Abstract
Zika virus (ZIKV) has emerged as major health concern, as ZIKV infection has been shown to be associated with microcephaly, severe neurological disease and possibly male sterility. As the largest protein component within the ZIKV replication complex, NS5 plays key roles in the life cycle and survival of the virus through its N‐terminal methyltransferase (MTase) and C‐terminal RNA‐dependent RNA polymerase (RdRp) domains. Here, we present the crystal structures of ZIKV NS5 MTase in complex with an RNA cap analogue (m7GpppA) and the free NS5 RdRp. We have identified the conserved features of ZIKV NS5 MTase and RdRp structures that could lead to development of current antiviral inhibitors being used against flaviviruses, including dengue virus and West Nile virus, to treat ZIKV infection. These results should inform and accelerate the structure‐based design of antiviral compounds against ZIKV.
Keywords: crystal structure, methyltransferase, polymerase, RNA capping, Zika virus
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Introduction
Zika virus (ZIKV), a mosquito‐borne flavivirus, has become a major public health concern over the past year (Calvet et al, 2016). ZIKV infection in pregnant women can cause congenital malformations including fetal and newborn microcephaly, and serious neurological complications, such as Guillain–Barré syndrome (Carteaux et al, 2016; Driggers et al, 2016; Mecharles et al, 2016; Mlakar et al, 2016; Zhang et al, 2016). In addition to transmission by mosquitoes, ZIKV has been shown to be able to establish long‐term persistent infection (Mansuy et al, 2016a) and be transmitted by sexual activity (Davidson et al, 2016; Deckard et al, 2016; Prisant et al, 2016). More importantly, ZIKV was detected in human semen and spermatozoa (Mansuy et al, 2016b), and ZIKV infections of male adult mice can cause testicular and epididymal damage, resulting in cell death and destruction of the seminiferous tubules (Govero et al, 2016; Ma et al, 2016). These findings pose new challenges for controlling outbreaks caused by this virus. However, there is currently no available drug approved to treat or prevent ZIKV infections. Given this urgent situation, an important strategy to prevent the spread of ZIKV would be to develop antivirals to inhibit viral protein activities, which are central to survival of the virus. In particular, antivirals could also be used to avoid fetal neurological disorders or even male sterility induced by ZIKV infections.
A good target for designing antiviral inhibitors is the viral replication machinery, inhibition of which could block viral replication. Like other flaviviruses, such as Dengue virus (DENV), Japanese encephalitis virus (JEV), Yellow fever virus (YFV), West Nile virus (WNV) and tickborne encephalitis virus (TBEV), ZIKV is an enveloped, single‐stranded, positive‐sense RNA virus carrying a cap‐1 structure (m7G0pppAm2′‐O‐G‐RNA) at its 5′ end. The RNA genome of ZIKV translates into a long polyprotein in the cytoplasm of the infected cells. The polyprotein is further cleaved and processed by either host or viral proteases into three structural and seven nonstructural (NS) proteins. The structural proteins are precursor membrane (prM) protein, envelope (E) protein, capsid (C) protein, which constitute the virus particle, and seven NS proteins include NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5, which perform essential functions in genome replication, polyprotein processing and manipulation of cellular processes for virus infections. RNA replication occurs within a multi‐protein replication complex (RC) comprising of both NS proteins and host cofactors, which assemble on endoplasmic reticulum (ER)‐derived membranes (Welsch et al, 2009). Among them, NS5 is the largest enzyme and the most conserved protein of the RC. NS5 carries two essential enzymatic activities, methyltransferase (MTase) and RNA‐dependent RNA polymerase (RdRp), which are strictly required for the viral replication, and therefore constitute promising targets for the development of antiviral compounds.
NS5 MTase locates at the N‐terminus of the NS5 protein, methylates guanosine N‐7 and ribose 2′‐O positions of the viral RNA cap, which is crucial for genome stability, efficient translation and evasion of the host immune response (Zhou et al, 2007). MTases from various flaviviruses perform the two cap methylations in a sequential manner: N‐7 methylation followed by 2′‐O‐methylation, that is, G0pppAG‐RNA–>m7G0pppAG‐RNA–>m7G0pppAm2′‐O‐G‐RNA, using S‐adenosyl‐L‐methionine (SAM) as the methyl donor and generate S‐adenosyl‐L‐homocysteine (SAH) as a by‐product (Zhao et al, 2015). N‐7‐MTase activity has been shown to be essential for viral replication by biochemical studies and reverse genetic analysis, while 2′‐O‐MTase defective viruses can replicate but are highly attenuated (Dong et al, 2010a; Zust et al, 2011, 2013). Therefore, suppression of both N7‐ and 2′‐O‐MTase activities is important for the development of antivirals. The C‐terminus of the NS5 protein is the RdRp that synthesizes the genome RNA in the absence of a primer strand, which is called a de novo initiation mechanism. Flavivirus RdRp contains functional nuclear localization sequence (NLS), which is the key region for interactions with other viral and host proteins. NS5 interacts with the NS3 protease‐helicase and several host proteins, including β‐importin (Yap et al, 2007; Tay et al, 2015). In addition to its enzymatic functions, NS5 acts as an antagonist of the host interferon response by interacting with and promoting the degradation of the signal transducer and activator of transcription 2 protein (Ashour et al, 2009), and the same mechanism has been proven in ZIKV recently (Grant et al, 2016; Kumar et al, 2016). The importance of NS5 in viral replication and host immune response modulation makes it an attractive target for developing broad‐spectrum antiviral inhibitors.
During the preparation of our manuscript, Javier Coloma et al have reported the high‐resolution crystal structures of NS5 MTase from the H/PF/2013 strain (isolated from French Polynesia) in complex with SAM or SAM and 7‐methyl guanosine diphosphate (m7Gpp) (Coloma et al, 2016). Here, we have studied the NS5 protein of the BeH819015 strain (isolated from Brazil in 2015, responsible for the ZIKV outbreak in South America), and determined the crystal structure of the NS5 MTase at a resolution of 2.5 Å in the presence of bound SAM and the complex structure of ZIKV MTase with both SAM and a bigger RNA analogue (m7GpppA) at a resolution of 2.6 Å, and the NS5 RdRp at a resolution of 1.8 Å. These studies provide important structural information to help guide the development of antiviral compounds against ZIKV NS5.
Results
Structure of ZIKV NS5 MTase bound to an RNA cap analogue m7GpppA
The crystal structure of ZIKV NS5 MTase was solved by molecular replacement to a resolution of 2.5 Å, with R work and R free values of 23.5 and 25.7%, respectively (Table 1). The ZIKV MTase has an overall globular fold and can be subdivided into three subdomains (Fig 1A). The subdomain 1 (N‐terminal extension) consists of a helix‐turn‐helix motif followed by a strand and a helix. The subdomain 2 forms the core structure, adopts the SAM‐dependent methyltransferase fold and folds into a seven‐stranded β‐sheet surrounded by four α‐helices. The subdomain 3 (C‐terminal extension) comprises an α‐helix and two β‐stands (Fig 1A and B). Although no SAM or SAH was added during purification or crystallization, SAM, the methyl donor for the methylation reaction of MTase, is co‐purified and clearly visible in the electron density in the active site (Fig 1A). This was also observed in other MTase structures (Mastrangelo et al, 2007; Bollati et al, 2009a,b; Jansson et al, 2009). The core domain contains a binding site for SAM, referred to here as the SAM‐binding pocket (Fig 1C). The SAM molecule binds to the SAM‐binding pocket mainly by hydrogen‐bonding interactions and van der Waals interactions, contacting with residues S56, G86, W87, K105, H110, D131, V132 and D146 (Figs 1C and 2), which are conserved among different flavivirus MTases (Fig 1D).
Table 1.
