated by PRRs, our genetic data with the cop1 toc1 prr5 prr7 prr9 quintuple mutant do not currently support this hypothesis, because the cop1 mutation is epistatic to the prr mutations with respect to CO protein abundance (Fig 4E and F). If the PRRs do suppress other mechanisms that regulate CO protein abundance independently of COP1, such as those proposed to act through HOS1 or TOE1 (Lazaro et al, 2012, 2015; Zhang et al, 2015), then their increased activity by the prr mutations would be expected to reduce the level of CO protein even in the cop1 mutant background.
Alleles of PRR genes contribute to QTLs for flowering time in a wide range of crop species. Ppd‐H1, Ppd‐1, BOLTING TIME CONTROL 1 (BvBTC1), PRR37, and SbPRR37 encode PRR proteins in barley, wheat, beet, rice, and sorghum, respectively, and allelic variation at these genes confers natural diversity in flowering time (Turner et al, 2005; Pin et al, 2009; Murphy et al, 2011; Campoli et al, 2012; Shaw et al, 2012; Koo et al, 2013). Interestingly, in contrast to A. thaliana PRRs that affect the time‐keeping mechanism associated with CO transcription, the PRRs identified in several crop species have weak or no effects on this timing mechanism, despite their strong effects on FT transcription (Turner et al, 2005; Campoli et al, 2012; Koo et al, 2013). In this study, we demonstrated a novel function of PRRs in A. thaliana, where they bind to and stabilize CO during the day to enable it to accumulate specifically under LDs and initiate floral transition. Whether these roles for PRRs are also conserved in other plants including crop species is an important issue to resolve. Further molecular‐genetic analyses using these crops may help us to understand how changes in the mechanisms of day‐length recognition facilitate adaptation to different latitudes in crops and has potential to improve performance in different environments.
Materials and Methods
Plant materials and growth conditions
We used toc1‐101, prr5‐11, prr7‐11, and prr9‐10 as prr mutants (Ito et al, 2003; Yamamoto et al, 2003; Salome et al, 2008). SUC2::CO and SUC2::HA:CO have been previously described (An et al, 2004; Jang et al, 2009). All of the single, double, triple, and quadruple prr mutants with SUC2::CO and SUC2::HA:CO were generated by genetic crossing. 35S::TOC1 was crossed to SUC2::HA:CO and used for ChIP experiments. For cop1 mutant analyses, cop1‐4 was used (McNellis et al, 1994). For fkf1 mutant analyses, fkf1‐2 was used (Imaizumi et al, 2003). SUC2::HA:CO fkf1‐2 was generated by genetic crossing. TMG, pPRR5::PRR5:GFP, pPRR7::FLAG:PRR7:GFP, and pPRR9::FLAG:PRR9:GFP plants have been previously described (Mas et al, 2003a; Fujiwara et al, 2008; Nakamichi et al, 2010). For plasmid construction of pCO::HA:CO, 35S promoter in the pAlligator2 vector was replaced by 2.5 kb CO promoter. Primers for amplifying the CO promoter are GCGGCTCTAGATTTTGATCATGCAACATAAACTTATAAGC and CGGCCCTCGAGAATAACTCAGATGTAGTAAGTTTGATGGTG. Full‐length CO cDNA was inserted into the vector by recombination following Invitrogen instructions. Arabidopsis thaliana transformation was performed by floral dip using Agrobacterium. GFP‐positive seeds were selected using a Leica MZFLIII stereomicroscope equipped with GFP filters.
For cDNA and protein analyses, plants were grown for 9 days on MS agar plate. In all expression analyses (except Fig 4A–D), FRET, co‐IP, and ChIP experiments plants were grown in climatic chambers with fluorescent lamps at 22°C. In experiments in Fig 4A–D, plants were grown in climate chambers with LED light of blue or far‐red at 22°C. For flowering‐time analyses, plants were grown on soil at 21°C under 100–150 μmol m−2 s−1 photosynthetic active radiation. The number of total leaves and the number of days after sowing when flower bolts were 2 cm in length was used as a measure of flowering.