Data collection and refinement statistics
| ZIKV NS5 MTase (PDB code 5WZ1) | ZIKV NS5 MTase with RNA analogue (PDB code 5WZ2) | ZIKV NS5 RdRp (PDB code 5WZ3) | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.97853 | 0.97907 | 0.72929 |
| Space group | P 21 | C 2 | P 21 |
| Cell dimensions | |||
| a, b, c (Å) | 107.55, 86.85, 107.56 | 75.13, 78.27, 136.38 | 63.53, 84.85, 69.20 |
| α, β, γ (°) | 90.00, 97.39, 90.00 | 90.00, 90.10, 90.00 | 90.00, 113.37, 90.00 |
| Resolution (Å) | 50.00–2.50 (2.59–2.50)a | 50.00–2.60 (2.69–2.60) | 50.00–1.80 (1.86–1.80) |
| R merge b | 0.195 (1.085) | 0.161 (0. 815) | 0.078 (0.612) |
| I/σI | 8.075 (1.409) | 9.362 (1.953) | 22.098 (3.064) |
| CC 1/2 | 0.997 (0.695) | 0.993 (0.640) | 0.994 (0.823) |
| Completeness (%) | 98.6 (98.2) | 99.8 (98.6) | 98.5 (92.5) |
| Redundancy | 5.7 (5.9) | 4.7 (4.5) | 6.1 (5.2) |
| Refinement | |||
| Resolution (Å) | 45.45–2.50 | 42.46–2.60 | 34.16–1.80 |
| No. reflections | 63,210 | 23,751 | 61,141 |
| R work/R free c | 0.235/0.257 | 0.202/0.247 | 0.178/0.206 |
| No. atoms | |||
| Protein | 16,168 | 6,297 | 4,641 |
| Ligand/ion | 216 | 234 | 2 |
| Water | 0 | 90 | 503 |
| B‐factors | |||
| Protein | 40.6 | 48.4 | 27.6 |
| Ligand/ion | 34.0 | 60.6 | 18.3 |
| Water | – | 40.4 | 34.2 |
| Rms. deviations | |||
| Bond lengths (Å) | 0.004 | 0.004 | 0.005 |
| Bond angles (°) | 0.719 | 0.913 | 0.958 |
| Ramachandran plot | |||
| Favored (%) | 95.57 | 96.76 | 97.30 |
| Allowed (%) | 4.23 | 2.72 | 2.52 |
| Outliers (%) | 0.19 | 0.52 | 0.18 |
Values in parentheses are for highest resolution shell.
R merge = ΣiΣhkl|I i−<I>|/ΣiΣhkl I i, where I i is the observed intensity and <I> is the average intensity from multiple measurements.
R work = Σ | |F o|−|F c| |/Σ |F o|, where F o and F c are the structure‐factor amplitudes from the data and the model, respectively. R free is the R factor for a subset (5%) of reflections that was selected prior to refinement calculations and was not included in the refinement.
Figure 1. Structure of ZIKV MTase in complex with the cofactor SAM .
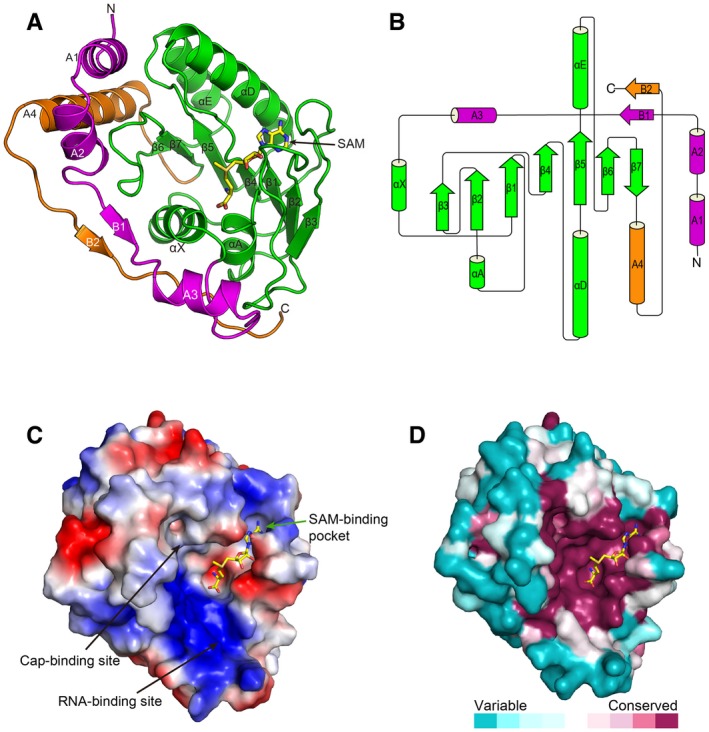
- Cartoon representation of ZIKV methyltransferase colored by three subdomains: subdomain 1 (magenta), subdomain 2 (green) and subdomain 3 (orange). The cofactor SAM is shown as sticks. The secondary structure element nomenclature corresponds to the DENV2 MTase (Egloff et al, 2002).
- Topology diagram for ZIKV MTase. The colors are coherent with the above representation of the structure.
- The electrostatic surface potential of ZIKV MTase. Red and blue colors indicate negative potential and positive potential, respectively. The cofactor SAM is shown as sticks colored in yellow.
Figure 2. Detailed interaction of ZIKV MTase bound to the cofactor SAM and the RNA analogue m7GpppA.
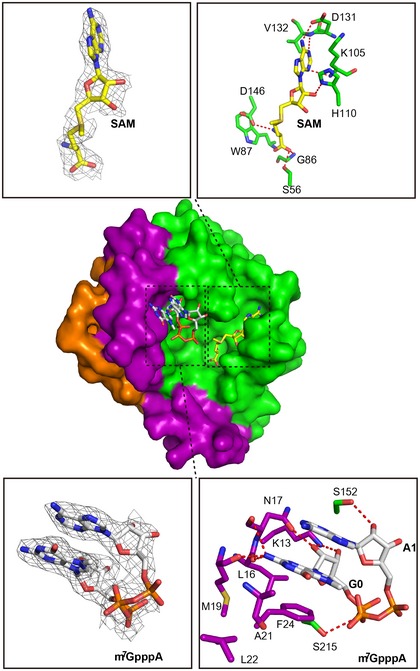
Overall structure showing ZIKV MTase in complex with the cofactor SAM and the RNA analogue (m7GpppA). The 2F o−F c electron density map for SAM or RNA analogue contoured at 1.0 sigma is represented, respectively, in gray. The hydrogen bond interactions of ZIKV MTase bound to SAM and m7GpppA are shown in dashed lines.
The overall structure of the ZIKV MTase is highly like other flaviviruses MTase structures, with root‐mean‐square differences (rmsd) of 0.41–0.83 Å (Fig EV1). Of note, among these structures, three regions, residues 48–52, 173–l77 and 245–249, show the greatest structural differences. The differences are partially due to the insertion or deletion of amino acid residues in these loops. Besides the SAM pocket, the core domain contains a pocket that binds to the GTP cap of the viral RNA, referred to as the cap‐binding site, and a positively charged groove between these two pockets referred to as the RNA‐binding site (Fig 1C). We compared the ZIKV MTase structure with all the other available flavivirus MTases structures, including DENV2, YFV, DENV3, WNV, Murray Valley encephalitis virus (MVEV), Wesselsbron virus (WESSV), Modoc virus (MODV), Meaban virus (MEAV), Yokose virus (YOKV) and JEV (Egloff et al, 2002; Assenberg et al, 2007; Mastrangelo et al, 2007; Zhou et al, 2007; Bollati et al, 2009a,b; Geiss et al, 2009; Jansson et al, 2009; Lu & Gong, 2013; Coutard et al, 2014). The electrostatic surface potential maps of these proteins show very similar patterns, especially the positively charged surface in the RNA‐binding site (Fig EV2). We then analyzed the conserved features based on the alignment of MTase sequences from 73 flaviviruses (Fig 1D). The most conserved regions are the three binding sites: the SAM‐binding pocket, cap‐binding pocket and RNA‐binding site, indicating a conserved methylation mechanism for different flaviviruses.
Figure EV1. Superposition of the ZIKV MTase structure with other flavivirus MTase structures.
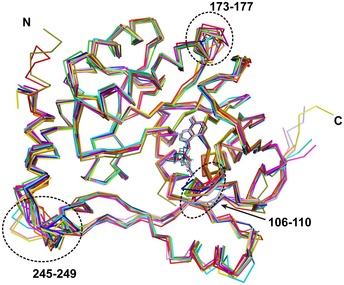
Ribbon representation of each structure is colored separately (ZIKV, green; DENV2, PDB: 1L9K, light blue; WNV, PDB: 2OY0, cyan; YFV, PDB: 3EVC, magenta; DENV3, PDB: 4CTJ, blue; MVEV, PDB: 2PX2, pink; WESSV, PDB: 3ELU, orange; MODV, PDB: 2WA1, deep olive; MEAV, PDB: 2OXT, red; YOKV, PDB: 3GCZ, purple; JEV, PDB: 4K6M, yellow). The loops that show significant differences are labeled.
Figure EV2. Comparison of the electrostatic surfaces of ZIKV MTase structure with other flavivirus MTase structures.
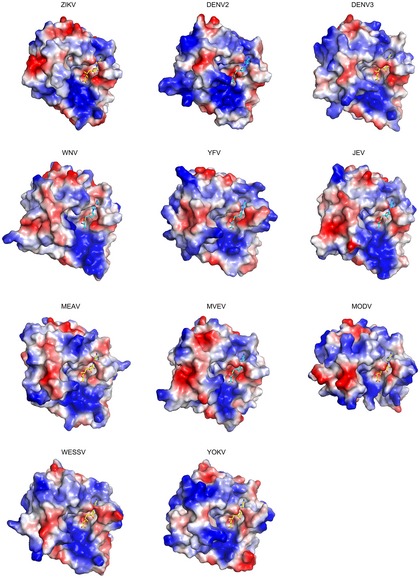
Red and blue colors indicate negative potential and positive potential, respectively. SAM and SAH are shown as sticks and colored in yellow and cyan, respectively. PDB codes of structures used for analysis are described in the Fig EV1 legend.
To elucidate the molecular mechanism of ZIKV MTase in recognizing capped RNA and to know the detailed mechanism of RNA analogue inhibition, we determined the crystal structure of ZIKV MTase in complex with a RNA analogue at 2.6 Å, with R work and R free values of 20.5 and 25.5%, respectively (Table 1). Little conformational changes of the protein were observed upon RNA analogue binding. The RNA analogue binds to the cap‐binding site, formed by helices A1, A2 and a loop (amino acids 208–218), in a stacked conformation (Fig 2). The second nucleobase A1 stacks against the first nucleobase 7mG0, displaying a hairpin‐like shape. The aromatic side chain of residue F24 arranges in parallel with the two base moieties of 7mG0 and A1, below the 7mG0, forming strong π–π interaction. The exocyclic 2′‐amine group of 7mG0 interacts via hydrogen bonding with the main‐chain carbonyl groups of residues L16, N17 and M19, while the 2′‐hydroxyl group is hydrogen‐bonded to the amino group of residue K13 side chain and the oxygen of residue N17 main‐chain. The amino group K13 side chain also forms a hydrogen bond with the 3′‐hydroxyl group of 7mG0 ribose. The 2′‐hydroxyl group of the A1 ribose interacts via hydrogen bond with the hydroxyl group of residue S152. In addition, the first phosphate of triphosphate forms a hydrogen bond with residue S215 (Fig 2).
Interestingly, we found that ZIKV MTase contains nonpolar hydrophobic residues A21 and L22 near the cap‐binding site, while in other flaviviruses they are all polar residues (DENV2: K22/S23; DENV3: R22/K23; WESSV: K21/Q22) at the same sites (Fig 3). Moreover, the shorter side chain of A21 in ZIKV MTase compared to Arg or Lys in other flaviviruses makes the ZIKV MTase cap‐binding pocket more shallow and “open”.
Figure 3. Comparison of the structures of ZIKV MTase with other flavivirus MTases in complex with SAM/SAH and the RNA molecule.
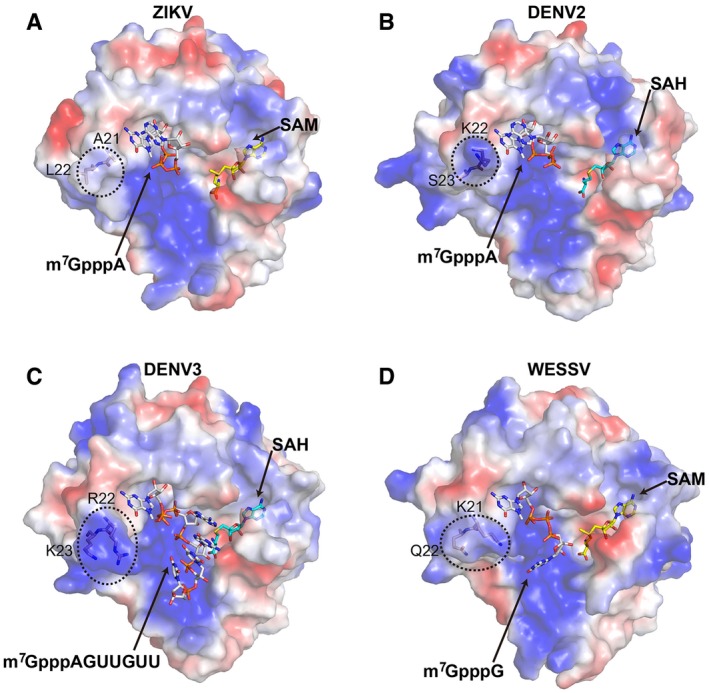
-
A–DThe ZIKV MTase in complex with the cofactor SAM and the RNA analogue m7GpppA (A), DENV2 MTase in complex with SAH and m7GpppA (PDB: 2P3O) (Egloff et al, 2007) (B), DENV3 MTase in complex with SAH and m7GpppAGUUGUU (PDB: 5DTO) (Zhao et al, 2015) (C) and WESSV MTase in complex with SAM and m7GpppG (PDB: 3EMB) (Bollati et al, 2009b) (D) are shown respectively. The RNA molecule, SAM, and SAH are shown as sticks and colored in white, yellow and cyan, respectively. The variable residues near the capping binding site are highlighted in dotted boxes and labeled.
As the host also has RNA MTases for methylation, we compared our ZIKV MTase structure with human RNA guanine‐7 methyltransferase (RNMT) structure in complex with its activating subunit, RNMT‐activating miniprotein (RAM) (Varshney et al, 2016), and mRNA 2′‐O‐methylation MTase CMTr1 (Smietanski et al, 2014) (Fig 4). For SAM‐binding pocket, ZIKV MTase has a conserved additional hydrophobic pocket located next to the binding site (Fig 4B). This pocket has been proven to be important both for viral replication and cap methylations and for specific inhibitor design targeting flavivirus MTase (Dong et al, 2010b). For the cap‐binding site, cellular and viral MTase interact with the guanosine cap in very different ways. In the structure of the human CMTr1, m7G is bound in a deep pocket, and the orientation and binding mode is quite different from m7G binding to ZIKV MTase (Fig 4E and F). Therefore, both the SAM‐binding pocket and cap‐binding site have diverse properties between the cellular and viral enzymes, making the ZIKV MTase an attractive target for the development of inhibitors.
Figure 4. Comparison of the structures of ZIKV MTase with human RNA MTases.