RNA isolation, cDNA analysis, and expression data analyses
RNA was isolated from 50 to 100 A. thaliana seedlings for each line grown on an agar plate by RNeasy Plant Mini Kit (Qiagen) with on‐column DNAase treatment. cDNA was synthesized by Superscript II reverse transcriptase (Life Technologies). Expression analyses were performed by real‐time PCR (Roche LightCycler). Primers used for the expression analyses are CO (GCATGTGTCACAACAGCTTCAC and ATGCCTTCCTCGAAGCATACC), FT (TCAGAGGGAGAGTGGCTG and TCACCGTTCGTTACTCGTATC), PP2a (CTTGGTGGAGCTAAGTGAAGACC and CGCCCAACGAACAAATCACAGA), PIF4 (TCCGACCGGTTTGCTAGA and ACCTAGTGGTCCAAACGAGAA), TOE1 (GTAACTGGGGATGGCAGAGA and TGTCCCCTTACAAGTTAAGGGT), TOE2 (TCTCATGAACCGCCCACAA and TGTGCTGAGCCAAACGATGA), and PRR9 (CCTCGAGTGAAAGGCCAGT and CAAAAGTTGCCCCAGTATCTCA).
For expression data analyses including both mRNA and proteins, the values of the expression levels in two or three biological data sets were averaged and combined. Prior to this process, the values of expression levels in each data set were pre‐normalized to the highest value in each data set, which was given the value of 1.
Analysis of HA:CO protein abundance
50–100 Arabidopsis seedlings for each line grown on an agar plate were ground in liquid nitrogen. Approximately 500 μl of leaf powder was suspended with the nuclear isolation buffer (20 mM Tris–HCl, 20 mM MgCl2, pH 6.8, 5% sucrose, 40% glycerol, 0.8% Triton X‐100, 0.08% β‐mercaptoethanol, 0.2% SIGMA plant protease inhibitor, 1 mM DTT, 1.3 mM PMSF). The samples were centrifuged at 3,000 g for 5 min, and the supernatant was removed. The pellets were washed four times with the nuclear isolation buffer with the same procedure. The pellets were suspended in the SDS sample buffer and thereafter heated at 95°C for 10 min, and 20 μl of the supernatants were loaded on acrylamide gel. For Western analyses, anti‐HA peroxidase high affinity (Roche) was used for detecting the HA:CO signal. Anti‐histone 3a was used for detecting the H3a signal as loading control. Signals were detected by LAS‐4000 (Fujifilm) and measured by the ImageJ software.
MG132 treatment and induction of HA:CO accumulation
Approximately 30 seedlings for each line were grown on MS agar media for 10 days and transferred at ZT5 to MS liquid media containing either 100 μM of MG132 or only the DMSO solvent. Samples were vacuum infiltrated for 10 min, transferred back to the growth cabinet with the liquid media, and harvested at ZT8. Nuclear protein was extracted with the method described above. HA:CO and H3a signals were detected by Western blot with antibodies indicated in the main method section.
FRET and co‐IP experiments
For FRET in A. thaliana cells, protoplasts from adult leaves of plants grown under 12‐h light/12‐h dark were isolated by previously described methods (Yoo et al, 2007; Wu et al, 2009). In these experiments, 9 μg of plasmids carrying 35S::PRR:CFP or 35S::CO:YFP was used for transfection to ~100 000 protoplasts. After transfection, the protoplasts were placed in a growth chamber with 12‐h light/12‐h dark with the low intensity of light and the next day FRET was performed. 20 and 10 cells for the first and second experiments, respectively, were imaged with the confocal laser scanning microscope SP8 (Leica), and after bleaching of the acceptor (CO:YFP), the change in donor (PRR:CFP) fluorescence was quantified by comparing pre‐ and post‐bleaching images. FRET efficiency was calculated with a formula “(pre‐bleaching – post‐bleaching)/pre‐bleaching”. For FRET using N. benthamiana leaves, Agrobacterium containing 35S::PRR:CFP or 35S::CO:YFP was infiltrated into Nicotiana benthamiana, and after 3 days, protein–protein interaction was measured by CFP‐YFP FRET by acceptor photobleaching. 30 and 10 Nicotiana benthamiana cells for the first and second experiments, respectively, were imaged with LSM 780 (Carl Zeiss), and after bleaching of the acceptor (CO:YFP), the change in donor (PRR:CFP) fluorescence was quantified by comparing pre‐ and post‐bleaching images.