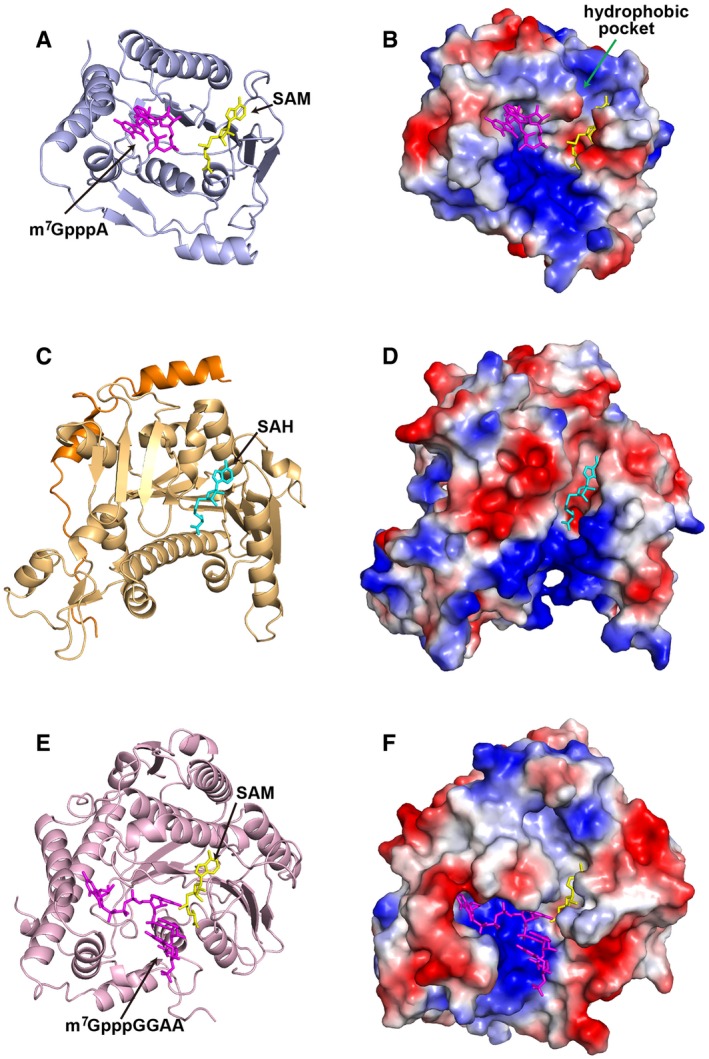
-
A–FThe ZIKV MTase in complex with the cofactor SAM and the RNA analogue m7GpppA shown in cartoon (A) and electrostatic surface (B). Human RNA guanine‐7 methyltransferase RNMT‐RAM in complex with SAH (PDB: 5E8J) (Varshney et al, 2016) shown in cartoon (C) and electrostatic surface (D). Human 2′‐O‐ribose MTase CMTr1 in complex with a methyl group donor SAM and a capped oligoribonucleotide (PDB: 4N48) (Smietanski et al, 2014) shown in cartoon (E) and electrostatic surface (F). The RNA molecule, SAM, and SAH are shown in sticks and colored in magenta, yellow and cyan, respectively.
Structure of ZIKV NS5 RdRp
The crystal structure of ZIKV NS5 RdRp was solved here by molecular replacement to a resolution of 1.8 Å, with R work and R free values of 17.8 and 20.6%, respectively (Table 1). The ZIKV RdRp displays an overall spherical fold, and has the canonical right‐hand conformation consisting of fingers (α1‐α9 from residues 275 to 498, and α12 to β3 from residues 544 to 607), palm (α10, α11 and 310 helix η1 from residues 499 to 543, and α14 to 310 helix η2 from residues 608 to 709) and thumb (β6 to α23 from residues 710 to 887) subdomains (Fig 5A). The ZIKV RdRp fold is analogous to that of other flaviviruses like DENV, WNV and JEV (Malet et al, 2007; Yap et al, 2007; Lu & Gong, 2013). One special characteristic of flavivirus RdRp is that it contains the NLS region, which is distributed between the fingers and thumb subdomains, consisting of βNLS and α/βNLS sequences. In ZIKV RdRp structure, the βNLS (residues 316–367) including a helix‐turn‐helix motif (α2‐α3) lies on top of the thumb subdomain, and the α/βNLS (residues 368–415) consisting of helices α5 and α6 are located between the fingers and the palm subdomain (Fig 5A). It has been shown that the NLS region is necessary for DENV NS5 structure stabilization (Yap et al, 2007). The NLS could be recognized by cellular factors such as β‐importin to transport NS5 to nucleus, which is helpful for viral replication (Uchil et al, 2006). This region is also thought to interact with NS3 helicase, which may modulate their respective enzymatic activities (Johansson et al, 2001).
Figure 5. Structure of ZIKV NS5 RdRp.
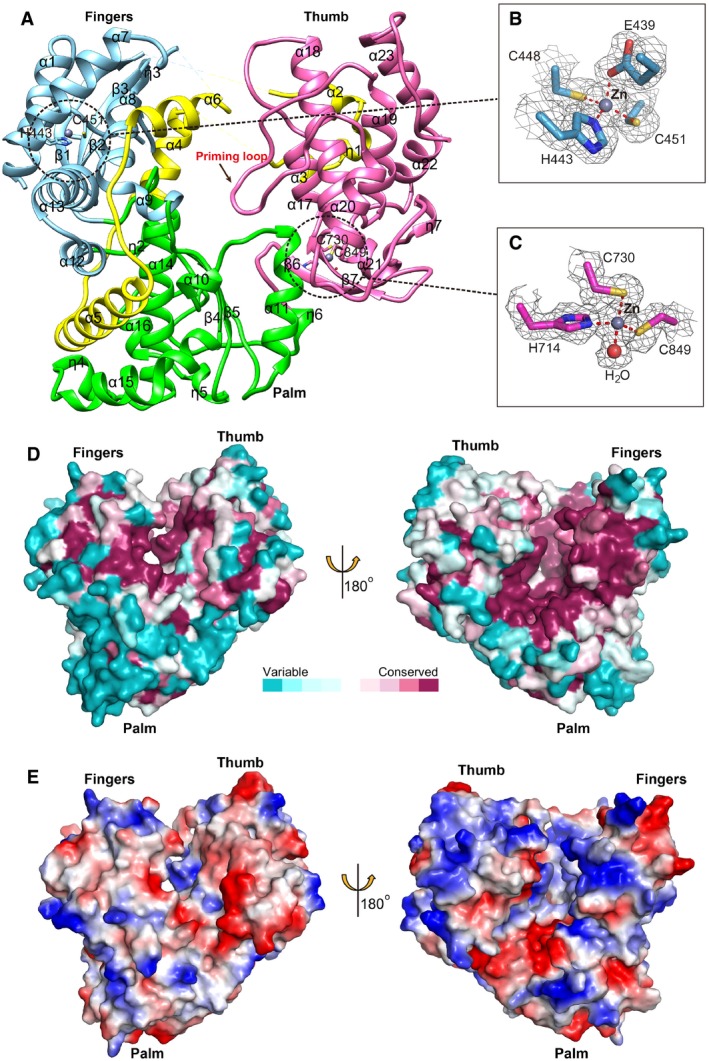
-
A–CRibbon representation of the overall fold with the three subdomains: fingers (sky blue), palm (green) and thumb (light pink) (A). The putative NLS region is colored in yellow. The priming loop is highlighted by an arrow. The catalytic residues are indicated by sticks. The two zinc‐binding sites are highlighted by dashed circles, and detailed interactions are shown as sticks (B and C). The zinc ions are displayed as gray spheres, and the water interacting with the zinc ion is displayed as pink sphere. The 2F o−F c electron density maps contoured at 1.0 sigma are represented respectively in gray.
- D
-
EThe electrostatic surface potential of ZIKV RdRp. Red and blue colors indicate negative potential and positive potential, respectively.
There are four flexible loops in the fingers subdomain (β1‐α2 loop, α3‐α4 loop, α6‐α7 loop and α7‐α8 loop), which are disordered in our structure and other flavivirus RdRp structures. They are important for the conformation modulation of RdRp during replication as the location of the template tunnel beneath these mobile loops. The catalytic active residues D665 and D666 are located in the turn between strands β4 and β5 in the palm subdomain. The priming loop locates between α19 and α20 in the subthumb domain, which is deployed inside the catalytic chamber to promote formation of the initiation complex. This loop is well‐ordered and inserting into a narrow RNA binding tunnel in our structure, implying that our ZIKV RdRp adopts a “closed” pre‐initiation state conformation (Noble et al, 2013, 2016; Lim et al, 2016).
There are two zinc‐binding pockets in our structure. These two pockets are observed in all the structure‐available flavivirus RdRps. One zinc atom is coordinated by E439, H443, C448 and C451 in the fingers subdomain (Fig 5B). The other zinc atom is coordinated by H714, C730, C849 and a water molecule in the thumb subdomain (Fig 5C). In DENV and JEV RdRp structures, the water‐mediated interaction is replaced by a histidine. However, in the same position, the histidine is replaced by asparagine or threonine in the ZIKV or WNV RdRp, respectively. As the second zinc‐binding site is near the two important residues S712 and R731 which are responsible for binding to the incoming rNTP, the second zinc ion could play a role in regulating conformational switch within the thumb subdomain.
From the conserved analysis based on an alignment of RdRp sequences from 73 flaviviruses, we found that the priming loop, the RNA tunnel and the zinc pocket sites are the most conserved regions (Fig 5D). We also found that the RNA tunnel displays a positively charged surface, which is favorable for binding of RNA (Fig 5E).
We compared the ZIKV RdRp structure with the available RdRp structures of DENV3, WNV and JEV (Malet et al, 2007; Lu & Gong, 2013; Noble et al, 2013). The thumb subdomain of ZIKV RdRp rotates more to the fingers subdomain, compared with DENV3, WNV and JEV (Fig 6). In this way, the RNA tunnel will be narrower in ZIKV RdRp. In addition, we found that the α8 helix of ZIKV RdRp is truncated, compared with the WNV and DENV3 RdRp structures (Fig 6A and B). Thus, the orientation of the flexible loop between α7 helix and α8 helix is different to that of WNV or DENV3 RdRp. When we compared with the full‐length JEV NS5 structure (Fig 6C), we found that the JEV RdRP also has a similar truncated α8 helix, and the α7/α8 loop is visible, interacting with the MTase. It indicates that full‐length ZIKV NS5 might have similar spatial organization to JEV NS5.
Figure 6. Comparison of the structures of ZIKV RdRp with other flavivirus RdRps.
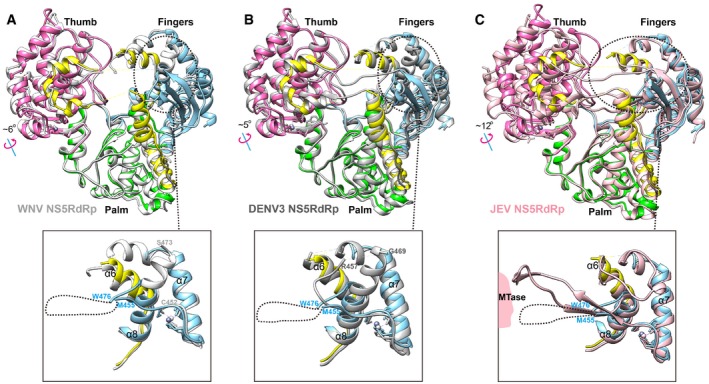
-
A–CSuperimposition of ZIKV RdRp with WNV RdRp (white, PDB: 2HCS) (Malet et al, 2007) (A), DENV3 (gray, PDB: 4HHJ) (Noble et al, 2013) (B), or JEV NS5 (light pink, PDB: 4K6M) (Lu & Gong, 2013) (C). ZIKV RdRp is colored by subdomains. The rotations of the thumbs are observed between the two structures. The variable regions in fingers subdomain are highlighted by dashed cycles and shown in large view. The disordered region in ZIKV RdRp (between M455 and W476) is shown as a dashed line.
Conserved drug target sites in both ZIKV MTase and RdRp compared to other flaviviruses
For the MTase, the GTP‐binding and SAM‐binding pockets are potential targets for antiviral development since both of them have been shown to bind to low molecular weight ligands. Previous studies on other flaviviruses have developed small molecules specifically targeting these two pockets. Targeting the SAM‐binding site, Sinefungin (SIN) was found displaying broad‐spectrum inhibition of MTase activity and can inhibit viral replication of WNV and DENV, but the potential cytotoxicity is a concern (Dong et al, 2010b). Structural identification of the additional hydrophobic pocket next to the SAM‐binding pocket has stimulated the design of SAH derivatives with substituents in the N6 adenine moiety that extend to the pocket (Dong et al, 2010b; Lim et al, 2011). One compound 10* was found to be potent against DENV MTase without suppressing host MTases. Binding of compound 10* in DENV MTase induced conformational changes in residues lining the pocket. The side chain of F133 rotated away from the pocket to accommodate the N6‐substituted benzylic ring of compound 10*, and the side chains of R163 shifted toward and stack onto the N6 benzylic ring of compound 10* to form a strong cation interaction (Fig 7A) (Lim et al, 2011). We have compared the corresponding site for compound 10* in our ZIKV MTase structure with that in DENV MTase structure, and found that all the residues involved in the interactions are conserved and have almost identical side chain conformation to the unbound DENV structure (Fig 7A). Recently, a novel broad‐spectrum inhibitor 12155 was identified by virtual screening (Brecher et al, 2015a). We compared the binding cavity of the 12155 with corresponding site in our ZIKV MTase structure and found that almost all residues involved in the interactions are conserved (Fig 7B). The ribavirin 5′‐triphosphate (RTP) molecule was reported to inhibit the 2′‐O‐MTase activity of DENV MTase by targeting the GTP‐binding pocket (Benarroch et al, 2004). We also compared the binding mode of RTP molecule with that of RNA analogue in our ZIKV MTase structure, and found that the binding mode of RTP is almost the same to that of the first nucleotide of RNA analogue in our ZIKV MTase structure, and almost all residues involved in the interactions are conserved (Fig 7C).
Figure 7. Conserved drug target sites in MTase.
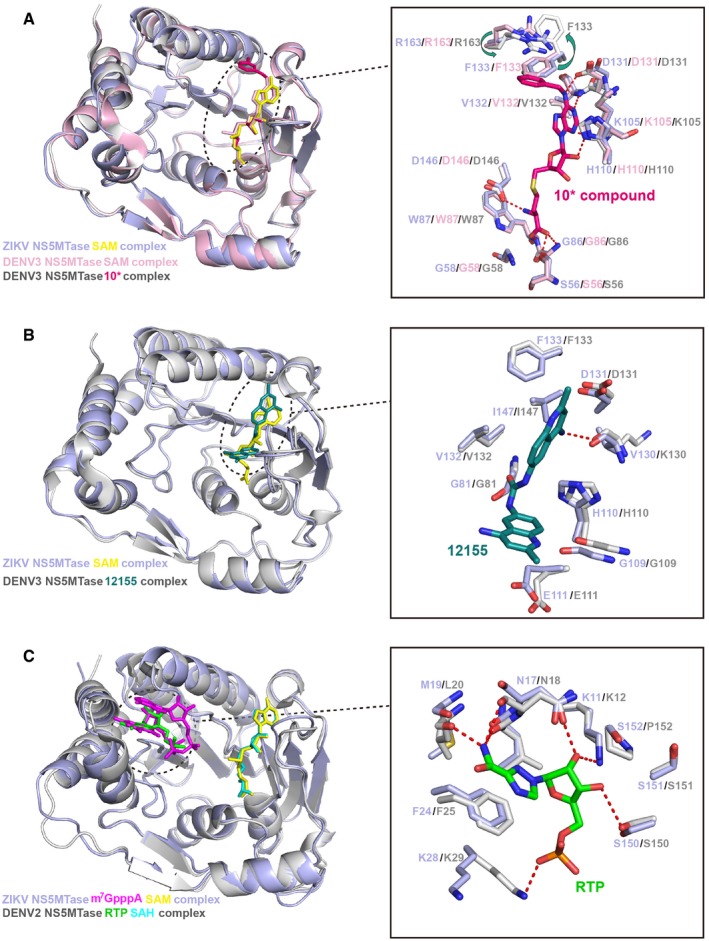
- Superimposition of the structure of ZIKV MTase (light blue) bound to SAM (yellow) with that of DENV3 MTase (light pink) bound to SAM (light pink) (PDB: 3P97) or DENV3 MTase (gray) bound to compound 10* (hot pink) (PDB: 3P8Z) (Lim et al, 2011). The detailed interactions are shown in the panel on the right side.
- Superimposition of the structure of ZIKV MTase (light blue) bound to SAM (yellow) with that of DENV3 MTase (gray) bound to the broad‐spectrum inhibitor 12155 (deep teal) (PDB: 5CUQ) (Brecher et al, 2015a). The detailed interactions are shown in the panel on the right side.
- Superimposition of the structure of ZIKV MTase (light blue) bound to SAM (yellow) and RNA analogue (magenta) with that of DENV2 MTase (gray) bound to ribavirin 5′‐triphosphate (RTP) molecule (green) (PDB: 1R6A) (Benarroch et al, 2004). The detailed interactions are shown in the panel on the right side.