For coIP experiment, Agrobacterium containing 35S::PRR:GFP or 35S::HA:CO was infiltrated into Nicotiana benthamiana, and after 3 days, the infiltrated leaves were harvested. The leaves were ground in liquid N2, and ~500 μl of the leaf powder was suspended in the nuclear isolation buffer (50 mM Tris–HCl pH 7.4, 0.5% NP‐40, 40% glycerol, 1 mM DTT, 0.2% SIGMA plant protease inhibitor, 1.3 mM PMSF, 50 μM MG132). The samples were centrifuged at 1,000 g for 5 min, and the supernatants were removed. Pellets were washed four times with the nuclear isolation buffer using the same procedure. After removing the supernatant, the pellets were suspended in 0.3 ml of the sonication buffer (50 mM Tris–HCl pH 7.4, 50 mM NaCl, 0.5% NP‐40, 1 mM DTT, 0.2% SIGMA plant protease inhibitor, 1.3 mM PMSF, 50 μM MG132), and the samples were sonicated for 2.5 min, 5 min, or 7.5 min for PRR9, PRR5/PRR7, or TOC1, respectively, with a bath sonicator (DIAGENODE BIORUPTOR) with “high” setting for 15 min with 15 s sonications separated by 15 s breaks to release PRR:GFP and HA:CO proteins from the nuclei. 0.7 ml of the sonication buffer and 20 μl of 5 M NaCl were added to the samples, and the samples were incubated for 15 min with mild agitation. The samples were centrifuged at 21,000 g for 15 min, and the supernatants were recovered. Immunoreaction was performed by adding the anti‐GFP antibody (Roche) to the supernatants and incubating these for 1 h, and afterward protein‐G sepharose (Roche) was added and the samples were incubated for 3 h. The sepharose was washed four times with the washing buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5% NP‐40, 1 mM DTT). The sepharose beads was incubated with 40 μl of the SDS sample buffer and heated at 95°C for 10 min, and a half of the supernatant was loaded on SDS acrylamide gel. For input samples, 7.5% of the sonicated protein samples were loaded on the gel. GFP and HA signals were detected with the anti‐GFP antibody (Abcam) and the anti‐HA peroxidase (Roche), respectively. For the co‐IP experiments between TOC1:HA and truncated CO:YFPs, 1–121 a.a. for the zinc fingers, 122–297 a.a. for the middle part, 122–373 a.a. for the middle‐CCT, and 298–373 a.a. for the CCT were fused to YFP.
ChIP assay
ChIP was performed as published previously (Hyun et al, 2016) with minor modifications. Plants were grown for 10 days in LDs and harvested at Zeitgeber time (ZT) 12. Before plant tissue was cross‐linked in 1% formaldehyde for 10 min, plants were infiltrated with 1 μM DSG for 10 min in a vacuum desiccator. Chromatin immunoprecipitation of HA:CO for toc1 prr5 prr7 prr9 SUC2::HA:CO and 35S::TOC1 SUC2::HA:CO was performed by using HA antibody (Abcam, ab9110). ChIP‐qPCR was analyzed, and the relative enrichment of the IP/Input at each genomic region tested was normalized to that of the reference locus, ACT8.
Author contributions
LS‐K performed a part of the experiments on flowering‐time measurement and co‐IP, as well as the experiment for checking SUC2 gene expression. LS‐K also prepared the A. thaliana pCO::HA:CO line that SJ previously created through vector construction and transformation. RR performed all the ChIP experiments. VF performed the experiment for checking effect of MG132 treatment on HA:CO accumulation. RH carried out all other experiments. GC and RH wrote this article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We are grateful to the following people for kindly supplying materials. Dr. David Somers provided Agrobacterium carrying 35S::PRR:GFP and plants carrying pPRR5::PRR5:GFP. Dr. Steve Kay provided TMG plants. Dr. Takeshi Mizuno provided 35S::PRR9 and 35S::PRR5 plants, and Dr. Norihito Nakamichi provided 35S::TOC1. Dr. Nam‐Hai Chua provided pPRR::FLAG:PRR:GFP plants. Dr. Takayuki Shindo provided Agrobacterium carrying 35S::YFP and 35S::CFP. Dr. Nabil Elrouby provided Agrobacterium carrying 35S::CO:YFP. Dr. Yasuyuki Takahashi created constructs for truncated CO proteins. Dr. Yue Zhou provided plasmids carrying 35S::AGL16:YFP, 35S::AGL16:CFP and 35S::TRB3:YFP. We thank Dr. Elmon Schmelzer gave advice on FRET. Finally, we thank all G.C. laboratory members for useful discussions. This work was funded by the DFG through SFB635, the ERC through Hylife, and work in the laboratory of GC is funded by a core grant from the Max Planck Society.