Previously, two conserved inhibitor‐binding sites have been identified for DENV RdRp (Noble et al, 2013, 2016; Lim et al, 2016; Yokokawa et al, 2016). One is targeting the RNA template entry tunnel (Noble et al, 2013), and the other pocket, called as the “N pocket”, located in the thumb subdomain, close to the enzyme active site (Lim et al, 2016; Noble et al, 2016; Yokokawa et al, 2016). By targeting the RNA tunnel, the compound NITD107 was first identified by enzyme activity‐based screening and the binding site was defined by complex structure (Noble et al, 2013). NITD107 forms a dimer, locates in the RNA binding groove and induces the conformational changes in RdRp after binding. We compared the binding cavity of NITD107 in DENV RdRp structure with corresponding site in our ZIKV RdRp structure and found that almost all residues involved in the interactions are conserved (Fig 8A). The carboxylic groups of both molecules are engaged in several hydrogen bonds with residues K401 (DENV3 RdRp numbering), N492 (molecule NITD107‐1) and T413 (molecule NITD107‐2), while the sulfonamide group of both molecules form several hydrogen bonds with residues S600, V604, T605 (molecule NITD107‐1) and V411, W795 (molecule NITD107‐2). NITD107 stabilizes the α6‐α7 loop (residues 410–419, DENV3 RdRp numbering). That loop is disordered both in the free DENV3 RdRp and our ZIKV RdRp structures, while it becomes ordered upon compound binding (Fig 8A). In addition, compound binding induces the shift of the β3‐α14 loop (residues 600–605, DENV3 RdRp numbering) and the conformational change of side chain of R481. NITD107, therefore, prevents viral RNA synthesis via competition with the RNA substrate.
Figure 8. Conserved drug target sites in RdRp.
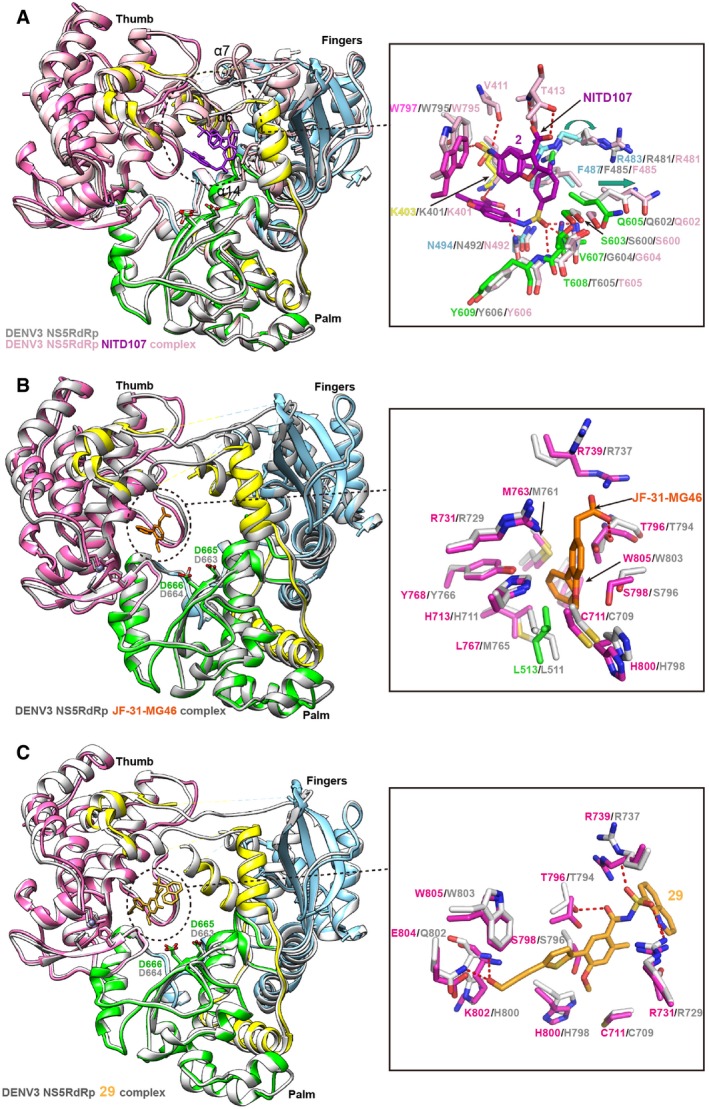
- Superimposition of ZIKV RdRp with DENV3 RdRp (gray) bound to JF‐31‐MG46 molecule (orange) (PDB: 5F3T) (Noble et al, 2016). The detailed interactions are shown in the panel on the right side.
To date, a series of anti‐DENV inhibitors have been designed to target the “N pocket” which is identified through fragment‐based screening via X‐ray crystallography (Noble et al, 2016). One of the inhibitor molecules called JF‐31‐MG46 displays good binding affinity and inhibitory activity. We then compared the binding cavity of JF‐31‐MG46 molecule with corresponding site in our ZIKV RdRp structure and found that almost all residues involved in the interactions are conserved (Fig 8B), indicating that JF‐31‐MG46 could bind to the ZIKV RdRp well. Subsequently, the optimization of this kind of molecules using the structure‐guided approach creates more potent inhibitors that bound to this pocket and inhibited DENV1‐4 viral replication across various cell‐based assays, for example, compound 29 (Lim et al, 2016; Yokokawa et al, 2016). We therefore compared the binding cavity of compound 29 with corresponding site in our ZIKV RdRp structure and found that almost all residues involved in the interactions are conserved (Fig 8C). The terminal hydroxyl group of propargyl alcohol arm forms hydrogen bond interactions with the backbone amide of H800 and the side chain of Q802 (DENV3 RdRp numbering). We should note that, in ZIKV RdRp, the residue equivalent to Q802 in DENV3 is E804, which can form hydrogen bond in a similar way. In addition, the acyl‐sulfonamide carbonyl moiety of compound 29 forms hydrogen bonds with the side chain of T794 and the acyl‐sulfonamide group interact with residues S710, R729 and R737, which is highly conserved in ZIKV RdRp structure.
Discussion
In this study, we have solved both the crystal structure of ZIKV NS5 MTase bound to SAM and the tertiary complex structure of ZIKV MTase with SAM and RNA analogue (m7GpppA), and we also solved the ZIKV NS5 RdRp structure (though the full‐length structure of NS5 has not been solved here, but the structures of two functional domains have been solved). Both ZIKV MTase and RdRp structures show conserved features with other flaviviruses. For MTase, we show that the RNA analogue binds to the cap‐binding site in a stacked conformation, and the second nucleobase A1 stacks against the first nucleobase 7mG0 and the aromatic residue F24. The binding mode of RNA analogue provides important clue for subsequent inhibitor design. Although the cap‐binding pocket are highly conserved, ZIKV MTase possesses a hydrophobic site (consisting of A21 and L22) near the binding pocket, in contrast to positively charged corresponding site in other flaviviruses (Fig 3), which should be taken into consideration for the antiviral inhibitor design. For RdRp, we found the thumb subdomain rotates more to the fingers subdomain, and has a tighter and more closed conformation compared to DENV3, WNV and JEV. Otherwise, ZIKV RdRp has a truncated α8 helix, which resembles more to JEV than to DENV or WNV RdRps.
The flavivirus NS5 MTases and RdRp have been attractive targets for antiviral development. A series of inhibitors have been identified through various approaches based on DENV or WNV (Dong et al, 2010b; Lim et al, 2011, 2016; Chen et al, 2013; Brecher et al, 2015b; Yokokawa et al, 2016). We analyzed the binding site of the representative inhibitors in our crystal structures, and found that the drug targeting sites in both MTase and RdRp are highly conserved between DENV and ZIKV. Therefore, we suggest these developed drugs against DENV have good chances to be developed to treat ZIKV infection, which should be tested as soon as possible. Our structural information of ZIKV MTase and RdRp would aid in the discovery and development of antiviral inhibitors against ZIKV in future.
Materials and Methods
Gene cloning, protein production and purification
ZIKV MTase fragment of NS5 (amino acid residues 5–274, accession number: AMA12085) fused at its N‐terminus with a hexa‐histidine tag was cloned into the pET‐21a vector (Novagen) with NdeI and XhoI restriction sites. Transformed Escherichia coli strain BL21 (DE3) clones were grown in LB medium to an A260 of 0.6–0.8 at 37°C. Expression of the recombinant proteins was induced by the addition of 0.5 mM isopropyl‐β‐D‐thiogalactopyranoside, and incubation was continued for a further 16 h at 16°C. Cells were harvested by centrifugation at 8,000 g for 15 min at 4°C, and then resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl and 10% glycerol) and lysed by homogenization. The lysate was clarified by centrifugation at 20,000 g for 60 min at 4°C. The supernatant was purified by metal affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare). Proteins were eluted using buffer A supplemented with 300 mM imidazole. The proteins were further purified by gel filtration chromatography using a HiLoad 16/60 Superdex® 75 PG (GE Healthcare) with a running buffer of 20 mM Tris–HCl, 500 mM NaCl (pH 8.0) and 10% glycerol, and the collected protein fractions were concentrated to 10 mg/ml using a membrane concentrator with a molecular weight cutoff of 10 kDa (Millipore).
ZIKV NS5 C‐terminal RdRp (amino acid residues 275–898) was constructed and expressed in the same way as MTase, respectively. After metal affinity chromatography, RdRp proteins were further purified by gel filtration chromatography in a buffer containing 20 mM Tris–HCl, pH 8.0, 250 mM NaCl and 10% glycerol on a Hiload 16/60 Superdex® 200 PG column (GE Healthcare). The collected protein fractions were concentrated to 5 mg/ml using a membrane concentrator with a molecular weight cutoff of 30 kDa (Millipore).
Crystallization, data collection and structure determination
Crystallization trials were set up with commercial crystallization kits (Molecular Dimensions) using sitting drop vapor diffusion. Normally, 1 μl protein was mixed with 1 μl reservoir solution. The resultant drop was then sealed, equilibrating against 100 μl reservoir solution at 4 or 18°C. Diffractable crystals of MTase were obtained in 0.2 M magnesium chloride, 0.1 M HEPES pH 7.0, 20% w/v PEG 6000 at 18°C. Diffractable crystals of RdRp were obtained in 20 mM d‐glucose, 20 mM d‐mannose, 20 mM d‐galactose, 20 mM l‐fucose, 20 mM d‐xylose, 20 mM N‐acetyl‐d‐glucosamine, 60.9 mM Tris (base), 39.1 mM Bicine, pH 8.5, 12% v/v ethylene glycol and 6% w/v PEG 8000. Crystals were flash‐cooled in liquid nitrogen after a brief soaking in reservoir solution with the addition of 17% (v/v) glycerol. The X‐ray diffraction data were collected under cryogenic conditions (100K) at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U or BL19U1, and indexed, integrated, and scaled with HKL2000 (Otwinowski & Minor, 1997). To obtain the ligand‐bound structure, MTase crystals were soaked for 4–6 h at 18°C in mother liquor solutions containing 2 mM RNA analogue (S1405, New England Biolabs) and then flash‐cooled in a nitrogen stream.
The ZIKV MTase structure was solved by the molecular replacement method using Phaser (McCoy et al, 2007) from the CCP4 program suite (Winn et al, 2011), with the structure of DENV2 MTase (PDB: 1L9K) (Egloff et al, 2002) as the search model. Initial restrained rigid‐body refinement and manual model building were performed using REFMAC5 (Murshudov et al, 2011) and COOT (Debreczeni & Emsley, 2012), respectively. The SAM molecule and RNA analogue (m7GpppA) were manually built using COOT based on the simulated annealing omit F o−F c maps and were further refined using Phenix (Adams et al, 2010). The ZIKV NS5 RdRp structure was solved by the molecular replacement method using Phaser, with the structure of DENV3 NS5273–900 (PDB: 2J7U) (Yap et al, 2007) as the search model. Final statistics for data collection and structure refinement are represented in Table 1.
Accession numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 5WZ1, 5WZ2 and 5WZ3.
Author contributions
GFG and YS designed and supervised the study. WD, HS, HW, and CS conducted the experiments. JQ and YC collected the data sets and solved the structures. HS, YS, and GFG analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
This work was supported by the Emergency Task‐Force Project (Grant No. 81641001) of the National Natural Science Foundation of China (NSFC), the China Ministry of Science and Technology National 973 Project (Grant No. 2014CB542503), the Task‐Force of ZIKV Research from the Chinese Academy of Sciences (CAS), the National Key Plan for Scientific Research and Development of China (2016YFD0500300 and 2016YFC1200305), Zika Special Project of the Ministry of Science and Technology Reform and Development Project and Strategic Priority Research Program of CAS (XDB08020100). This work is also supported in part by the External Cooperation Program of CAS (153211KYSB20160001). We thank the staff of BL17U beamline at SSRF and BL19U1 beamline at SSRF for assistance during data collection. Y.S. is supported by the Excellent Young Scientist Program from the NSFC (No. 81622031), the Excellent Young Scientist Program of CAS and the Youth Innovation Promotion Association CAS (2015078). H.S. is supported by the Youth Innovation Promotion Association CAS (2017117). G.F.G. is supported partly as a leading principal investigator of the NSFC Innovative Research Group (Grant No. 81621091).
The EMBO Journal (2017) 36: 919–933
Contributor Information
Yi Shi, Email: shiyi@im.ac.cn.
George F Gao, Email: gaof@im.ac.cn.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben‐Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44: W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour J, Laurent‐Rolle M, Shi PY, Garcia‐Sastre A (2009) NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83: 5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenberg R, Ren J, Verma A, Walter TS, Alderton D, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, Grimes JM (2007) Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J Gen Virol 88: 2228–2236 [DOI] [PubMed] [Google Scholar]
- Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B (2004) A structural basis for the inhibition of the NS5 dengue virus mRNA 2′‐O‐methyltransferase domain by ribavirin 5′‐triphosphate. J Biol Chem 279: 35638–35643 [DOI] [PubMed] [Google Scholar]
- Bollati M, Milani M, Mastrangelo E, de Lamballerie X, Canard B, Bolognesi M (2009a) Crystal structure of a methyltransferase from a no‐known‐vector flavivirus. Biochem Biophys Res Commun 382: 200–204 [DOI] [PubMed] [Google Scholar]
- Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, Nonnis S, Decroly E, Selisko B, de Lamballerie X, Coutard B, Canard B, Bolognesi M (2009b) Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA‐capping mechanisms in Flavivirus. J Mol Biol 385: 140–152 [DOI] [PubMed] [Google Scholar]
- Brecher M, Chen H, Li Z, Banavali NK, Jones SA, Zhang J, Kramer LD, Li H (2015a) Identification and characterization of novel broad‐spectrum inhibitors of the flavivirus methyltransferase. ACS Infect Dis 1: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M, Chen H, Liu B, Banavali NK, Jones SA, Zhang J, Li Z, Kramer LD, Li H (2015b) Novel broad spectrum inhibitors targeting the flavivirus methyltransferase. PLoS One 10: e0130062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet GA, Santos FB, Sequeira PC (2016) Zika virus infection: epidemiology, clinical manifestations and diagnosis. Curr Opin Infect Dis 29: 459–466 [DOI] [PubMed] [Google Scholar]
- Carteaux G, Maquart M, Bedet A, Contou D, Brugieres P, Fourati S, Cleret de Langavant L, de Broucker T, Brun‐Buisson C, Leparc‐Goffart I, Mekontso Dessap A (2016) Zika virus associated with meningoencephalitis. N Engl J Med 374: 1595–1596 [DOI] [PubMed] [Google Scholar]
- Chen H, Liu L, Jones SA, Banavali N, Kass J, Li Z, Zhang J, Kramer LD, Ghosh AK, Li H (2013) Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antiviral Res 97: 232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloma J, Jain R, Rajashankar KR, Garcia‐Sastre A, Aggarwal AK (2016) Structures of NS5 methyltransferase from Zika virus. Cell Rep 16: 3097–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Decroly E, Li C, Sharff A, Lescar J, Bricogne G, Barral K (2014) Assessment of Dengue virus helicase and methyltransferase as targets for fragment‐based drug discovery. Antiviral Res 106: 61–70 [DOI] [PubMed] [Google Scholar]
- Davidson A, Slavinski S, Komoto K, Rakeman J, Weiss D (2016) Suspected female‐to‐male sexual transmission of Zika virus ‐ New York City, 2016. MMWR Morb Mortal Wkly Rep 65: 716–717 [DOI] [PubMed] [Google Scholar]
- Debreczeni JE, Emsley P (2012) Handling ligands with Coot. Acta Crystallogr D Biol Crystallogr 68: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckard DT, Chung WM, Brooks JT, Smith JC, Woldai S, Hennessey M, Kwit N , Mead P (2016) Male‐to‐male sexual transmission of Zika virus‐Texas, January 2016. MMWR Morb Mortal Wkly Rep 65: 372–374 [DOI] [PubMed] [Google Scholar]
- Dong H, Chang DC, Xie X, Toh YX, Chung KY, Zou G, Lescar J, Lim SP, Shi PY (2010a) Biochemical and genetic characterization of dengue virus methyltransferase. Virology 405: 568–578 [DOI] [PubMed] [Google Scholar]