The EMBO Journal (2017) 36: 904–918
References
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis ‐regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis . Plant Cell 22: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez‐Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis . Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M (2012) Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS . Plant J 69: 868–880 [DOI] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF III (2014) A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis . Plant Cell 26: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Farré EM, Kay SA (2007) PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis . Plant J 52: 548–560 [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Franklin KA (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post‐translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo‐response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda‐Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA‐binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, Turck F, de Meaux J (2014) miR824‐regulated AGAMOUS‐LIKE16 contributes to flowering time repression in Arabidopsis . Plant Cell 26: 2024–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Perez‐Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez‐Gallegos R, Porri A, Coupland G (2016) Multi‐layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell 37: 254–266 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic‐specific light signalling in Arabidopsis . Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1F‐box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis . Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) Characterization of the APRR9 pseudo‐response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana . Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Ito S, Nakamichi N, Kiba T, Yamashino T, Mizuno T (2007) Rhythmic and light‐inducible appearance of clock‐associated pseudo‐response regulator protein PRR9 through programmed degradation in the dark in Arabidopsis thaliana . Plant Cell Physiol 48: 1644–1651 [DOI] [PubMed] [Google Scholar]
- Ito S, Niwa Y, Nakamichi N, Kawamura H, Yamashino T, Mizuno T (2008) Insight into missing genetic links between two evening‐expressed pseudo‐response regulator genes TOC1 and PRR5 in the circadian clock‐controlled circuitry in Arabidopsis thaliana . Plant Cell Physiol 49: 201–213 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G (2009) Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis . Plant J 60: 614–625 [DOI] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO‐RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana . Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Hicks KA, Somers DE (2005) Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 139: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang Z, Li J, Li Z, Paek NC (2013) Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant 6: 1877–1888 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Pineiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Mouriz A, Pineiro M, Jarillo JA (2015) Red light‐mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis . Plant Cell 27: 2437–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome‐interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1‐mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis . Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek P, Whitelam G, Fankhauser C (2008) Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis . Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana . Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo‐response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Misera S, Deng XW (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Yamashino T, Mizuno T (2004) Characterization of circadian‐associated APRR3 pseudo‐response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana . Plant Cell Physiol 45: 645–650 [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO‐RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana . Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock‐associated pseudo‐response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS‐dependent photoperiodic pathway. Plant Cell Physiol 48: 822–832 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO‐RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Deng XW (1998) Multiple photoreceptors mediate the light‐induced reduction of GUS‐COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light‐regulated development of Arabidopsis . Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus‐induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Farre EM, Imaizumi T, Pruneda‐Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS‐box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Zhang W, Vogt SH, Dally N, Buttner B, Schulze‐Buxloh G, Jelly NS, Chia TY, Mutasa‐Gottgens ES, Dohm JC, Himmelbauer H, Weisshaar B, Kraus J, Gielen JJ, Lommel M, Weyens G, Wahl B, Schechert A, Nilsson O, Jung C et al (2009) The role of a pseudo‐response regulator gene in life cycle adaptation and domestication of beet. Curr Biol 22: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Salome PA, Xie Q, McClung CR (2008) Circadian timekeeping during early Arabidopsis development. Plant Physiol 147: 1110–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid‐Krebs L, Panigrahi KC, Fornara F, Takahashi Y, Hayama R, Jang S, Tilmes V, Valverde F, Coupland G (2015) Phosphorylation of CONSTANS and its COP1‐dependent degradation during photoperiodic flowering of Arabidopsis . Plant J 84: 451–463 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day‐length measurement in Arabidopsis . Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrumpfova PP, Vychodilova I, Dvorackova M, Majerska J, Dokladal L, Schorova S, Fajkus J (2014) Telomere repeat binding proteins are functional components of Arabidopsis telomeres and interact with telomerase. Plant J 77: 770–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Turner AS, Laurie DA (2012) The impact of photoperiod insensitive Ppd‐1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J 71: 71–84 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock‐associated PAS protein from Arabidopsis . Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth‐Schultz HA, Imaizumi T (2014) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66: 441–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, Gonzalez‐Schain N, Martin G, Diaz C, Sentandreu M, Al‐Sady B, Quail PH, Monte E (2016) Molecular convergence of clock and photosensory pathways through PIF3‐TOC1 interaction and co‐occupancy of target promoters. Proc Natl Acad Sci USA 113: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suarez‐Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis . Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis ‐element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Torii KU, McNellis TW, Deng XW (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J 17: 5577–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo‐response regulator Ppd‐H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdan PD, Amasino RM (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C‐mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape‐Arabidopsis Sandwich – a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T (2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yamashino T, Ito S, Niwa Y, Kunihiro A, Nakamichi N, Mizuno T (2008) Involvement of Arabidopsis clock‐associated pseudo‐response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol 49: 1839–1850 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis . Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hartwig B, James GV, Schneeberger K, Turck F (2016) Complementary activities of TELOMERE REPEAT BINDING proteins and polycomb group complexes in transcriptional regulation of target genes. Plant Cell 28: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J‐Y, Oh E, Wang T, Wang Z‐Y (2016) TOC1‐PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis . Nat Commun 7: 13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light‐dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis . Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