- Dong H, Liu L, Zou G, Zhao Y, Li Z, Lim SP, Shi PY, Li H (2010b) Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J Biol Chem 285: 32586–32595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, Timofeev J, Rodriguez FJ, Levanov L, Razak J, Iyengar P, Hennenfent A, Kennedy R, Lanciotti R, du Plessis A, Vapalahti O (2016) Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med 374: 2142–2151 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B (2002) An RNA cap (nucleoside‐2′‐O‐)‐methyltransferase in the flavivirus RNA polymerase xml: crystal structure and functional characterization. EMBO J 21: 2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B (2007) Structural and functional analysis of methylation and 5′‐RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol 372: 723–736 [DOI] [PubMed] [Google Scholar]
- Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, Peersen OB (2009) Analysis of flavivirus NS5 methyltransferase cap binding. J Mol Biol 385: 1643–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, Moley KH, Diamond MS (2016) Zika virus infection damages the testes in mice. Nature 540: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez‐Seco MP, Evans MJ, Best SM, Garcia‐Sastre A (2016) Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19: 882–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson AM, Jakobsson E, Johansson P, Lantez V, Coutard B, de Lamballerie X, Unge T, Jones TA (2009) Structure of the methyltransferase domain from the Modoc virus, a flavivirus with no known vector. Acta Crystallogr D Biol Crystallogr 65: 796–803 [DOI] [PubMed] [Google Scholar]
- Johansson M, Brooks AJ, Jans DA, Vasudevan SG (2001) A small region of the dengue virus‐encoded RNA‐dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin‐beta and the viral helicase, NS3. J Gen Virol 82: 735–745 [DOI] [PubMed] [Google Scholar]
- Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC (2016) Zika virus inhibits type‐I interferon production and downstream signaling. EMBO Rep 17: 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR et al (2011) Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem 286: 6233–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Noble CG, Seh CC, Soh TS, El Sahili A, Chan GK, Lescar J, Arora R, Benson T, Nilar S, Manjunatha U, Wan KF, Dong H, Xie X, Shi PY, Yokokawa F (2016) Potent allosteric Dengue virus NS5 polymerase inhibitors: mechanism of action and resistance profiling. PLoS Pathog 12: e1005737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Gong P (2013) Crystal structure of the full‐length Japanese encephalitis virus NS5 reveals a conserved methyltransferase‐polymerase interface. PLoS Pathog 9: e1003549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Li S, Ma S, Jia L, Zhang F, Zhang Y, Zhang J, Wong G, Zhang S, Lu X, Liu M, Yan J, Li W, Qin C, Han D, Qin C, Wang N, Li X, Gao GF (2016) Zika virus causes testis damage and leads to male infertility in mice. Cell 167: 1511–1524.e1510 [DOI] [PubMed] [Google Scholar]
- Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non‐structural protein 5. J Biol Chem 282: 10678–10689 [DOI] [PubMed] [Google Scholar]
- Mansuy JM, Dutertre M, Mengelle C, Fourcade C, Marchou B, Delobel P, Izopet J, Martin‐Blondel G (2016a) Zika virus: high infectious viral load in semen, a new sexually transmitted pathogen? Lancet Infect Dis 16: 405 [DOI] [PubMed] [Google Scholar]
- Mansuy JM, Suberbielle E, Chapuy‐Regaud S, Mengelle C, Bujan L, Marchou B, Delobel P, Gonzalez‐Dunia D, Malnou CE, Izopet J, Martin‐Blondel G (2016b) Zika virus in semen and spermatozoa. Lancet Infect Dis 16: 1106–1107 [DOI] [PubMed] [Google Scholar]
- Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, de Lamballerie X, Bolognesi M (2007) Structural bases for substrate recognition and activity in Meaban virus nucleoside‐2′‐O‐methyltransferase. Protein Sci 16: 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecharles S, Herrmann C, Poullain P, Tran TH, Deschamps N, Mathon G, Landais A, Breurec S, Lannuzel A (2016) Acute myelitis due to Zika virus infection. Lancet 387: 1481 [DOI] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak‐Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T (2016) Zika virus associated with microcephaly. N Engl J Med 374: 951–958 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyomrattanakit P, Chen YL, Dong H, Yin Z, Qing M, Glickman JF, Lin K, Mueller D, Voshol H, Lim JY, Nilar S, Keller TH, Shi PY (2010) Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol 84: 5678–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Lim SP, Chen YL, Liew CW, Yap L, Lescar J, Shi PY (2013) Conformational flexibility of the Dengue virus RNA‐dependent RNA polymerase revealed by a complex with an inhibitor. J Virol 87: 5291–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Lim SP, Arora R, Yokokawa F, Nilar S, Seh CC, Wright SK, Benson TE, Smith PW, Shi PY (2016) A conserved pocket in the dengue virus polymerase identified through fragment‐based screening. J Biol Chem 291: 8541–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Prisant N, Bujan L, Benichou H, Hayot PH, Pavili L, Lurel S, Herrmann C, Janky E, Joguet G (2016) Zika virus in the female genital tract. Lancet Infect Dis 16: 1000–1001 [DOI] [PubMed] [Google Scholar]
- Smietanski M, Werner M, Purta E, Kaminska KH, Stepinski J, Darzynkiewicz E, Nowotny M, Bujnicki JM (2014) Structural analysis of human 2′‐O‐ribose methyltransferases involved in mRNA cap structure formation. Nat Commun 5: 3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay MY, Saw WG, Zhao Y, Chan KW, Singh D, Chong Y, Forwood JK, Ooi EE, Gruber G, Lescar J, Luo D, Vasudevan SG (2015) The C‐terminal 50 amino acid residues of dengue NS3 protein are important for NS3‐NS5 interaction and viral replication. J Biol Chem 290: 2379–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Kumar AV, Satchidanandam V (2006) Nuclear localization of flavivirus RNA synthesis in infected cells. J Virol 80: 5451–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D, Petit AP, Bueren‐Calabuig JA, Jansen C, Fletcher DA, Peggie M, Weidlich S, Scullion P, Pisliakov AV, Cowling VH (2016) Molecular basis of RNA guanine‐7 methyltransferase (RNMT) activation by RAM. Nucleic Acids Res 44: 10423–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero‐Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse‐Locker J, Bartenschlager R (2009) Composition and three‐dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Song H, Qi J, Liu Y, Wang H, Su C, Shi Y, Gao GF (2016) Contribution of intertwined loop to membrane association revealed by Zika virus full‐length NS1 structure. EMBO J 35: 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J (2007) Crystal structure of the dengue virus RNA‐dependent RNA polymerase catalytic domain at 1.85‐angstrom resolution. J Virol 81: 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa F, Nilar S, Noble CG, Lim SP, Rao R, Tania S, Wang G, Lee G, Hunziker J, Karuna R, Manjunatha U, Shi PY, Smith PW (2016) Discovery of potent non‐nucleoside inhibitors of Dengue viral RNA‐dependent RNA polymerase from a fragment hit using structure‐based drug design. J Med Chem 59: 3935–3952 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen W, Wong G, Bi Y, Yan J, Sun Y, Chen E, Yan H, Lou X, Mao H, Xia S, Gao GF, Shi W, Chen Z (2016) Highly diversified Zika viruses imported to China, 2016. Protein Cell 7: 461–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Soh TS, Lim SP, Chung KY, Swaminathan K, Vasudevan SG, Shi PY, Lescar J, Luo D (2015) Molecular basis for specific viral RNA recognition and 2′‐O‐ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc Natl Acad Sci USA 112: 14834–14839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H (2007) Structure and function of flavivirus NS5 methyltransferase. J Virol 81: 3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes‐Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V (2011) Ribose 2′‐O‐methylation provides a molecular signature for the distinction of self and non‐self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K (2013) Rational design of a live attenuated dengue vaccine: 2′‐O‐methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9: e1003521 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File


